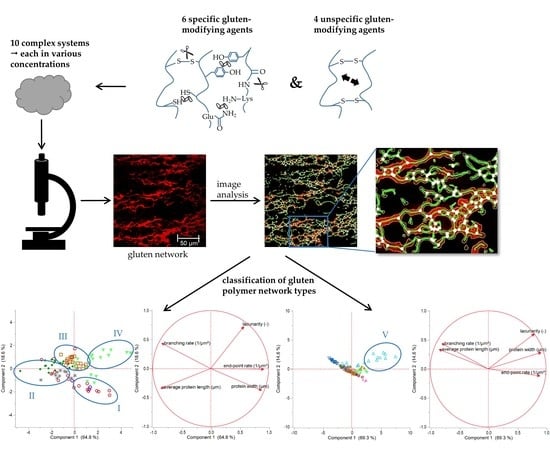
Gluten Polymer Networks—A Microstructural Classification in Complex Systems
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Flour-Water-Systems
2.3. Microstructure Analysis by Confocal Laser Scanning Microscopy
2.4. Image Processing and Analysis
2.5. Statistical Analysis
3. Results and Discussion
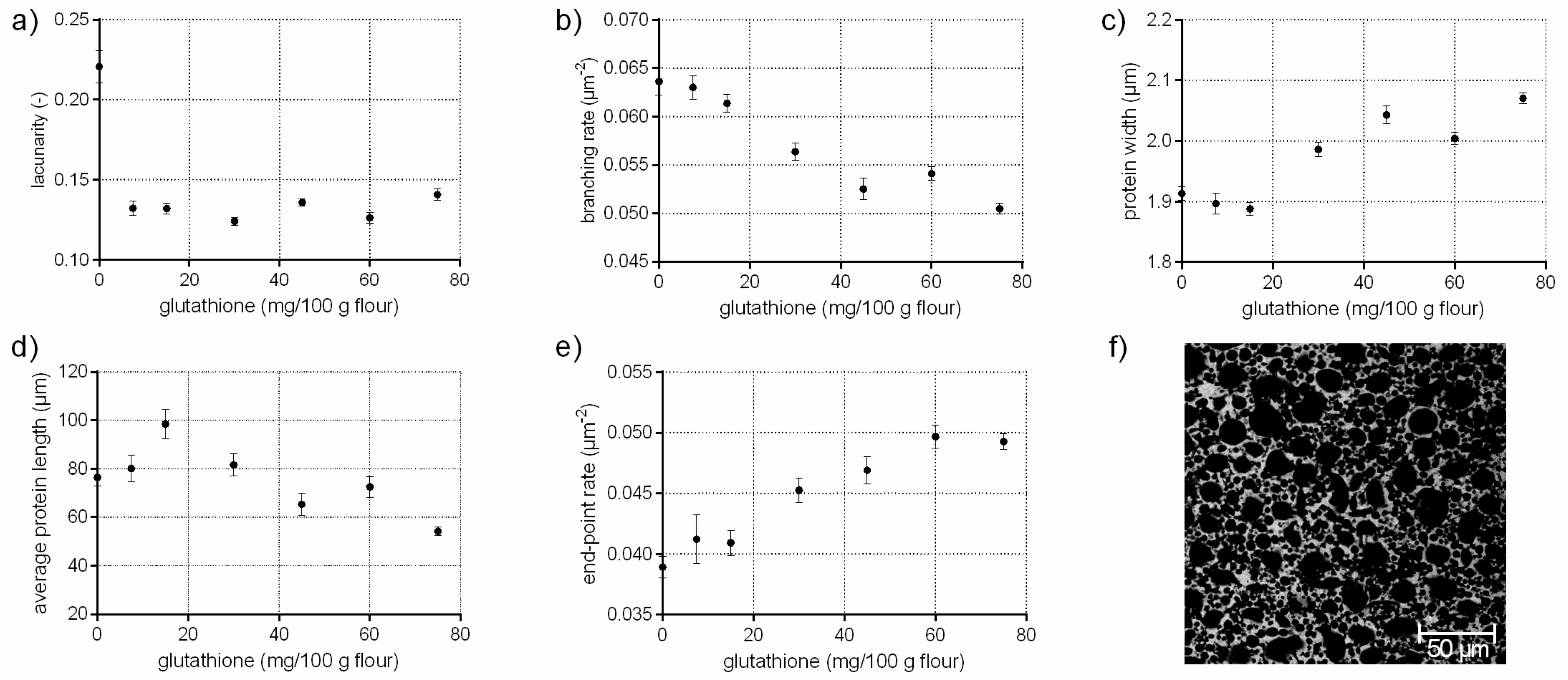
3.1. Protein Network Formation Modified by Glutathione
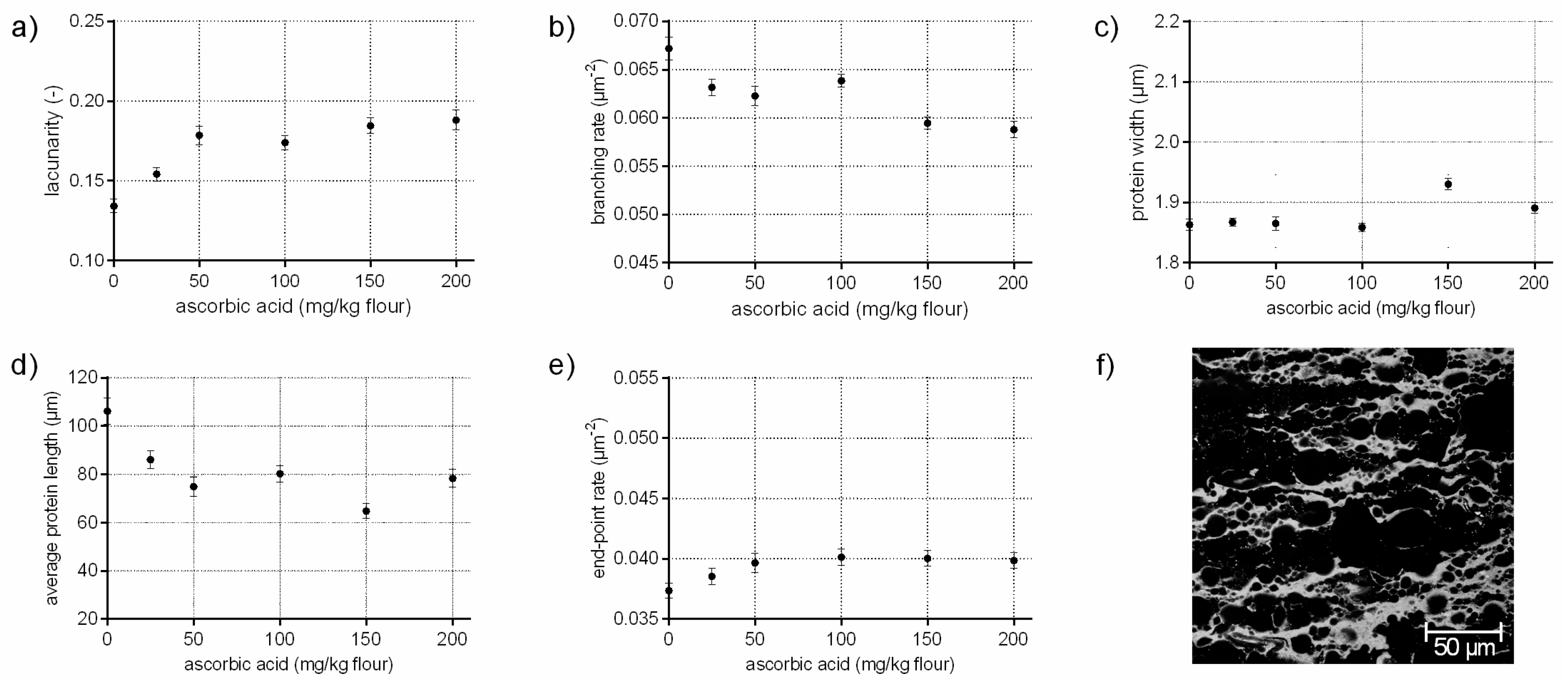
3.2. Protein Network Formation Modified by Ascorbic Acid
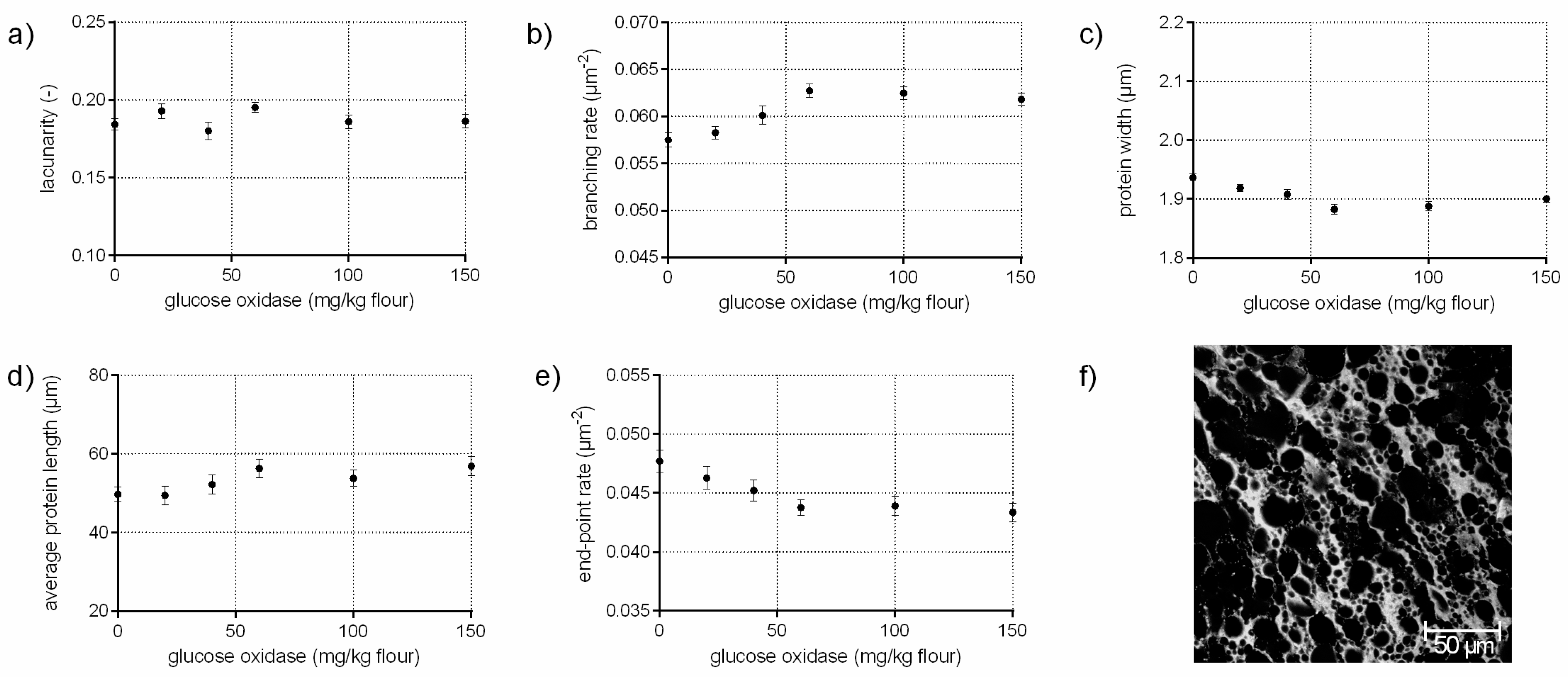
3.3. Protein Network Formation Modified by Glucose Oxidase
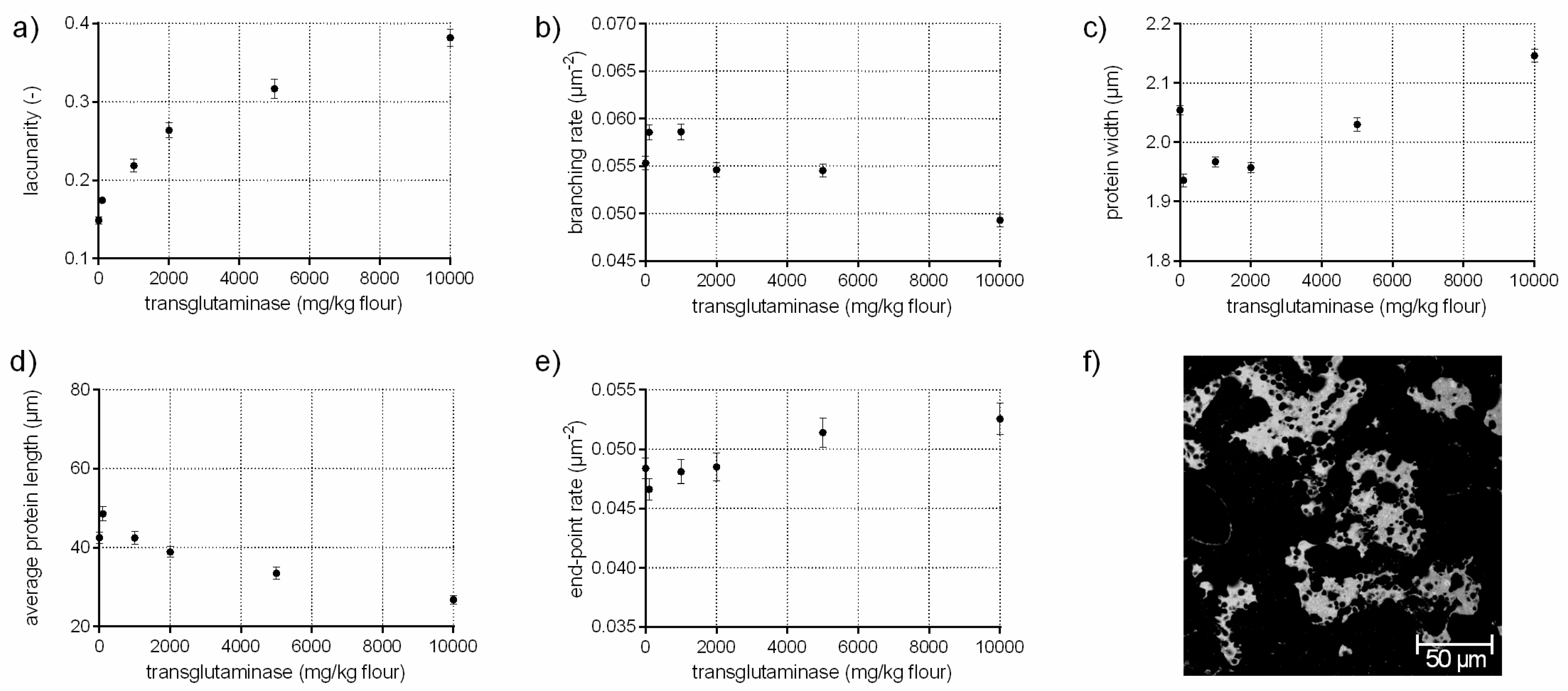
3.4. Protein Network Formation Modified by Transglutaminase
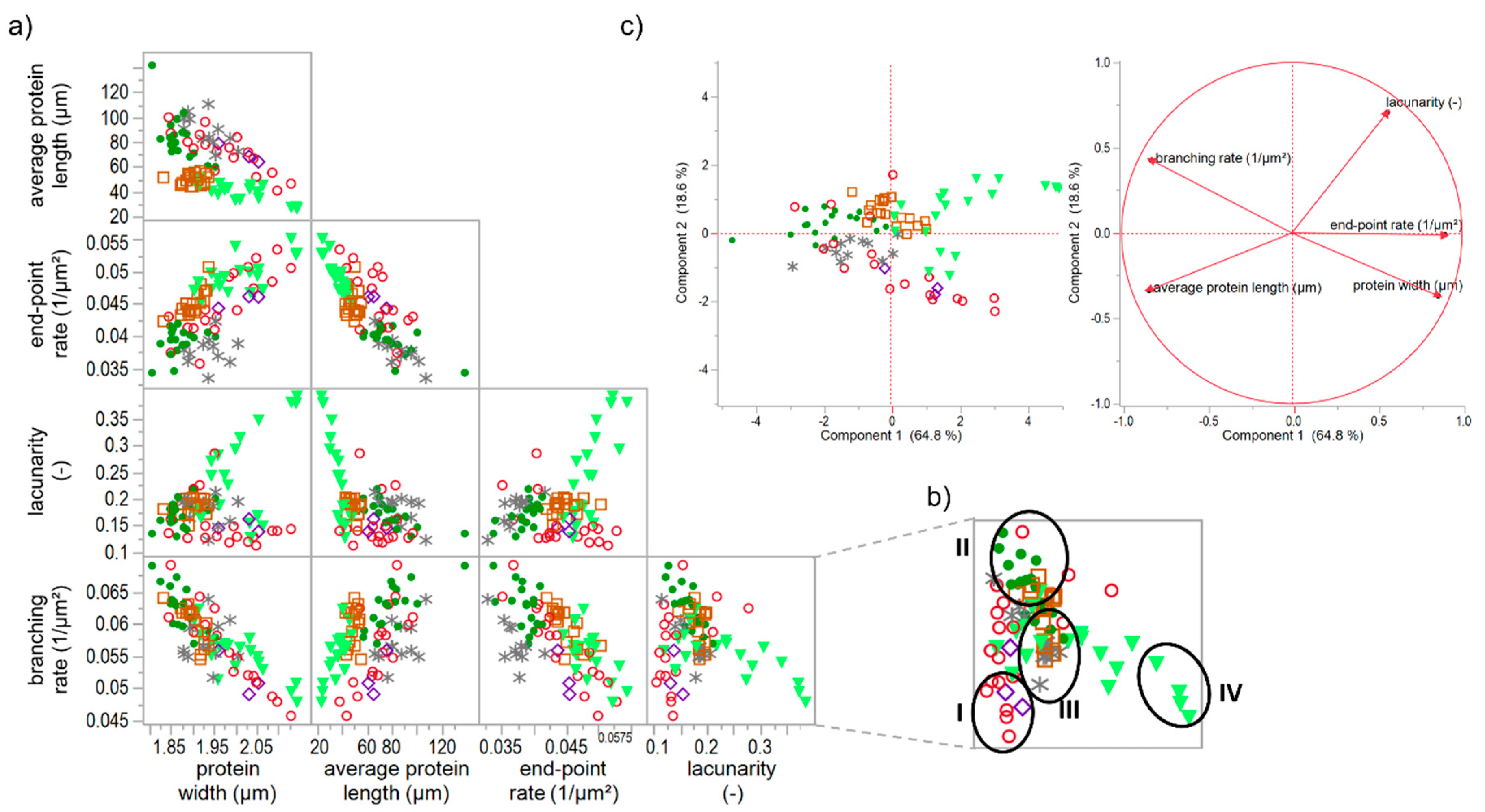
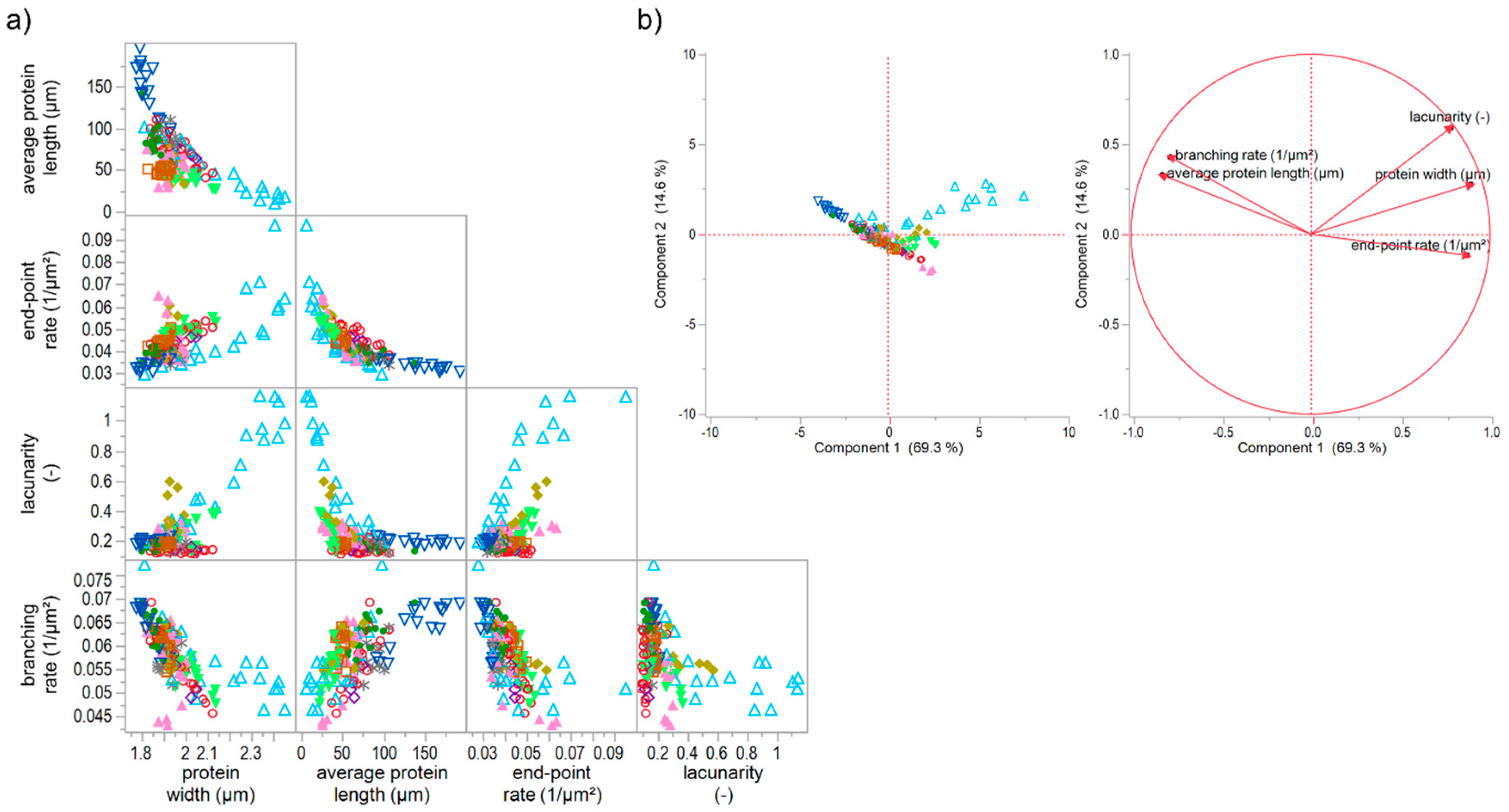
3.5. Classification of Gluten Polymer Networks
3.5.1. Effect of Specific Gluten-Modifying Agents
- A network with a low lacunarity (0–0.16), a very low branching rate, very high end-point rate as well as a high protein width, such as for flour-water-systems with glutathione (c.f. Section 3.1) or bromelain addition (c.f. supplementary data 1).
