Immobilization of Arylmalonate Decarboxylase
Abstract
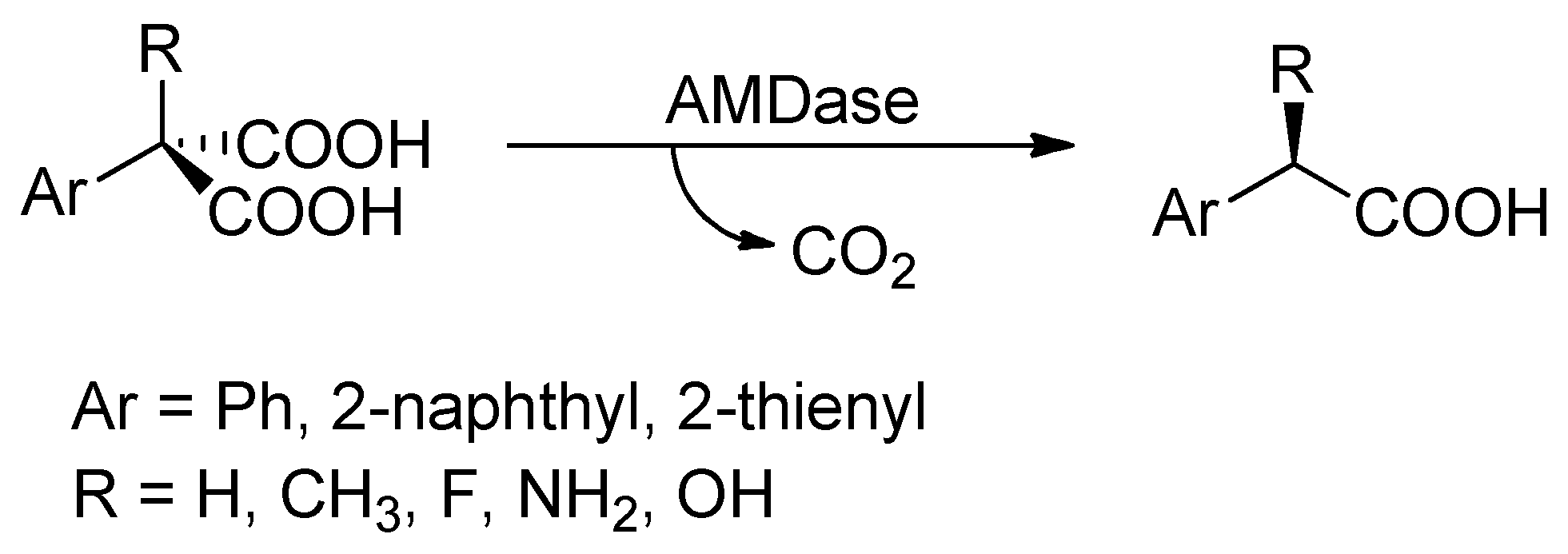
