Influence of Substituent Chain Branching on the Transfection Efficacy of Cyclopropenium-Based Polymers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Procedure for the Synthesis of 1,2-Bis(dibutylamino)-3-chlorocyclopropenium Chloride (BACBu)
2.2.1. Synthesis of 2,3-Bis(dibutylamino)-1-cyclopropenone
2.2.2. Synthesis of 1,2-Bis(dibutylamino)-3-chlorocyclopropenium Chloride (BACBu)
2.3. Synthesis of PMAS(Bu)
2.4. Synthesis of PEI(Bu)
2.5. Synthesis of 1,2-Bis(diisopropylamino)-3-chlorocyclopropenium Chloride (BACiP), PMAS(iP), and PEI(iP)
2.6. Cell Culture
2.7. Cell Viability
2.8. Cell Transfection and Luciferase Expression
2.9. Hydrodynamic Size and Zeta Potential Measurement
2.10. Gel Electrophoresis Shift Assay
3. Results and Discussion
3.1. Cationic Polymer Synthesis
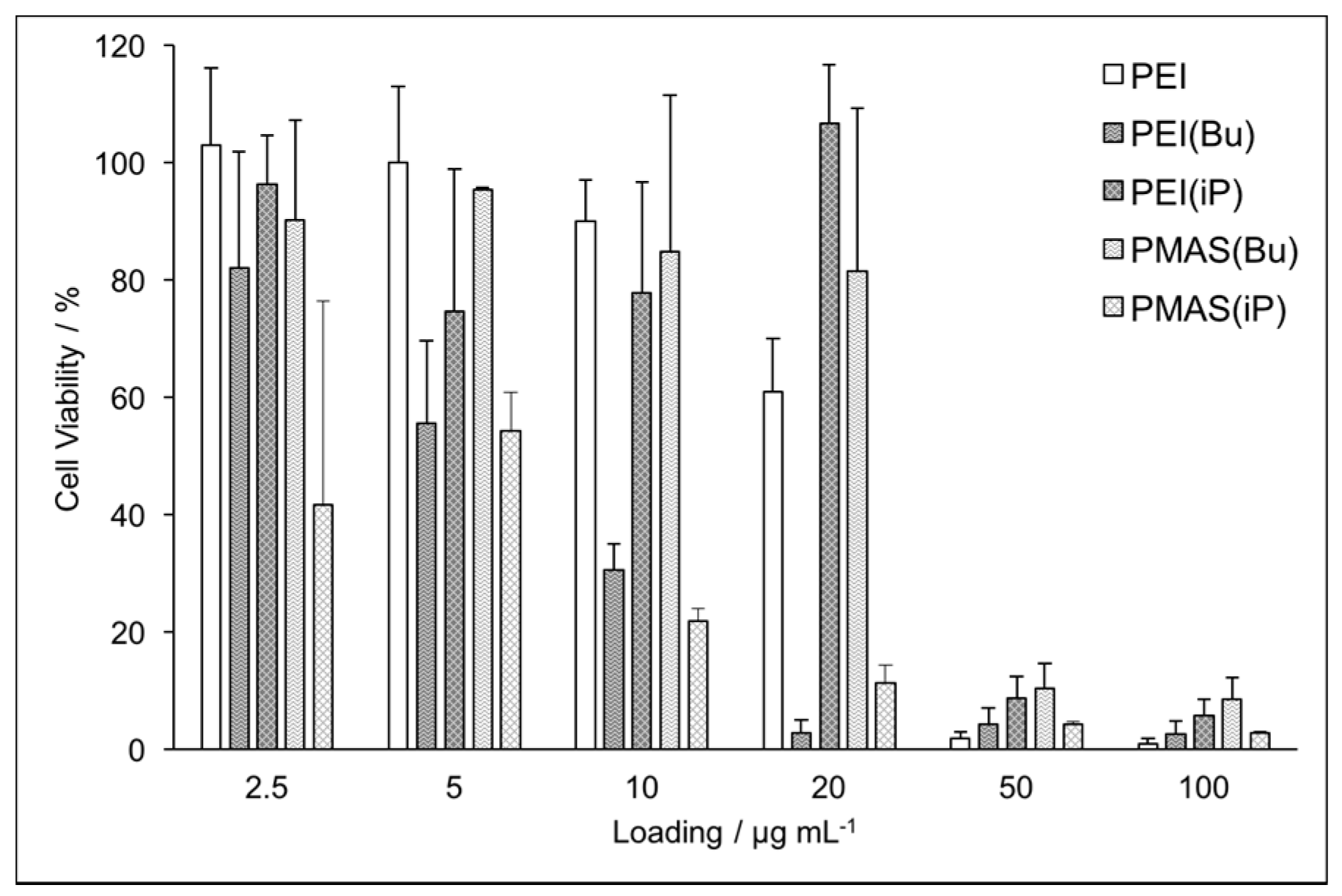
3.2. Biocompatibility Studies
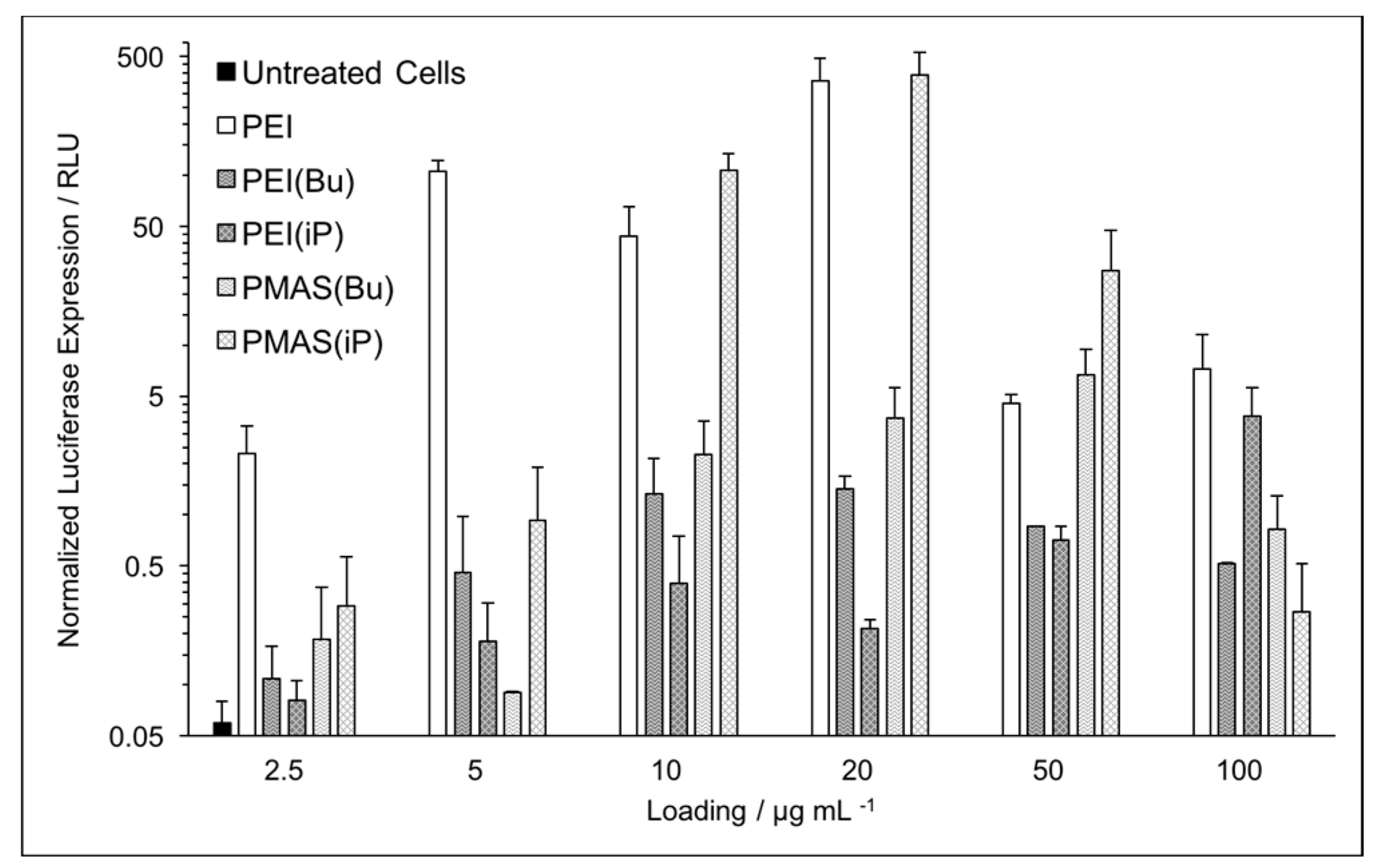
3.3. DNA Binding and In Vitro Gene Transfection
3.4. Hydrodynamic Size and Zeta Potential Measurements
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Mintzer, M.A.; Simanek, E.E. Nonviral vectors for gene delivery. Chem. Rev. 2009, 109, 259–302. [Google Scholar] [CrossRef] [PubMed]
- Bender, E. Gene therapy: Industrial strength. Nature 2016, 537, S57–S59. [Google Scholar] [CrossRef] [PubMed]
- Li, S.D.; Huang, L. Non-viral is superior to viral gene delivery. J. Control. Release 2007, 3, 181–183. [Google Scholar] [CrossRef] [PubMed]
- Kay, M.A. State-of-the-art gene-based therapies: The road ahead. Nat. Rev. Genet. 2011, 5, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Kostarelos, K.; Miller, A.D. Synthetic, self-assembly ABCD nanoparticles: A structural paradigm for viable synthetic non-viral vectors. Chem. Soc. Rev. 2005, 34, 970–994. [Google Scholar] [CrossRef] [PubMed]
- Samal, S.K.; Dash, M.; Van Vlierberghe, S.; Kaplan, D.L.; Chiellini, E.; van Blitterswijk, C.; Moroni, L.; Dubruel, P. Cationic polymers and their therapeutic potential. Chem. Soc. Rev. 2012, 21, 7147–7194. [Google Scholar] [CrossRef] [PubMed]
