Effects of Returning Different Organic Materials in Combination with Inorganic Fertilizers on the Diversity of Eukaryotic Microorganisms in Semi-Arid Northern China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Field Description and Experimental Design
2.2. Soil Sampling
2.3. Soil Physicochemical Analytical Procedures
2.4. Soil DNA Extraction and High-Throughput Sequencing
2.5. Statistical Analysis
2.5.1. Bioinformatics Analysis
2.5.2. Significance Analysis
3. Results
3.1. Soil Physicochemical Characteristics
3.2. Alpha-Diversity of Eukaryotic Microorganisms
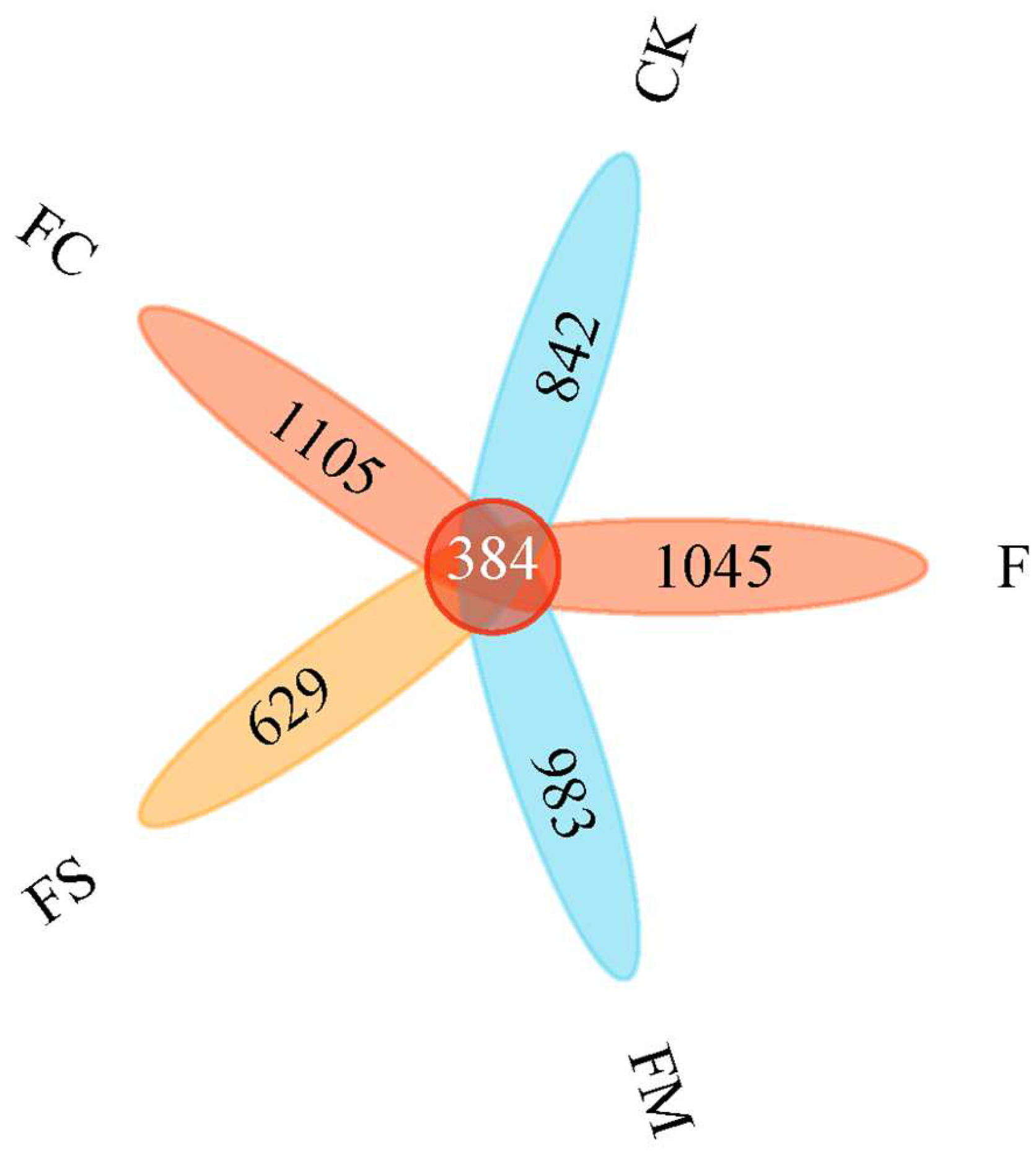
3.3. Composition of Eukaryotic Microbial Community under Different Treatments
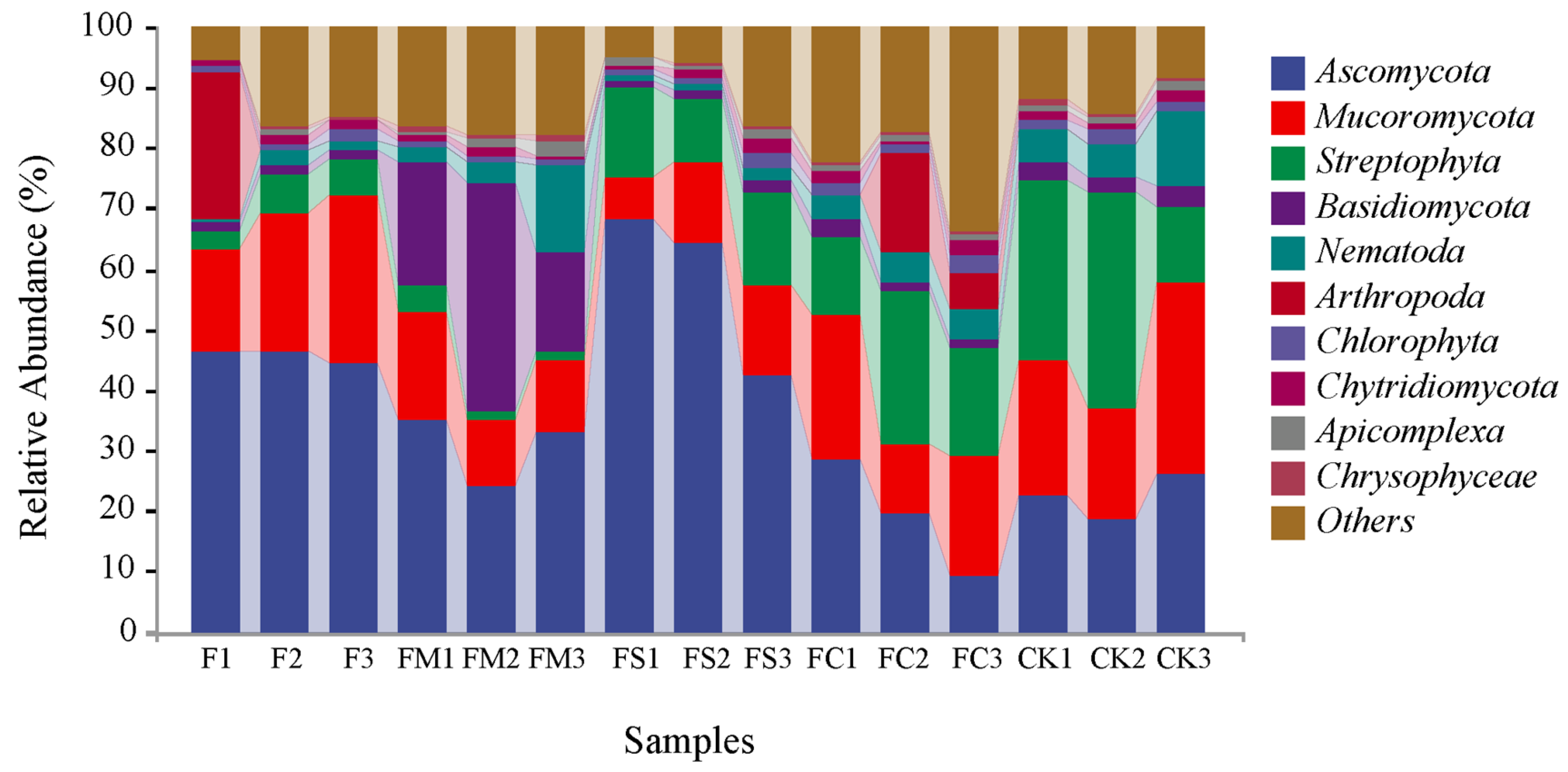
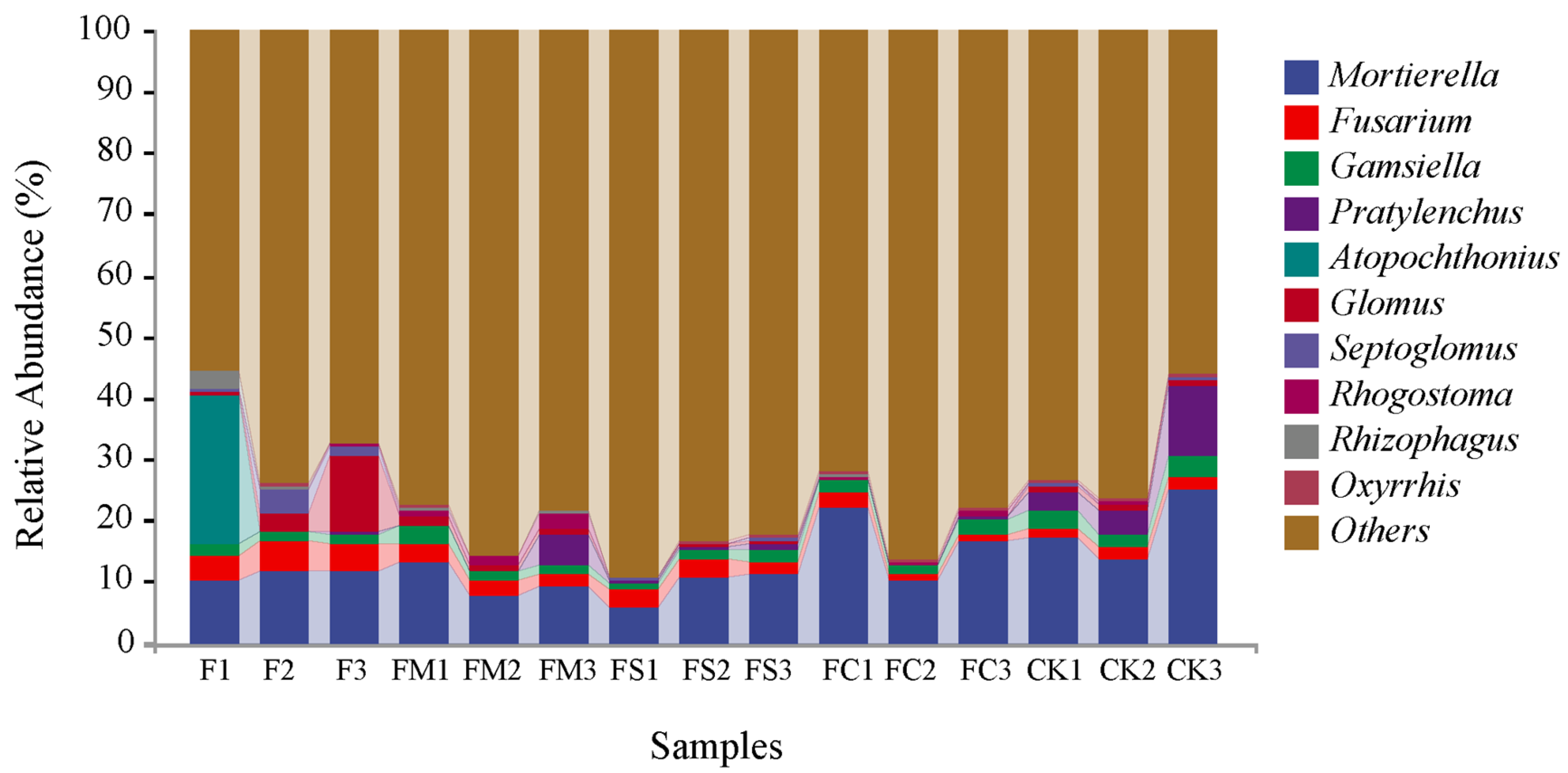
3.4. Taxonomic Composition Analysis at the Phylum and Genus Level
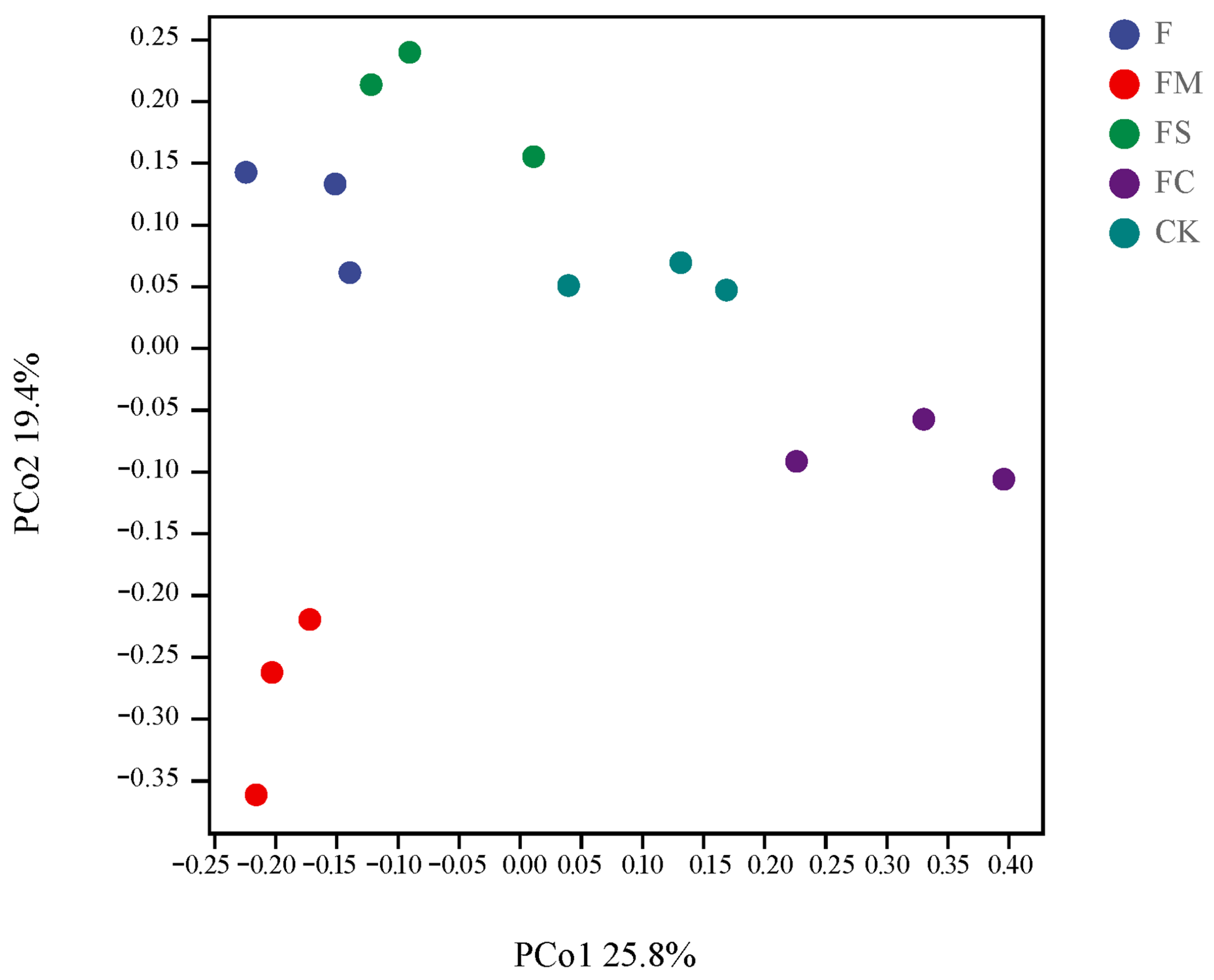
3.5. Beta-Diversity of Eukaryotic Microorganisms
3.6. Correlations
4. Discussion
4.1. Effects of Treatments on the Soil Physicochemical Properties
4.2. Effects of Different Treatments on the Eukaryotic Microorganisms Diversity and Community Composition
4.3. Relationship between Soil Eukaryotic Microorganisms and Environmental Factors
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chaparro, J.M.; Sheflin, A.M.; Manter, D.K.; Vivanco, J.M. Manipulating the soil microbiome to increase soil health and plant fertility. Biol. Fert. Soils 2012, 48, 489–499. [Google Scholar] [CrossRef]
- Chen, H.; Liang, Q.; Gong, Y.; Kuzyakov, Y.; Fan, M.; Plante, A.F. Reduced tillage and increased residue retention increase enzyme activity and carbon and nitrogen concentrations in soil particle size fractions in a long-term field experiment on Loess Plateau in China. Soil Till. Res. 2019, 194, 104–296. [Google Scholar] [CrossRef]
- Li, X.B.; He, H.B.; Zhang, X.D.; Yan, X.Y.; Six, J.; Cai, Z.; Barthel, M.; Zhang, J.; Necpalova, M.; Ma, Q. Distinct responses of soil fungal and bacterial nitrate immobilization to land conversion from forest to agriculture. Soil Biol. Biochem. 2019, 134, 81–89. [Google Scholar] [CrossRef]