- A network with a median lacunarity (0.17–0.26), very high branching rate, low end-point rate and a low protein width, such as for a standard flour-water-systems or with glucose oxidase addition (c.f. Section 3.3).
- A network with a median lacunarity (0.17–0.26), low/median branching rate, high end-point rate as well as a high protein width, such as for flour-water-systems with high ascorbic acid (c.f. Section 3.2) or potassium bromate (c.f. supplementary data 2) concentrations.
- A network with a high lacunarity (>0.27), low branching rate, very high end-point rate as well as a high protein width, such as for flour-water-systems with high transglutaminase concentration (<1000 mg transglutaminase/kg four; c.f. Section 3.4).
3.5.2. Effect of Unspecific and Specific Gluten-Modifying Agents
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Blomfeldt, T.O.J.; Kuktaite, R.; Johansson, E.; Hedenqvist, M.S. Mechanical properties and network structure of wheat gluten foams. Biomacromolecules 2011, 12, 1707–1715. [Google Scholar] [CrossRef] [PubMed]
- Hammann, F.; Schmid, M. Determination and quantification of molecular interactions in protein films: A review. Materials 2014, 7, 7975–7996. [Google Scholar] [CrossRef] [PubMed]
- Pommet, M.; Redl, A.; Morel, M.H.; Domenek, S.; Guilbert, S. Thermoplastic processing of protein-based bioplastics: Chemical engineering aspects of mixing, extrusion and hot molding. In Macromolecular Symposia; Wiley: Hoboken, NJ, USA, 2003; Volume 197, pp. 207–218. [Google Scholar]
- Lagrain, B.; Goderis, B.; Brijs, K.; Delcour, J.A. Molecular basis of processing wheat gluten toward biobased materials. Biomacromolecules 2010, 11, 533–541. [Google Scholar] [CrossRef] [PubMed]
- Wieser, H. Chemistry of gluten proteins. Food Microbiol. 2007, 24, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Tilley, K.A.; Benjamin, R.E.; Bagorogoza, K.E.; Okot-Kotber, B.M.; Prakash, O.; Kwen, H. Tyrosine cross-links: Molecular basis of gluten structure and function. J. Agric. Food Chem. 2001, 49, 2627–2632. [Google Scholar] [CrossRef] [PubMed]
- Belton, P.S. Mini review: On the elasticity of wheat gluten. J. Cereal Sci. 1999, 29, 103–107. [Google Scholar] [CrossRef]
- Delcour, J.A.; Joye, I.J.; Pareyt, B.; Wilderjans, E.; Brijs, K.; Lagrain, B. Wheat gluten functionality as a quality determinant in cereal-based food products. Annu. Rev. Food Sci. Technol. 2012, 3, 469–492. [Google Scholar] [CrossRef] [PubMed]
- Dobraszczyk, B.J.; Morgenstern, M.P. Rheology and the breadmaking process. J. Cereal Sci. 2003, 38, 229–245. [Google Scholar] [CrossRef]
