pH-Triggered Sheddable Shielding System for Polycationic Gene Carriers
Abstract
:1. Introduction
2. Experimental Section
2.1. Materials
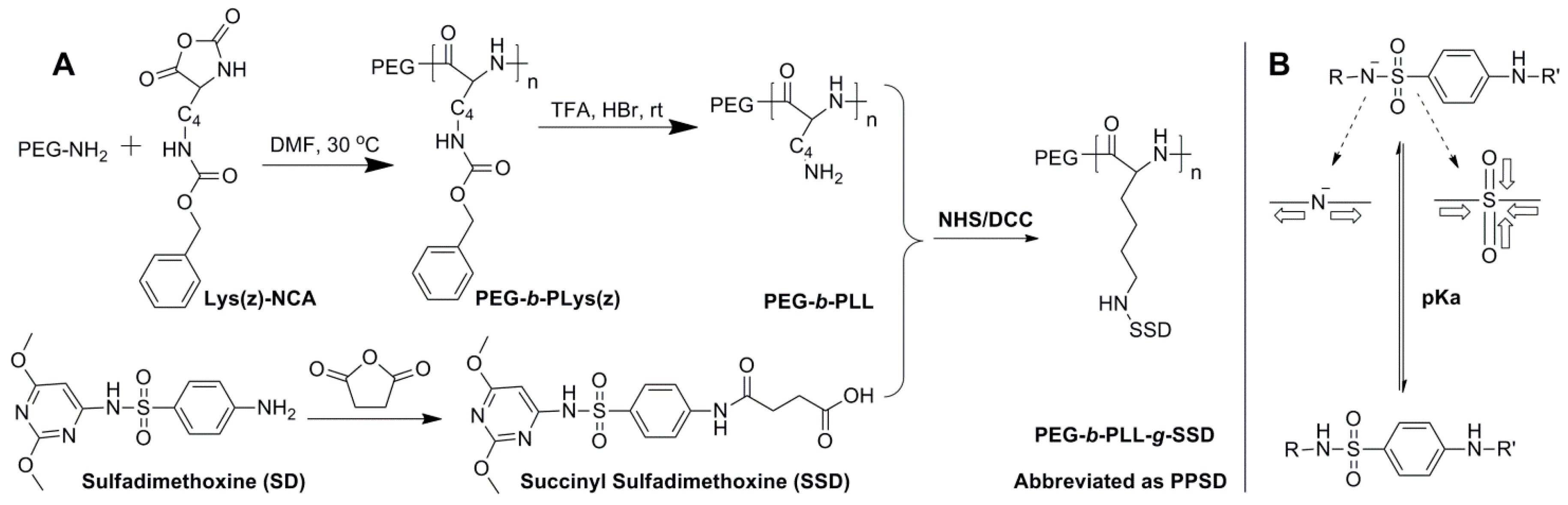
2.2. Synthesis and Characterization of PPSD
2.3. Preparation and Characterization of DNA/PEI/PPSD Ternary Complexes
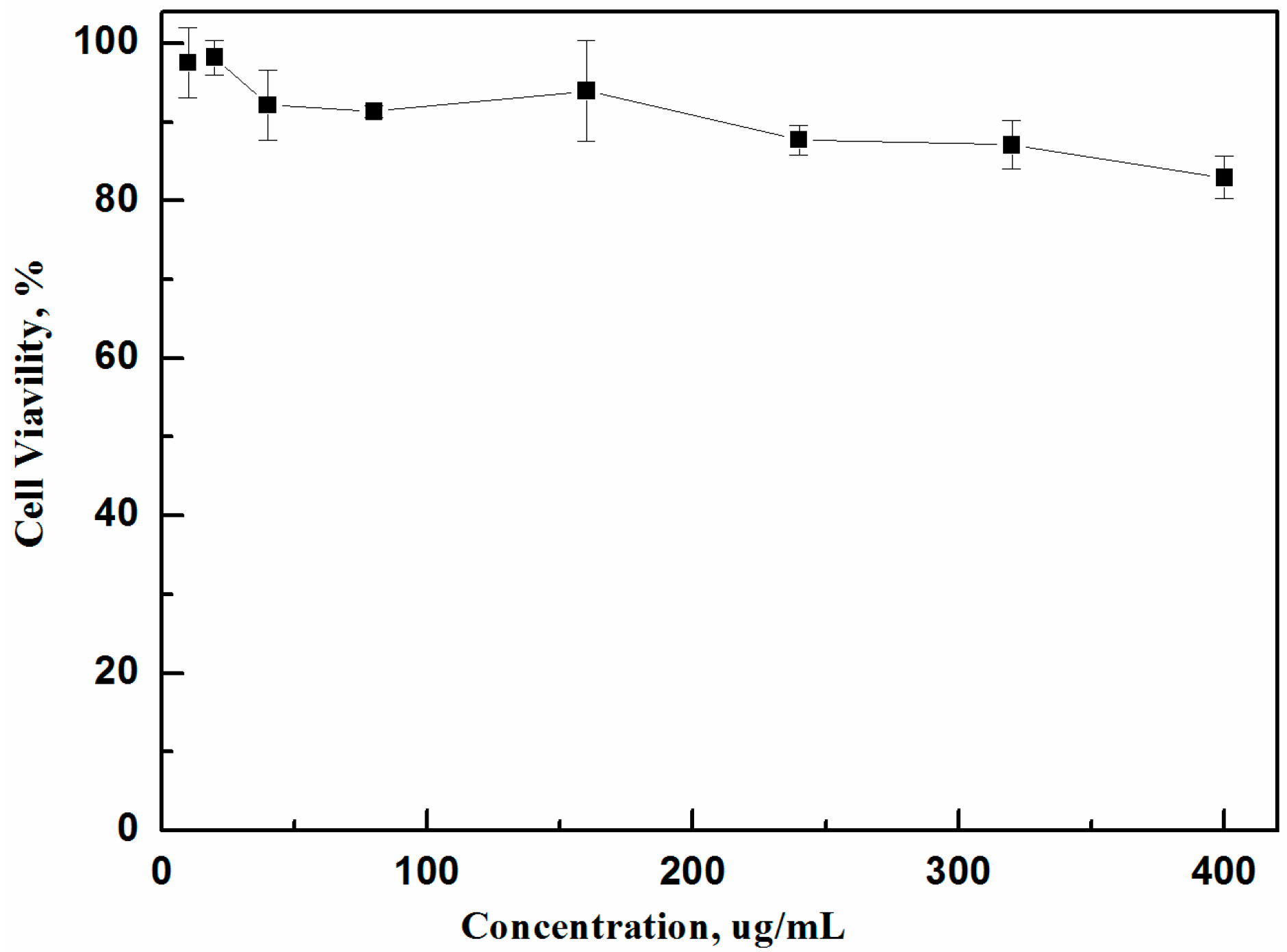
2.4. Cytotoxicity Assay
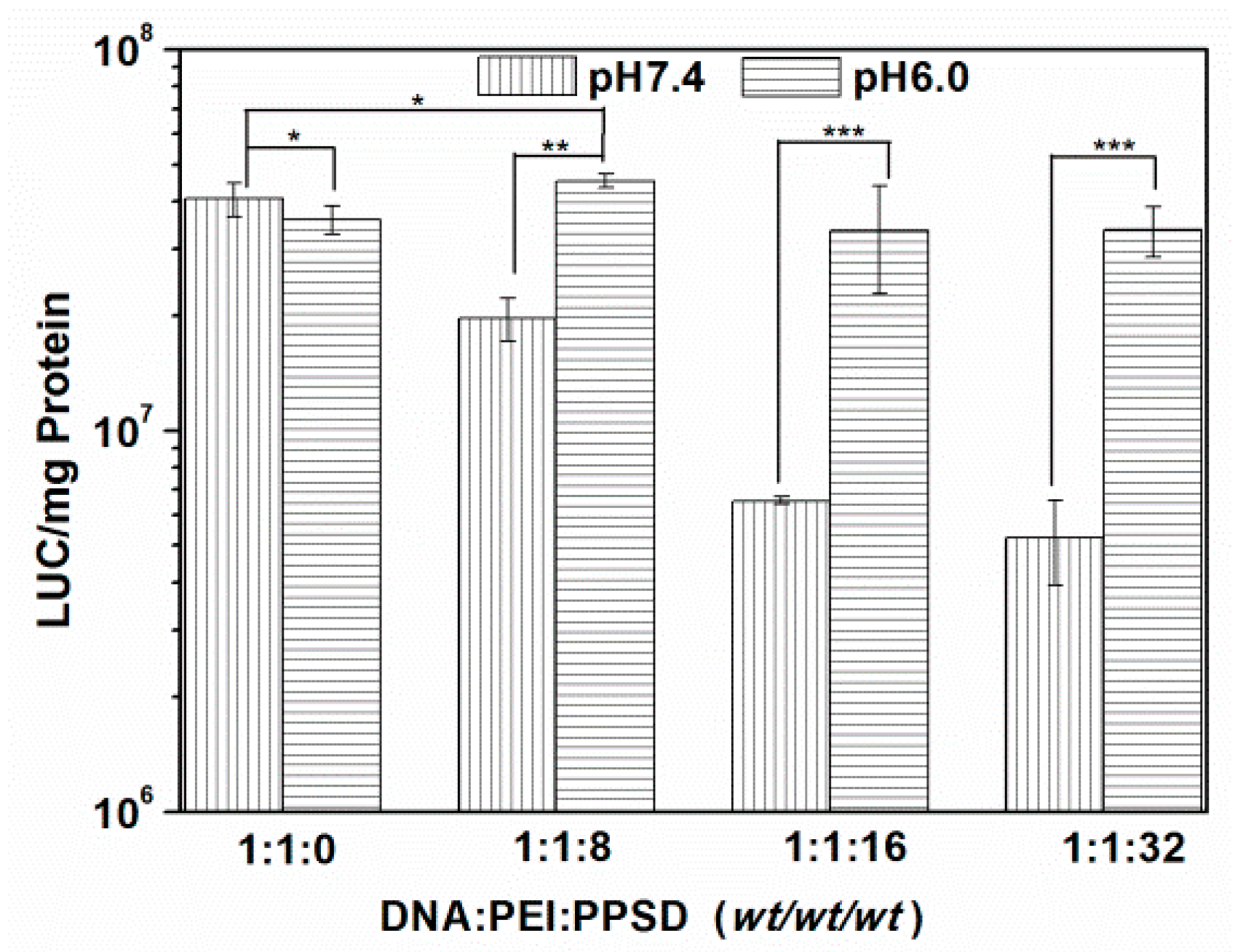
2.5. In Vitro Gene Transfection at Different pH Values
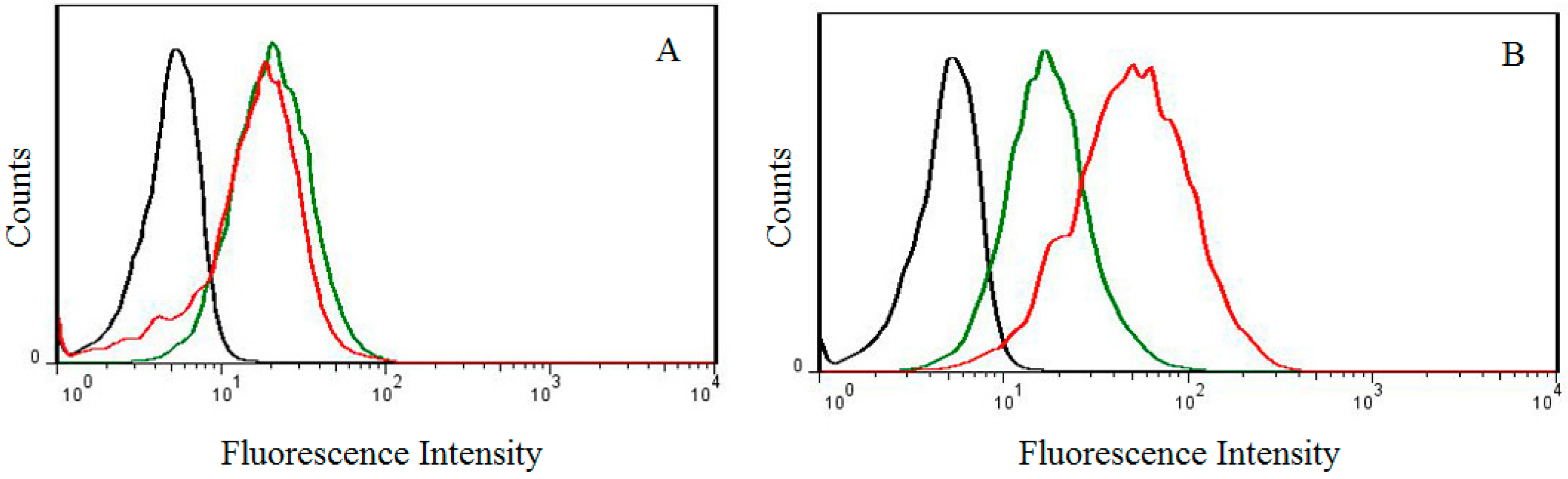
2.6. Cellular Uptake at Different pH Values
3. Results and Discussion
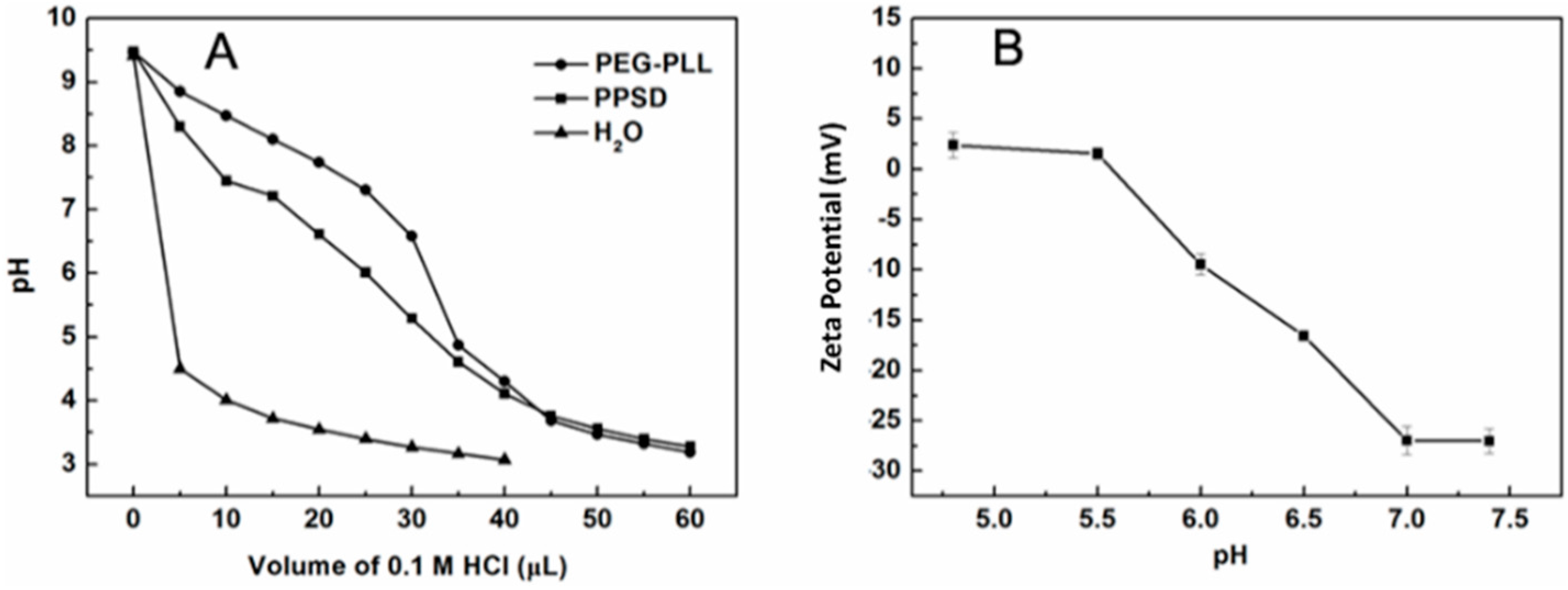
3.1. Synthesis and Characterization of PPSD
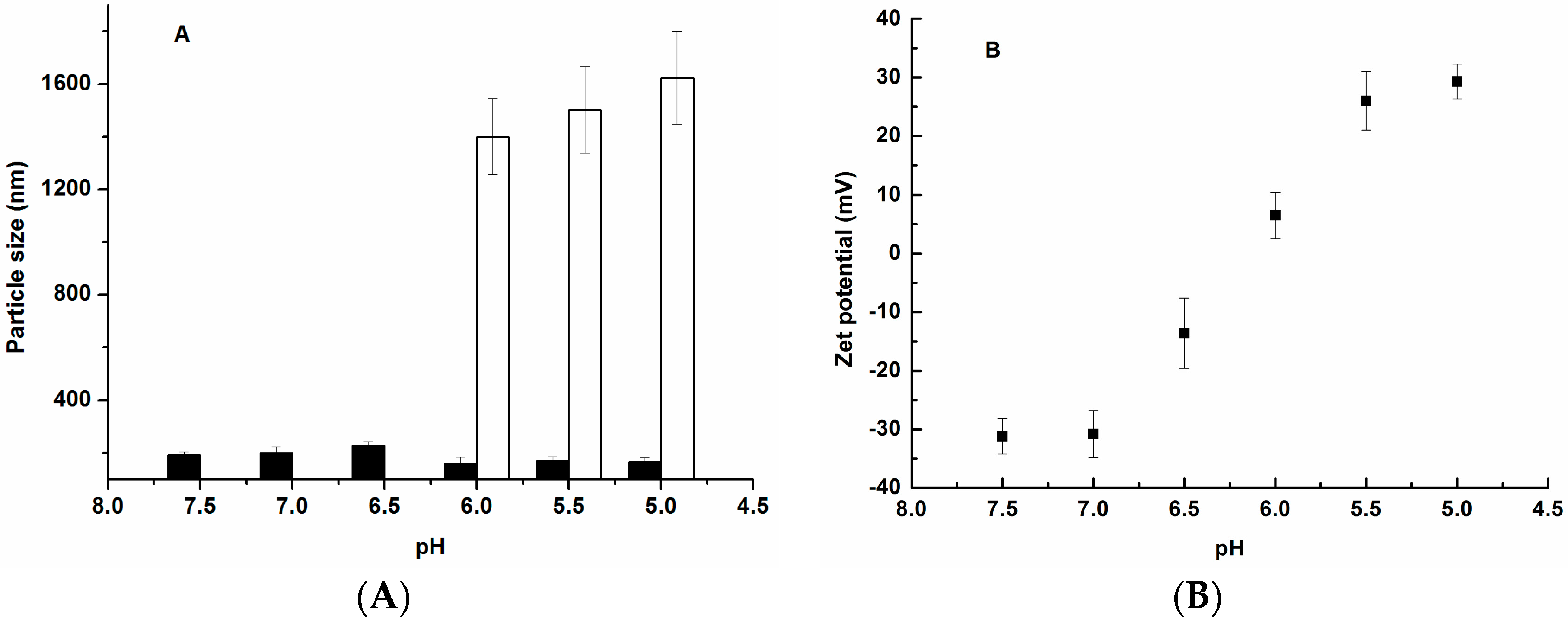
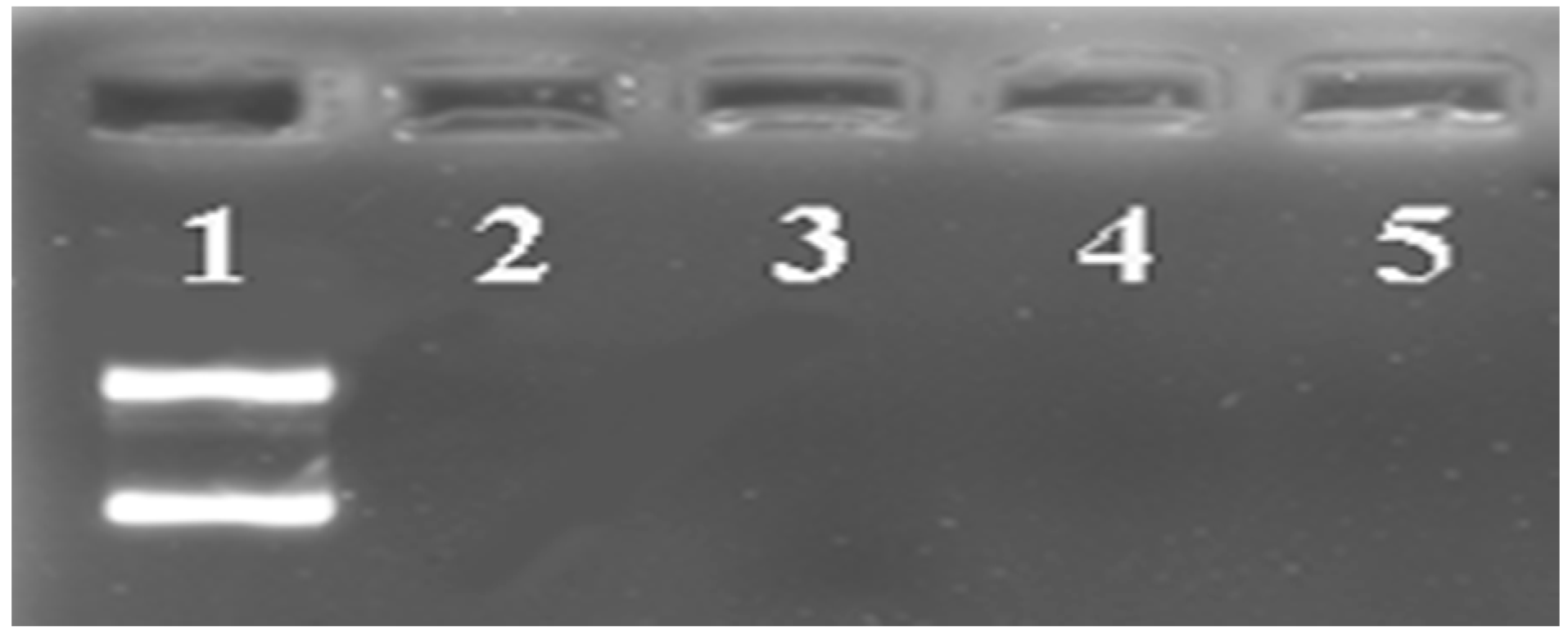
3.2. Characterization of Ternary Complexes
3.3. Cytotoxicity Assay, In Vitro Transfection, and Cellular Uptake Assay
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yan, X.; Blacklock, J.; Li, J.; Moehwald, H. One-pot synthesis of polypeptide-gold nanoconjugates for in vitro gene transfection. ACS Nano 2012, 6, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Dash, P.R.; Read, M.L.; Barrett, L.B.; Wolfert, M.; Seymour, L.W. Factors affecting blood clearance and in vivo distribution of polyelectrolyte complexes for gene delivery. Gene Ther. 1999, 6, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Rong, L.; Lei, Q.; Cao, P.X.; Qin, S.Y.; Zheng, D.W.; Jia, H.Z.; Zhu, J.Y.; Cheng, R.X.; Zhang, X.Z. A surface charge-switchable and folate modified system for co-delivery of proapoptosis peptide and p53 plasmid in cancer therapy. Biomaterials 2016, 77, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Tseng, W.C.; Su, L.Y.; Fang, T.Y. pH responsive PEGylation through metal affinity for gene delivery mediated by histidine-grafted polyethylenimine. J. Biomed. Mater. Res. B 2013, 101B, 375–386. [Google Scholar] [CrossRef] [PubMed]
- He, Y.Y.; Cheng, G.; Xie, L.; Nie, Y.; He, B.; Gu, Z.W. Polyethyleneimine/DNA polyplexes with reduction-sensitive hyaluronic acid derivatives shielding for targeted gene delivery. Biomaterials 2013, 34, 1235–1245. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.X.; Li, M.; Liu, C.X.; Liu, Y.J.; Zhang, N. pH-sensitive self-assembled carboxymethyl chitosan-modified DNA/polyethylenimine complexes for efficient gene delivery. J. Biomed. Nanotechnol. 2014, 10, 3397–3406. [Google Scholar] [CrossRef] [PubMed]
- Maeda, Y.; Pittella, F.; Nomoto, T.; Takemoto, H.; Nishiyama, N.; Miyata, K.; Kataoka, K. Fine-tuning of charge-conversion polymer structure for efficient endosomal escape of siRNA-loaded calcium phosphate hybrid micelles. Macromol. Rapid Commun. 2014, 35, 1211–1215. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Shen, G.; Chen, C.; Xing, R.; Zou, Q.; Ma, G.; Yan, X. Nanoengineering of stimuli-responsive protein-based biomimetic protocells as versatile drug delivery tools. Chem. Eur. J. 2014, 20, 6880–6887. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Fei, J.; Yan, X.; Wang, A.; Li, J. Enzyme-responsive release of doxorubicin from monodisperse dipeptide-based nanocarriers for highly efficient cancer treatment in vitro. Adv. Funct. Mater. 2015, 25, 1193–1204. [Google Scholar] [CrossRef]