- Assress, H.A.; Selvarajan, R.; Nyoni, H.; Ntushelo, K.; Mamba, B.B.; Msagati, T.A.M. Diversity, co-occurrence and implications of fungal communities in wastewater treatment plants. Sci. Rep. 2019, 9, 14056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jing, H.; Zhang, Y.; Li, Y.; Zhu, W.; Liu, H. Spatial variability of picoeukaryotic communities in the Mariana Trench. Sci. Rep. 2018, 8, 15357. [Google Scholar] [CrossRef] [Green Version]
- Falkowski, P.G.; Fenchel, T.; Delong, E.F. The microbial engines that drive Earth’s biogeochemical cycles. Science 2008, 320, 1034–1039. [Google Scholar] [CrossRef] [Green Version]
- Delong, E.F. The microbial ocean from genomes to biomes. Nature 2009, 459, 200–206. [Google Scholar] [CrossRef] [Green Version]
- Brussaard, L.; Ruiter, P.C.D.; Brown, G.G. Soil biodiversity for agricultural sustainability. Agric. Ecosyst. Environ. 2007, 121, 233–244. [Google Scholar] [CrossRef]
- Wang, X.L.; Ma, K.; Fu, Y.Z.; Wang, Z.Q.; An, Y.Y. Effects of no-tillage, mulching, and organic fertilization on soil fungal community composition and diversity. Chin. J. Appl. Ecol. 2020, 31, 890–898. [Google Scholar] [CrossRef]
- Nemergut, D.R.; Costello, E.K.; Hamady, M.; Lozupone, C.; Jiang, L.; Schmidt, S.K.; Fierer, N.; Townsend, A.R.; Cleveland, C.C.; Stanish, L.; et al. Global patterns in the biogeography of bacterial taxa. Environ. Microbiol. 2011, 13, 135–144. [Google Scholar] [CrossRef]
- Fenchel, T. Microbiology. Biogeography for bacteria. Science 2003, 301, 925–926. [Google Scholar] [CrossRef]
- Hartmann, M.; Frey, B.; Mayer, J.; Mder, P.; Widmer, F. Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 2014, 9, 1177–1194. [Google Scholar] [CrossRef] [Green Version]
- Luo, P.; Han, X.; Wang, Y.; Han, M.; Shi, H.; Liu, N.; Bai, H. Influence of long-term fertilization on soil microbial biomass, dehydrogenase activity, and bacterial and fungal community structure in a brown soil of northeast China. Annu. Microbiol. 2015, 65, 533–542. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.P.; Cui, Z.L.; Fan, M.S.; Vitousek, P.; Zhao, M.; Ma, W.Q.; Wang, Z.L.; Zhang, W.J.; Yan, X.Y.; Yang, J.C.; et al. Producing more grain with lower environmental costs. Nature 2014, 514, 486–489. [Google Scholar] [CrossRef]