- Steffolani, M.E.; Ribotta, P.D.; Pérez, G.T.; León, A.E. Effect of glucose oxidase, transglutaminase, and pentosanase on wheat proteins: Relationship with dough properties and bread-making quality. J. Cereal Sci. 2010, 51, 366–373. [Google Scholar] [CrossRef]
- Hanft, F.; Koehler, P. Studies on the effect of glucose oxidase in bread making. J. Sci. Food Agric. 2006, 86, 1699–1704. [Google Scholar] [CrossRef]
- Grosch, W.; Wieser, H. Redox reactions in wheat dough as affected by ascorbic acid. J. Cereal Sci. 1999, 29, 1–16. [Google Scholar] [CrossRef]
- Wikström, K.; Eliasson, A.-C. Effects of enzymes and oxidizing agents on shear stress relaxation of wheat flour dough: Additions of protease, glucose oxidase, ascorbic acid, and potassium bromate. Cereal Chem. 1998, 75, 331–337. [Google Scholar] [CrossRef]
- Lindahl, L.; Eliasson, A. Influence of added enzymes on the rheological properties of a wheat flour dough. Cereal Chem. 1992, 69, 542–546. [Google Scholar]
- Rasheed, F. Production of Sustainable Bioplastic Materials from Wheat Gluten Proteins; Faculty of Landscape Planning, Horticulture and Agriculture Science, Swedish University of Agriculture Sciences: Uppsala, Sweden, 2011. [Google Scholar]