- Merdan, T.; Kopeček, J.; Kissel, T. Prospects for cationic polymers in gene and oligonucleotide therapy against cancer. Adv. Drug Deliv. Rev. 2002, 54, 715–758. [Google Scholar] [CrossRef]
- Boussif, O.; Lezoualc’h, F.; Zanta, M.A.; Mergny, M.D.; Scherman, D.; Demeneix, B.; Behr, J.P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proc. Natl. Acad. Sci. USA 1995, 92, 7297–7301. [Google Scholar] [CrossRef] [PubMed]
- Godbey, W.T.; Wu, K.K.; Mikos, A.G. Size matters: Molecular weight affects the efficiency of poly(ethylenimine) as a gene delivery vehicle. J. Biomed. Mater. Res. 1998, 3, 268–275. [Google Scholar] [CrossRef]
- Fischer, D.; Bieber, T.; Li, Y.; Elsässer, H.P.; Kissel, T. A novel non-viral vector for DNA delivery based on low molecular weight, branched polyethylenimine: Effect of molecular weight on transfection efficiency and cytotoxicity. Pharm. Res. 1999, 8, 1273–1279. [Google Scholar] [CrossRef]
- Layman, J.M.; Ramirez, S.M.; Green, M.D.; Long, T.E. Influence of polycation molecular weight on poly(2-dimethylaminoethyl methacrylate)-mediated DNA delivery in vitro. Biomacromolecules 2009, 10, 1244–1252. [Google Scholar] [CrossRef] [PubMed]
- Bishop, C.J.; Abubaker-Sharif, B.; Guiriba, T.; Tzeng, S.Y.; Green, J.J. Gene delivery polymer structure-function relationships elucidated via principal component analysis. Chem. Commun. 2015, 51, 12134–12137. [Google Scholar] [CrossRef] [PubMed]
- Breitenkamp, R.B.; Emrick, T. Pentalysine-grafted ROMP polymers for DNA complexation and delivery. Biomacromolecules 2008, 9, 2495–2500. [Google Scholar] [CrossRef] [PubMed]
- Synatschke, C.V.; Schallon, A.; Jérôme, V.; Freitag, R.; Müller, A.H.E. Influence of polymer architecture and molecular weight of poly(2-(dimethylamino)ethyl methacrylate) polycations on transfection efficiency and cell viability in gene delivery. Biomacromolecules 2011, 12, 4247–4255. [Google Scholar] [CrossRef] [PubMed]
- Raup, A.; Stahlschmidt, U.; Jérôme, V.; Synatschke, C.V.; Müller, A.H.E.; Freitag, R. Influence of polyplex formation on the performance of star-shaped polycationic transfection agents for mammalian cells. Polymers 2016, 8, 224. [Google Scholar] [CrossRef]