- Ibrahim, A.; Abaidoo, R.C.; Fatondji, D.; Opoku, A. Hill placement of manure and fertilizer micro-dosing improves yield and water use efficiency in the Sahelian low input millet-based cropping system. Field Crops Res. 2015, 180, 29–36. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, P.; Zeng, Z. Dynamics of Bacterial Communities in a 30-Year Fertilized Paddy Field under Different Organic-Inorganic Fertilization Strategies. Agronomy 2019, 9, 14. [Google Scholar] [CrossRef] [Green Version]
- Lu, F.; Wang, X.K.; Han, B.; Ouyang, Z.Y.; Zheng, H. Straw return to rice paddy: Soil carbon sequestration and increased methane emission. Chin. J. Appl. Ecol. 2010, 21, 99–108. [Google Scholar] [CrossRef]
- Wang, W.; Luo, X.; Chen, Y.; Ye, X.; Wang, H.; Cao, Z.; Ran, W.; Cui, Z. Succession of Composition and Function of Soil Bacterial Communities During Key Rice Growth Stages. Front. Microbiol. 2019, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Chang, A.R.; Li, J.; Zhang, S.; Zhan, J.W.; Wei, D.H.; Wang, P.W.; Yu, J.J. Analysis of bacterial community structure in rhizosphere soil of tobacco based on the metagenomics 16S rDNA sequencing technology. J. Agric. Sci. Technol. 2017, 19, 43–50. [Google Scholar] [CrossRef]
- Sun, R.; Zhang, X.X.; Guo, X.; Wang, D.; Chu, H. Bacterial diversity in soils subjected to long-term chemical fertilization can be more stably maintained with the addition of livestock manure than wheat straw. Soil Biol. Biochem. 2015, 88, 9–18. [Google Scholar] [CrossRef]
- MacLean, D.; Jones, J.D.; Studholme, D.J. Application of next-generation sequencing technologies to microbial genetics. Nat. Rev. Microbiol. 2009, 7, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Caron, D.A.; Countway, P.D.; Jones, A.C.; Kim, D.Y.; Schnetzer, A. Marine protistan diversity. Ann. Rev. Mar. Sci. 2011, 4, 467–493. [Google Scholar] [CrossRef] [Green Version]