- Bernklau, I.; Lucas, L.; Jekle, M.; Becker, T. Protein network analysis—A new approach for quantifying wheat dough microstructure. Food Res. Int. 2016, 89, 812–819. [Google Scholar] [CrossRef] [PubMed]
- Lucas, I.; Stauner, B.; Jekle, M.; Becker, T. Staining methods for dough systems—Impact on microstructure and functionality. LWT Food Sci. Technol. 2018, 88, 139–145. [Google Scholar] [CrossRef]
- Bernklau, I.; Neußer, C.; Moroni, A.V.; Gysler, C.; Spagnolello, A.; Chung, W.; Jekle, M.; Becker, T. Structural, textural and sensory impact of sodium reduction on long fermented pizza. Food Chem. 2017, 234, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Meister, A. [1] Glutathione metabolism. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1995; Volume 251, pp. 3–7. [Google Scholar]
- Verheyen, C.; Albrecht, A.; Herrmann, J.; Strobl, M.; Jekle, M.; Becker, T. The contribution of glutathione to the destabilizing effect of yeast on wheat dough. Food Chem. 2015, 173, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Hüttner, S.; Wieser, H. Studies on the distribution and binding of endogenous glutathione in wheat dough and gluten. II. Binding sites of endogenous glutathione in glutenins. Eur. Food Res. Technol. 2001, 213, 460–464. [Google Scholar] [CrossRef]