:1. Introduction
2. Results and Discussion
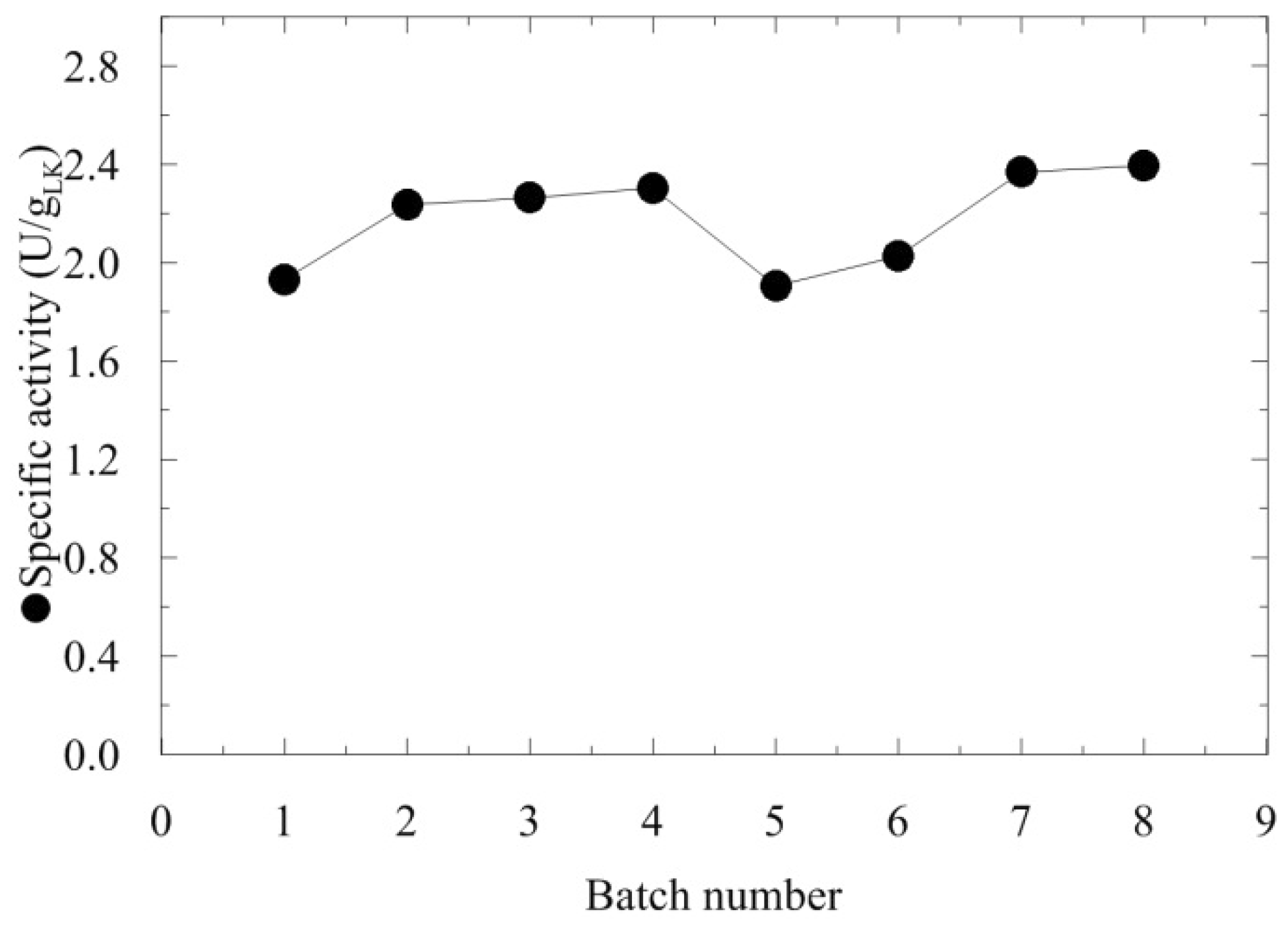