- Xia, J.L.; Feng, Z.C.; Yang, H.Y.; Lin, S.Q.; Han, B. Acidity-activated shielding strategies of cationic gene delivery for cancer therapy. Curr. Pharm. Biotechnol. 2016, 17, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Du, J.Z.; Mao, C.Q.; Yuan, Y.Y.; Yang, X.Z.; Wang, J. Tumor extracellular acidity-activated nanoparticles as drug delivery systems for enhanced cancer therapy. Biotechnol. Adv. 2014, 32, 789–803. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Miura, S.; Na, K.; Bae, Y.H. pH-responsive and charge shielded cationic micelle of poly(l-histidine)-block-short branched PEI for acidic cancer treatment. J. Control. Release 2013, 172, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.C.; Huh, K.M.; Bae, Y.H. Polymeric nucleic acid carriers: current issues and novel design approaches. J. Control. Release 2012, 164, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.I.; Bae, Y.H. pH-induced solubility transition of sulfonamide-based polymers. J. Control. Release 2002, 80, 145–155. [Google Scholar] [CrossRef]
- Sethuraman, V.A.; Na, K.; Bae, Y.H. pH-responsive sulfonamide/PEI system for tumor specific gene delivery: An in vitro study. Biomacromolecules 2006, 7, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Miyata, K.; Oba, M.; Ishii, T.; Fukushima, S.; Han, M.; Koyama, H.; Nishiyama, N.; Kataoka, K. Charge-conversion ternary polyplex with endosome disruption moiety: A technique for efficient and safe gene delivery. Angew. Chem. Int. Ed. 2008, 47, 5163–5166. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.Y.; Mao, C.Q.; Du, X.J.; Du, J.Z.; Wang, F.; Wang, J. Surface charge switchable nanoparticles based on zwitterionic polymer for enhanced drug delivery to tumor. Adv. Mater. 2012, 24, 5476–5480. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.Z.; Du, J.Z.; Dou, S.; Mao, C.Q.; Long, H.Y.; Wang, J. Sheddable ternary nanoparticles for tumor acidity-targeted siRNA delivery. ACS Nano 2012, 6, 771–781. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Zhou, Z.; Sui, M.; Tang, J.; Xu, P.; van Kirk, E.A.; Murdoch, W.J.; Fan, M.; Radosz, M. Charge-reversal polyamidoamine dendrimer for cascade nuclear drug delivery. Nanomedicine-UK 2010, 5, 1205–1217. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Shen, Y.; Tang, J.; Fan, M.; Kirk, E.A.V.; Murdoch, W.J.; Radosz, M. Charge-reversal drug conjugate for targeted cancer cell nuclear drug delivery. Adv. Funct. Mater. 2009, 19, 3580–3589. [Google Scholar] [CrossRef]
- Guan, X.W.; Li, Y.H.; Jiao, Z.X.; Lin, L.L.; Chen, J.; Guo, Z.P.; Tian, H.Y.; Chen, X.S. Codelivery of antitumor drug and gene by a pH-sensitive charge-conversion system. ACS Appl. Mate. Interfaces 2015, 7, 3207–3215. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, X.Z.; Tian, H.Y.; Zhu, X.J.; Chen, X.S. Application of zwitterionic polymers in the treatment of malignant tumors. Chem. J. Chin. Univ. 2015, 36, 2148–2156. [Google Scholar]
- Chen, J.; Dong, X.; Feng, T.S.; Lin, L.; Guo, Z.P.; Xia, J.L.; Tian, H.Y.; Chen, X.S. Charge-conversional zwitterionic copolymer as pH-sensitive shielding system for effective tumor treatment. Acta. Biomater. 2015, 26, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.Y.; Guo, Z.P.; Lin, L.; Jiao, Z.X.; Chen, J.; Gao, S.Q.; Zhu, X.J.; Chen, X.S. pH-responsive zwitterionic copolypeptides as charge