- Logares, R.; Haverkamp, T.H.A.; Kumar, S.; Lanzén, A.; Nederbragt, A.J.; Quince, C.; Kauserud, H. Environmental microbiology through the lens of high-throughput DNA sequencing: Synopsis of current platforms and bioinformatics approaches. J. Microbiol. Methods 2012, 91, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Lie, A.A.Y.; Liu, Z.; Hu, S.K.; Jones, A.C.; Kim, D.Y.; Countway, P.D.; Amaral-Zettler, L.A.; Cary, S.C.; Sherr, E.B.; Sherr, B.F. Investigating microbial eukaryotic diversity from a global census: Insights from a comparison of pyrotag and full-length sequences of 18S rRNA genes. Appl. Environ. Microbiol. 2014, 80, 4363–4373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahé, F.; Mayor, J.; Bunge, J.; Chi, J.; Siemensmeyer, T. Comparing high-throughput platforms for sequencing the V4 region of SSU-rDNA in environmental microbial eukaryotic diversity surveys. J. Eukaryot. Microbiol. 2015, 62, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Johan, D.G.; Weedon, J.T.; Stéphane, B.; Steven, D.; Fernandez-Garberí, P.; Geisen, S.; Motte, L.G.D.L.; Heinesch, B.; Janssens, T.A.; Leblans, N.; et al. Patterns of local, intercontinental and interseasonal variation of soil bacterial and eukaryotic microbial communities. FEMS Microbiol. Ecol. 2020, 96, fiaa018. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, P.; Yu, X.F.; Shu, W.; Xu, Q.Y. Structure and Composition of Sediment-Associated Bacterial and Eukaryotic Communities in the River-Lake System of Poyang Lake, China. Geomicrobiol. J. 2019, 36, 727–736. [Google Scholar] [CrossRef]
- Zhang, R.; Liu, Y.; Zhao, X.F.; Zhang, H.L.; Zhao, Z.H.; Shang, Z.Z.; Lan, W.S. Eukaryotic communities in coastal water from Shenzhen in South China. Ecotoxicology 2020, 12, 1644–1651. [Google Scholar] [CrossRef]
- ISS-CAS, Institute of Soil Science, Chinese Academy of Science Scientific Data Base. China Soil Data Base. Available online: http://www.soil.csdb.cn/ (accessed on 10 March 2021).
- IUSS Working Group WRB. World Reference Base for Soil Resources 2006. In A Framework for International Classification, Correlation and Communication, 2nd ed.; World Soil Resources Reports No. 103; FAO: Rome, Italy, 2006. [Google Scholar]
- Bao, S.D. Analysis Method of Soil and Agricultural Chemistry, 3rd ed.; China Agricultural Press: Beijing, China, 2000; pp. 25–108. [Google Scholar]