2.1. AMDase production
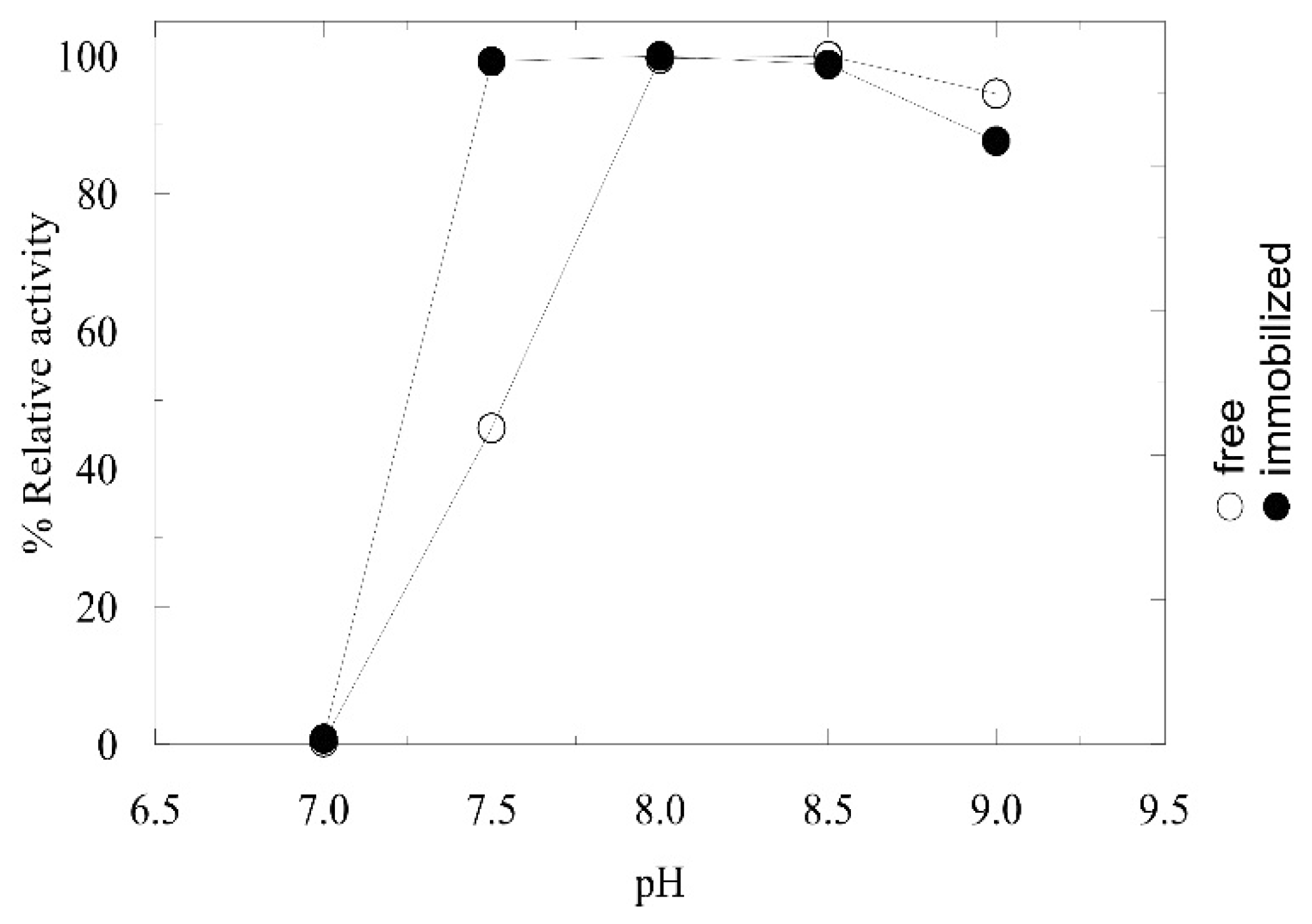