- Lagrain, B.; Thewissen, B.G.; Brijs, K.; Delcour, J.A. Impact of redox agents on the extractability of gluten proteins during bread making. J. Agric. Food Chem. 2007, 55, 5320–5325. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Hoseney, R. Effects of certain breadmaking oxidants and reducing agents on dough rheological properties. Cereal Chem. 1995, 72, 58–63. [Google Scholar]
- Koehler, P. Concentrations of low and high molecular weight thiols in wheat dough as affected by different concentrations of ascorbic acid. J. Agric. Food Chem. 2003, 51, 4948–4953. [Google Scholar] [CrossRef] [PubMed]
- Hanft, F.; Koehler, P. Quantitation of dityrosine in wheat flour and dough by liquid chromatography—Tandem mass spectrometry. J. Agric. Food Chem. 2005, 53, 2418–2423. [Google Scholar] [CrossRef] [PubMed]
- Rasiah, I.; Sutton, K.; Low, F.; Lin, H.-M.; Gerrard, J. Crosslinking of wheat dough proteins by glucose oxidase and the resulting effects on bread and croissants. Food Chem. 2005, 89, 325–332. [Google Scholar] [CrossRef]
- Niu, M.; Xiong, L.; Zhang, B.; Jia, C.; Zhao, S. Comparative study on protein polymerization in whole-wheat dough modified by transglutaminase and glucose oxidase. LWT 2018, 90, 323–330. [Google Scholar] [CrossRef]