- Logares, R.; Audic, S.; Bass, D.; Bittner, L.; Boutte, C.; Christen, R.; Claverie, J.M.; Decelle, J.; Dolan, J.R.; Dunthorn, M. Patterns of rare and abundant marine microbial eukaryotes. Curr. Biol. 2014, 24, 813–821. [Google Scholar] [CrossRef] [Green Version]
- Varsos, C.; Patkos, T.; Oulas, A.; Pavloudi, C.; Gougousis, A.; Zeeshan Ijaz, U.; Filiopoulou, I.; Pattakos, N.; Vanden Berghe, E.; Fernández-Guerra, A.; et al. Optimized R functions for analysis of ecological community data using the R virtual laboratory(Rvlab). Biodivers. Data J. 2016, 4, 8357–8383. [Google Scholar] [CrossRef]
- Guillou, L.; Bachar, D.; Audic, S.; Bass, D.; Berney, C.; Bittner, L.; Boutte, C.; Burgaud, G.; De, V.C.; Decelle, J.; et al. The protist ribosomal reference database (PR2): A catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 2013, 41, 597–604. [Google Scholar] [CrossRef] [Green Version]
- Ramette, A. Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 2007, 62, 142–160. [Google Scholar] [CrossRef] [Green Version]
- McArdle, B.H.; Anderson, M.J. Fitting multivariate models to community data: A comment on distance-based redundancy analysis. Ecology 2001, 82, 290–297. [Google Scholar] [CrossRef]
- Tian, D.; Niu, S. A global analysis of soil acidification caused by nitrogen addition. Environ. Res. Lett. 2015, 10, 1714–1721. [Google Scholar] [CrossRef]
- Schroder, J.L.; Zhang, H.; Girma, K.; Raun, W.R.; Penn, C.J.; Payton, M.E. Soil acidification from long-term use of nitrogen fertilizers on winter wheat. Soil Sci. Soc. Am. J. 2011, 75, 957–964. [Google Scholar] [CrossRef]
- Hu, X.J.; Liu, J.J.; Wei, D.; Zhu, P.; Wang, G. Soil bacterial communities under different long-term fertilization regimes in three locations across the black soil region of Northeast China. Pedosphere 2018, 28, 751–763. [Google Scholar] [CrossRef]
- Wang, J.D.; Xu, X.J.; Ning, Y.W.; Zhang, H.; Hong-Bo, M.A.; Zhang, Y.C. Progresses in agriculral driving factors on accelerated acidification of soils. Soils 2015, 47, 627–633. [Google Scholar] [CrossRef]