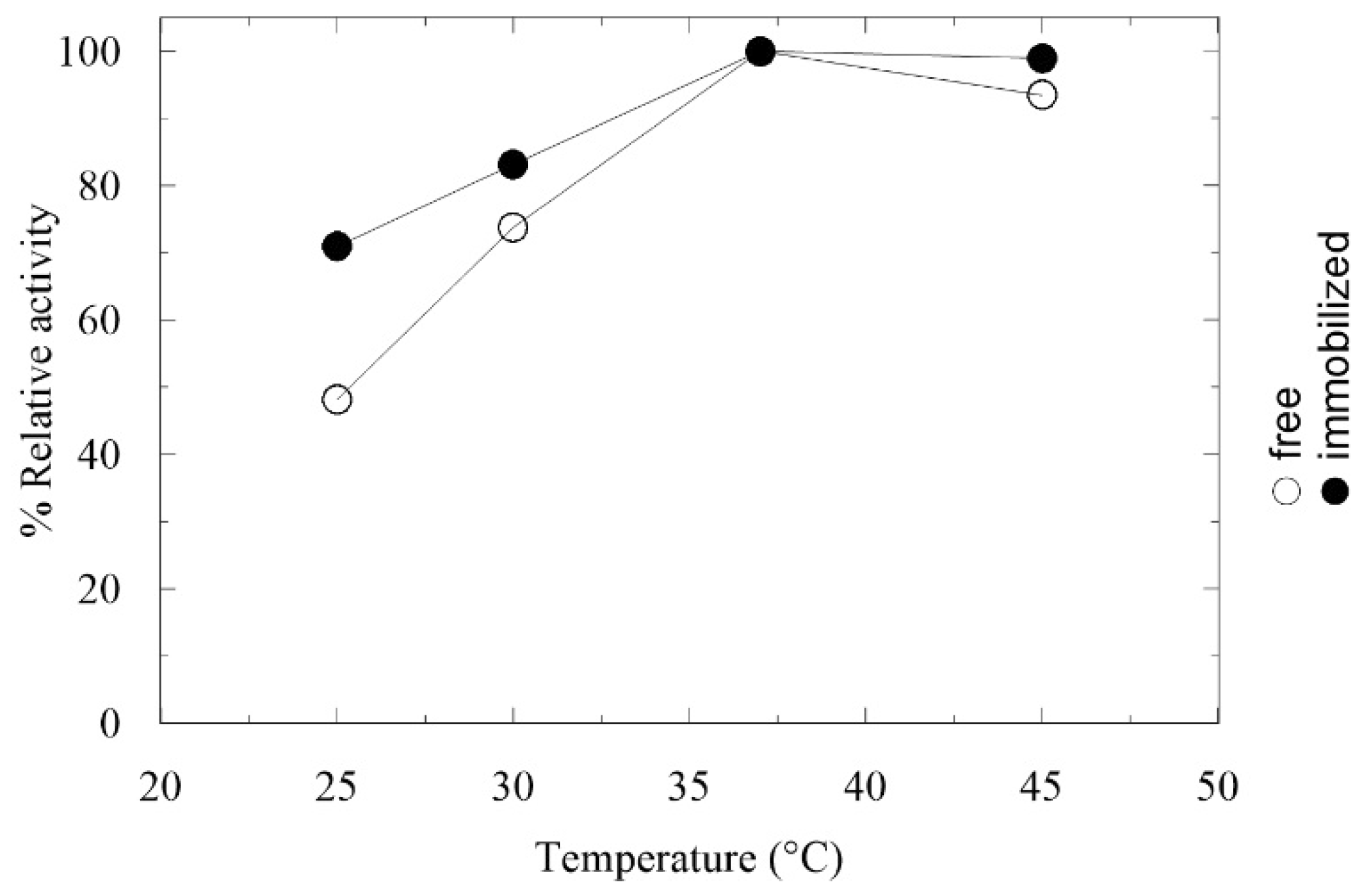
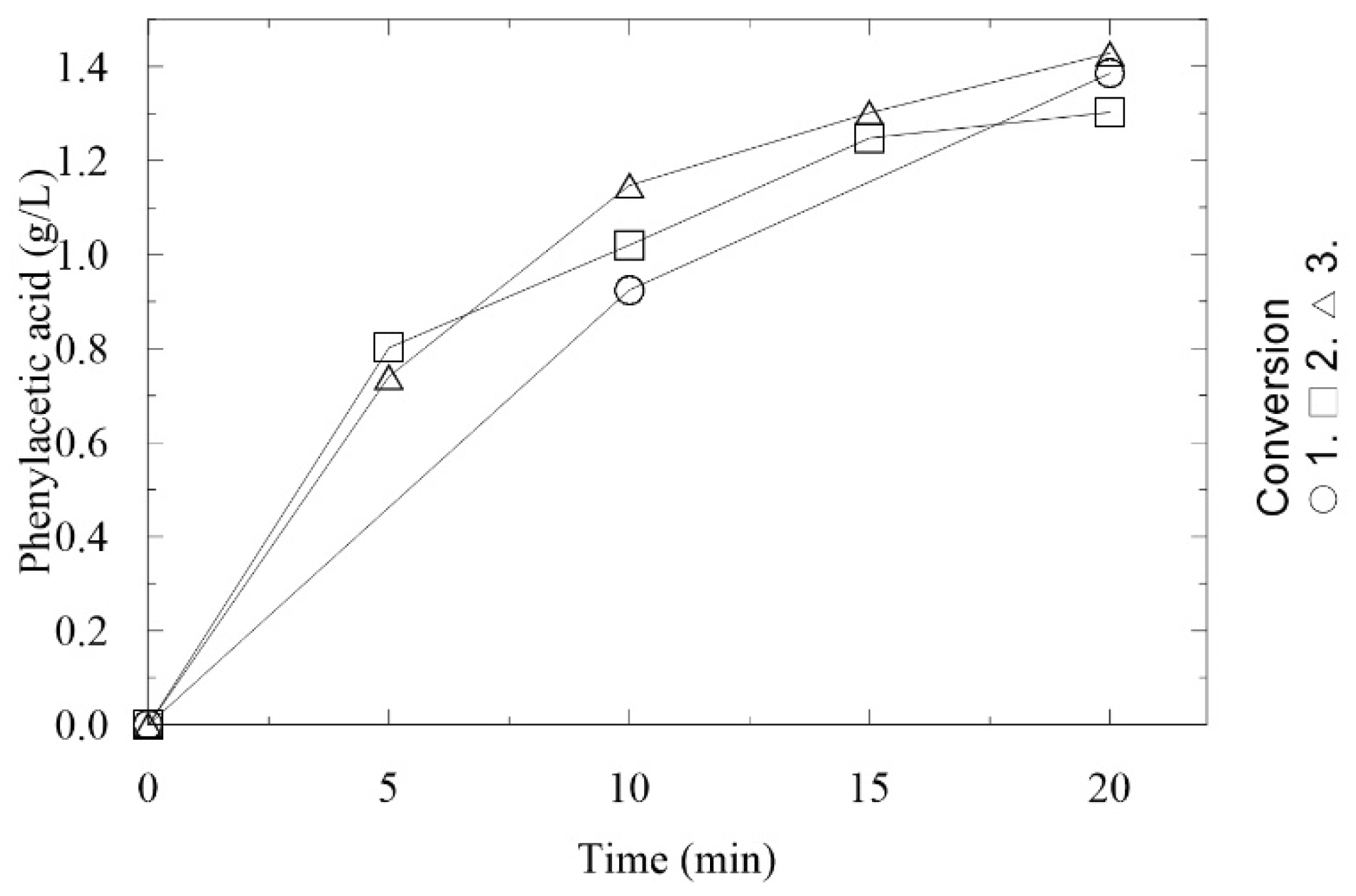
2.2. Immobilization