- Zhou, D.; Cutlar, L.; Gao, Y.; Wang, W.; O’Keeffe-Ahern, J.; McMahon, S.; Duarte, B.; Larcher, F.; Rodriguez, B.J.; Greiser, U.; et al. The transition from linear to highly branched poly(β-amino ester)s: Branching matters for gene delivery. Sci. Adv. 2016, 2, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Cutlar, L.; Zhou, D.; Gao, Y.; Zhao, T.; Greiser, U.; Wang, W.; Wang, W. Highly branched poly(β-amino esters): Synthesis and application in gene delivery. Biomacromolecules 2015, 16, 2609–2617. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Gao, Y.; O’Keeffe Ahern, J.; Xu, Q.; Huang, X.; Greiser, U.; Wang, W. Development of branched poly(5-amino-1-pentanol-co-1,4-butanediol diacrylate) with high gene transfection potency across diverse cell types. ACS Appl. Mater. Interfaces 2016, 8, 34218–34226. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Gao, Y.; Aied, A.; Cutlar, L.; Igoucheva, O.; Newland, B.; Alexeeve, V.; Greiser, U.; Uitto, J.; Wang, W. Highly branched poly(β-amino ester)s for skin gene therapy. J. Control. Release 2016, 244, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Gao, Y.; Zhou, D.; Greiser, U.; Guo, T.; Guo, R.; Wang, W. Biodegradable highly branched poly(β-amino ester)s for targeted cancer cell gene transfection. ACS Biomater. Sci. Eng. 2016, 18, 440–451. [Google Scholar] [CrossRef]
- Freyer, J.L.; Brucks, S.D.; Gobieski, G.S.; Russell, S.T.; Yozwiak, C.E.; Sun, M.; Chen, Z.; Jiang, Y.; Bandar, J.S.; Stockwell, B.R.; et al. Clickable poly(ionic liquids): A materials platform for transfection. Angew. Chem. Int. Ed. 2016, 55, 12382–12386. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Chen, L.; Zhang, Z.; Gu, W.; Li, Y. Linear cationic click polymer for gene delivery: Synthesis, biocompatibility, and in vitro transfection. Biomacromolecules 2010, 11, 3102–3111. [Google Scholar] [CrossRef] [PubMed]