- Langarica-Fuentes, A.; Zafar, U.; Heyworth, A.; Brown, T.; Fox, G.; Robson, G.D. Fungal succession in an in-vessel composting system characterized using 454 pyrosequencing. FEMS Microbiol. Ecol. 2014, 88, 296–308. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Zeng, C.L.; Gao, F.; Shi, A.P.; Mao, H.P.; Wei, W. Characteristic analysis of microbial diversity in crud fertilizer from compost of rice straw. Trans. Chin. Soc. Agric. Mach. 2018, 49, 228–234. [Google Scholar] [CrossRef]
- Zhao, H.; Li, X.; Zhang, Z.; Zhao, Y.; Chen, P.; Zhu, Y. Drivers and assemblies of soil eukaryotic microbes among different soil habitat types in a semi-arid mountain in China. PeerJ 2018, 6, e6042. [Google Scholar] [CrossRef]
- Zheng, B.H.; Chen, Z.J.; Li, Y.Y.; Fohrer, N.; Zhang, Y.; Wu, D.Y.; Yan, X.Y.; Li, B.L. Structural characteristics and driving factors of planktonic eukaryotic community in the Danjiangkou Reservoir, China. Water 2020, 12, 3499. [Google Scholar] [CrossRef]
- Meadow, J.F.; Zabinski, C.A. Spatial heterogeneity of eukaryotic microbial communities in an unstudied geothermal diatomaceous biological soil crust: Yellow stone National Park, WY, USA. FEMS Microbiol. Ecol. 2012, 82, 182–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, T.; Ayres, E.; Bardgett, R.D.; Wall, D.H.; Garey, J.R. Molecular study of worldwide distribution and diversity of soil animals. Proc. Natl. Acad. Sci. USA 2011, 108, 17720–17725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, C.C.; Liang, W.J.; Shi, Y.; Lin, X.G.; Zhang, H.Y.; Wu, X.; Xie, G.; Chain, P.; Grogan, P.; Chu, H.Y. Contrasting elevational diversity patterns between eukaryotic soil microbes and plants. Ecol. Soc. Am. 2014, 95, 3190–3202. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.Y.; Xu, M.G.; Zhou, B.K.; Ma, X.; Duan, Y.H. Response and Driving Factors of Bacterial and Fungal Community to Long-Term Fertilization in Black Soil. Sci. Agric. Sin. 2018, 51, 914–925. [Google Scholar] [CrossRef]
- Tsyganov, A.N.; Milbau, A.; Beyens, L. Environmental factors influencing soil testate amoebae in herbaceous and shrubby vegetation along an altitudinal gradient in subarctic tundra (Abisko, Sweden). Eur. J. Protistol. 2013, 49, 238–248. [Google Scholar] [CrossRef]