- Autio, K.; Kruus, K.; Knaapila, A.; Gerber, N.; Flander, L.; Buchert, J. Kinetics of transglutaminase-induced cross-linking of wheat proteins in dough. J. Agric. Food Chem. 2005, 53, 1039–1045. [Google Scholar] [CrossRef] [PubMed]
- Basman, A.; Köksel, H.; Ng, P.K. Effects of increasing levels of transglutaminase on the rheological properties and bread quality characteristics of two wheat flours. Eur. Food Res. Technol. 2002, 215, 419–424. [Google Scholar] [CrossRef]
- Steffolani, M.E.; Pérez, G.T.; Ribotta, P.D.; Puppo, M.C.; León, A.E. Effect of transglutaminase on properties of glutenin macropolymer and dough rheology. Cereal Chem. 2008, 85, 39–43. [Google Scholar] [CrossRef]
- Joye, I.J.; Lagrain, B.; Delcour, J.A. Use of chemical redox agents and exogenous enzymes to modify the protein network during breadmaking—A review. J. Cereal Sci. 2009, 50, 11–21. [Google Scholar] [CrossRef]
| Floura | Flourb | Flourc | Flourd | |||||||
| Ascorbic acid * (3 mg/ kg flour) | yes | yes | no | no | ||||||
| Protein (g/100 g dry flour) | 12.70 ± 0.04 | 11.49 ± 0.04 | 11.85 ± 0.04 | 12.50 ± 0.02 | ||||||
| Ash (g/100 g dry flour) | 0.63 ± 0.01 | 0.65 ± 0.01 | 0.58 ± 0.00 | 0.64 ± 0.00 | ||||||
| Falling number (s) | 407.0 ± 14.4 | 437.3 ± 8.1 | 434.3 ± 16.8 | 488.5 ± 18.9 | ||||||