- Hemp, S.T.; Allen, M.H., Jr.; Green, M.D.; Long, T.E. Phosphonium-containing polyelectrolytes for nonviral gene delivery. Biomacromolecules 2012, 13, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Jarzębińska, A.; Pasewald, T.; Lambrecht, J.; Mykhaylyk, O.; Kümmerling, L.; Beck, P.; Hasenpusch, G.; Rudolph, C.; Plank, C.; Dohmen, C. A Single methylene group in oligoalkylamine-based cationic polymers and lipids promotes enhanced mRNA delivery. Angew. Chem. Int. Ed. 2016, 55, 9591–9595. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Freyer, J.L.; Cotanda, P.; Brucks, S.D.; Killops, K.L.; Bandar, J.S.; Torsitano, C.; Balsara, N.P.; Lambert, T.H.; Campos, L.M. The evolution of cyclopropenium ions into functional polyelectrolytes. Nat. Commun. 2015, 6, 5950. [Google Scholar] [CrossRef] [PubMed]
- Vaidyanathan, S.; Orr, B.G.; Banaszak Holl, M.M. Role of cell membrane-vector interactions in successful gene delivery. Acc. Chem. Res. 2016, 49, 1486–1493. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.L.; Oliveira, H.; Pandita, D.; Rodrigues, J.; Pêgo, A.P.; Granja, P.L.; Tomás, H. Functionalization of poly(amidoamine) dendrimers with hydrophobic chains for improved gene delivery in mesenchymal stem cells. J. Control. Release 2010, 144, 55–64. [Google Scholar] [CrossRef] [PubMed]

| Transfection agent 1 | MM 2 (kDa) | Charge Ratio 3 | DH (nm) | ζ Potential (mV) |
|---|---|---|---|---|
| PEI(Bu) | 215 | 6:1 | 110 ± 40 | 45 ± 7 |
| PEI(iP) | 182 | 35:1 | 100 ± 40 | 58 ± 5 |
| PMAS(Bu) | 25 | 12:1 | 150 ± 50 | 31 ± 8 |
| PMAS(iP) | 21 | 5:1 | 160 ± 40 | 44 ± 6 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brucks, S.D.; Freyer, J.L.; Lambert, T.H.; Campos, L.M. Influence of Substituent Chain Branching on the Transfection Efficacy of Cyclopropenium-Based Polymers. Polymers 2017, 9, 79. https://doi.org/10.3390/polym9030079
Brucks SD, Freyer JL, Lambert TH, Campos LM. Influence of Substituent Chain Branching on the Transfection Efficacy of Cyclopropenium-Based Polymers. Polymers. 2017; 9(3):79. https://doi.org/10.3390/polym9030079
Chicago/Turabian StyleBrucks, Spencer D., Jessica L. Freyer, Tristan H. Lambert, and Luis M. Campos. 2017. "Influence of Substituent Chain Branching on the Transfection Efficacy of Cyclopropenium-Based Polymers" Polymers 9, no. 3: 79. https://doi.org/10.3390/polym9030079