- Lauber, C.L.; Strickland, M.S.; Bradford, M.A.; Fierer, N. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol. Biochem. 2008, 40, 2407–2415. [Google Scholar] [CrossRef]
- Cai, H.B.; Feng, W.W.; Dong, Y.H.; Ma, Z.L.; Cao, H.J.; Sun, J.D.; Zhang, B.G. Microbial community succession in industrial composting with livestock manure and peach branches and relations with environmental factors. Environ. Sci. 2020, 41, 997–1004. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Polme, S.; Koljalg, U.; Yorou, N.S.; Wijesundera, R.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef] [Green Version]
- Dupont, A.; Griffiths, R.; Bell, T.; Bass, D. Differences in soil micro-eukaryotic communities over soil pH gradients are strongly driven by parasites and saprotrophs. Environ. Microbiol. 2016, 18, 2010–2024. [Google Scholar] [CrossRef]
- Li, D.D.; Luo, P.Y.; Han, X.R.; Yang, J.F.; Cai, F.F.; Liu, T.C. Influence of long-term fertilization on structures of arbuscular mycorrhizal fungi community in a brown soil. J. Plant Nutr. Fert. 2018, 24, 651–660. [Google Scholar] [CrossRef]


| Soil Layer (cm) | pH | Organic Matter (g·kg−1) | Total N (g·kg−1) | Total P (g·kg−1) | Total K (g·kg−1) | Alkali-Hydrolyzable N (mg·kg−1) | Available P (mg·kg−1) | Available K (mg·kg−1) |
|---|---|---|---|---|---|---|---|---|
| 0–20 | 8.49 | 11.92 | 1.07 | 0.67 | 23.55 | 88.65 | 8.09 | 104.98 |
| 20–40 | 8.54 | 7.48 | 0.81 | 0.50 | 24.22 | 54.16 | 6.36 | 75.2 |
| Treatment | Chemical Fertilizer (kg/hm2) | Organic Fertilizer (kg/hm2) | Total Nutrient (kg/hm2) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| N | P2O5 | K2O | N | P2O5 | K2O | N | P2O5 | K2O | |
| F | 225 | 75 | 75 | 0 | 0 | 0 | 225 | 75 | 75 |
| FM | 225 | 75 | 75 | 70 | 45 | 145 | 295 | 120 | 220 |
| FS | 225 | 75 | 75 | 45 | 25 | 80 | 270 | 100 | 155 |
| FC | 225 | 75 | 75 | 60 | 105 | 85 | 285 | 180 | 160 |
| CK | 0 | 0 | 0 | 0 | 0 | 00 | 0 | 0 | 0 |
| Treatment | pH | Organic Matter (g·kg−1) | Total N (g·kg−1) | Total P (g·kg−1) | Total K (g·kg−1) | Alkali-Hydrolyzable N (mg·kg−1) | Available P (mg·kg−1) | Available K (mg·kg−1) |
|---|---|---|---|---|---|---|---|---|