| Kneading time ** (s) to 500 FU | 180 | 180 | 300 | 240 | ||||||
| Flour-Water-Systems | ||||||||||
| Floura | Flourb | Flourc | Flourd | |||||||
| Variations | IHL | ROI | SHO | GLU | RHL | ASC | TG | GOX | KBrO3 | BRN |
| Moisture ** (g/100 g flour) | 14.17 ± 0.03 | 13.92 ± 0.01 | 13.92 ± 0.01 | 13.91 ± 0.02 | 14.13 ± 0.3 | 14.86 ± 0.07 | 14.28 ± 0.15 | 14.14 ± 0.08 | 13.86 ± 0.04 | 13.86 ± 0.04 |
| Water addition (mL/100 g flour) | 59.18 | 58.32 | 58.32 | 57.76 | 57.83 | 56.17 | 60.49 | 61.06 | 61.36 | 61.36 |
| Kneading time *** (s) to 500 FU | 180 | 180 | 180 | 180 | 180 | 300 | 250 | 250 | 240 | 240 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lucas, I.; Becker, T.; Jekle, M. Gluten Polymer Networks—A Microstructural Classification in Complex Systems. Polymers 2018, 10, 617. https://doi.org/10.3390/polym10060617
Lucas I, Becker T, Jekle M. Gluten Polymer Networks—A Microstructural Classification in Complex Systems. Polymers. 2018; 10(6):617. https://doi.org/10.3390/polym10060617
Chicago/Turabian StyleLucas, Isabelle, Thomas Becker, and Mario Jekle. 2018. "Gluten Polymer Networks—A Microstructural Classification in Complex Systems" Polymers 10, no. 6: 617. https://doi.org/10.3390/polym10060617