conversional shielding system for gene carriers. J. Control. Release 2014, 174, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.L.; Tian, H.Y.; Chen, J.; Guo, Z.P.; Lin, L.; Yang, H.Y.; Han, B. Polyglutamic acid based polyanionic shielding system for polycationic gene carriers. Chin. J. Polym. Sci. 2016, 34, 316–323. [Google Scholar] [CrossRef]
- Guo, S.T.; Huang, Y.Y.; Zhang, W.D.; Wang, W.W.; Wei, T.; Lin, D.S.; Xing, J.F.; Deng, L.D.; Du, Q.; Liang, Z. CTernary complexes of amphiphilic polycaprolactone-graft-poly(N,N-dimethylaminoethyl methaaylate), DNA and polyglutamic acid-graft-poly (ethylene glycol) for gene delivery. Biomaterials 2011, 32, 4283–4292. [Google Scholar] [CrossRef] [PubMed]
- Kurosaki, T.; Kitahara, T.; Fumoto, S.; Nishida, K.; Nakamura, J.; Niidome, T.; Kodama, Y.; Nakagawa, H.; To, H.; Sasaki, H. Ternary complexes of pDNA, polyethylenimine, and γ-polyglutamic acid for gene delivery systems. Biomaterials 2009, 30, 2846–2853. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.L.; Chen, J.; Tian, H.Y.; Chen, X.S. Synthesis and characterization of a pH-sensitive shielding system for polycation gene carriers. Sci. China Chem. 2010, 53, 502–507. [Google Scholar] [CrossRef]
- Park, J.S.; Yi, S.W.; Kim, H.J.; Park, K.H. Receptor-mediated gene delivery into human mesenchymal stem cells using hyaluronic acid-shielded polyethylenimine/pDNA nanogels. Carbohydr. Polym. 2016, 136, 791–802. [Google Scholar] [CrossRef] [PubMed]


© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, J.; Tian, H.; Chen, J.; Lin, L.; Guo, Z.; Han, B.; Yang, H.; Feng, Z. pH-Triggered Sheddable Shielding System for Polycationic Gene Carriers. Polymers 2016, 8, 141. https://doi.org/10.3390/polym8040141
Xia J, Tian H, Chen J, Lin L, Guo Z, Han B, Yang H, Feng Z. pH-Triggered Sheddable Shielding System for Polycationic Gene Carriers. Polymers. 2016; 8(4):141. https://doi.org/10.3390/polym8040141
Chicago/Turabian StyleXia, Jialiang, Huayu Tian, Jie Chen, Lin Lin, Zhaopei Guo, Bing Han, Hongyan Yang, and Zongcai Feng. 2016. "pH-Triggered Sheddable Shielding System for Polycationic Gene Carriers" Polymers 8, no. 4: 141. https://doi.org/10.3390/polym8040141