| F | 8.28 ± 0.01 a | 10.56 ± 0.99 d | 1.02 ± 0.01 a | 0.71 ± 0.01 b | 21.48 ± 1.28 c | 91.33 ± 7.95 c | 9.62 ± 1.11 d | 75.88 ± 2.23 e |
| FM | 8.10 ± 0.02 b | 13.03 ± 1.02 a | 1.08 ± 0.01 a | 0.83 ± 0.01 ab | 23.07 ± 1.40 a | 103.57 ± 9.91 a | 21.18 ± 2.14 b | 156.63 ± 9.92 a |
| FS | 8.25 ± 0.01 a | 11.17 ± 1.23 c | 0.98 ± 0.01 a | 0.86 ± 0.02 a | 22.49 ± 1.22 b | 78.75 ± 1.19 e | 11.25 ± 1.82 c | 83.94 ± 8.00 c |
| FC | 8.27 ± 0.03 a | 12.17 ± 0.90 b | 1.08 ± 0.02 a | 0.72 ± 0.02 b | 20.76 ± 1.99 d | 97.20 ± 5.81 b | 31.92 ± 2.18 a | 113.02 ± 5.98 b |
| CK | 8.31 ± 0.01 a | 11.13 ± 0.75 c | 0.95 ± 0.01 a | 0.58 ± 0.01 c | 18.00 ± 1.20 e | 87.83 ± 3.27 d | 7.87 ± 1.90 e | 82.53 ± 2.59 d |
| Treatment | Chao1 | Observed_Species | Shannon | Simpson |
|---|---|---|---|---|
| F | 1302.99 ± 93.29 a | 1215.77 ± 97.41 a | 7.0417 ± 0.8380 a | 0.9610 ± 0.0315 a |
| FM | 1227.52 ± 176.91 a | 1167.53 ± 147.06 a | 6.8935 ± 0.6906 a | 0.9418 ± 0.0451 a |
| FS | 1031.93 ± 195.72 a | 968.43 ± 162.85 a | 6.3081 ± 0.4664 a | 0.9420 ± 0.0171 a |
| FC | 1275.53 ± 177.55 a | 1211.47 ± 167.38 a | 7.2898 ± 0.5858 a | 0.9715 ± 0.0148 a |
| CK | 1218.75 ± 21.89 a | 1161.00 ± 6.88 a | 7.2049 ± 0.1519 a | 0.9708 ± 0.0110 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Zhou, H.; Xie, W.; Yang, Z.; Zhang, P. Effects of Returning Different Organic Materials in Combination with Inorganic Fertilizers on the Diversity of Eukaryotic Microorganisms in Semi-Arid Northern China. Agronomy 2022, 12, 3116. https://doi.org/10.3390/agronomy12123116
Liu Z, Zhou H, Xie W, Yang Z, Zhang P. Effects of Returning Different Organic Materials in Combination with Inorganic Fertilizers on the Diversity of Eukaryotic Microorganisms in Semi-Arid Northern China. Agronomy. 2022; 12(12):3116. https://doi.org/10.3390/agronomy12123116
Chicago/Turabian StyleLiu, Zhiping, Huaiping Zhou, Wenyan Xie, Zhenxing Yang, and Pengfei Zhang. 2022. "Effects of Returning Different Organic Materials in Combination with Inorganic Fertilizers on the Diversity of Eukaryotic Microorganisms in Semi-Arid Northern China" Agronomy 12, no. 12: 3116. https://doi.org/10.3390/agronomy12123116