and Characterization of AMDase in LentiKats®
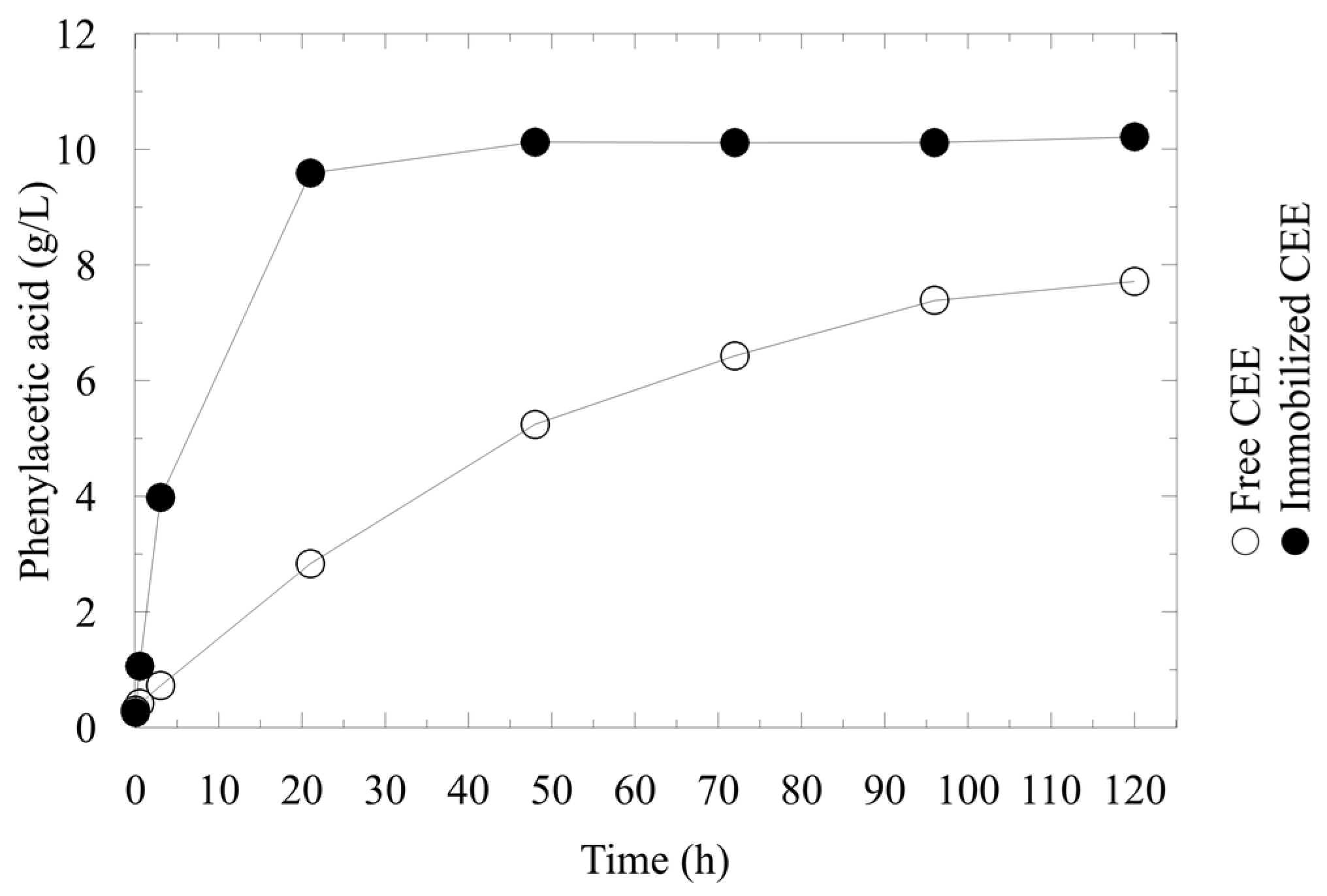
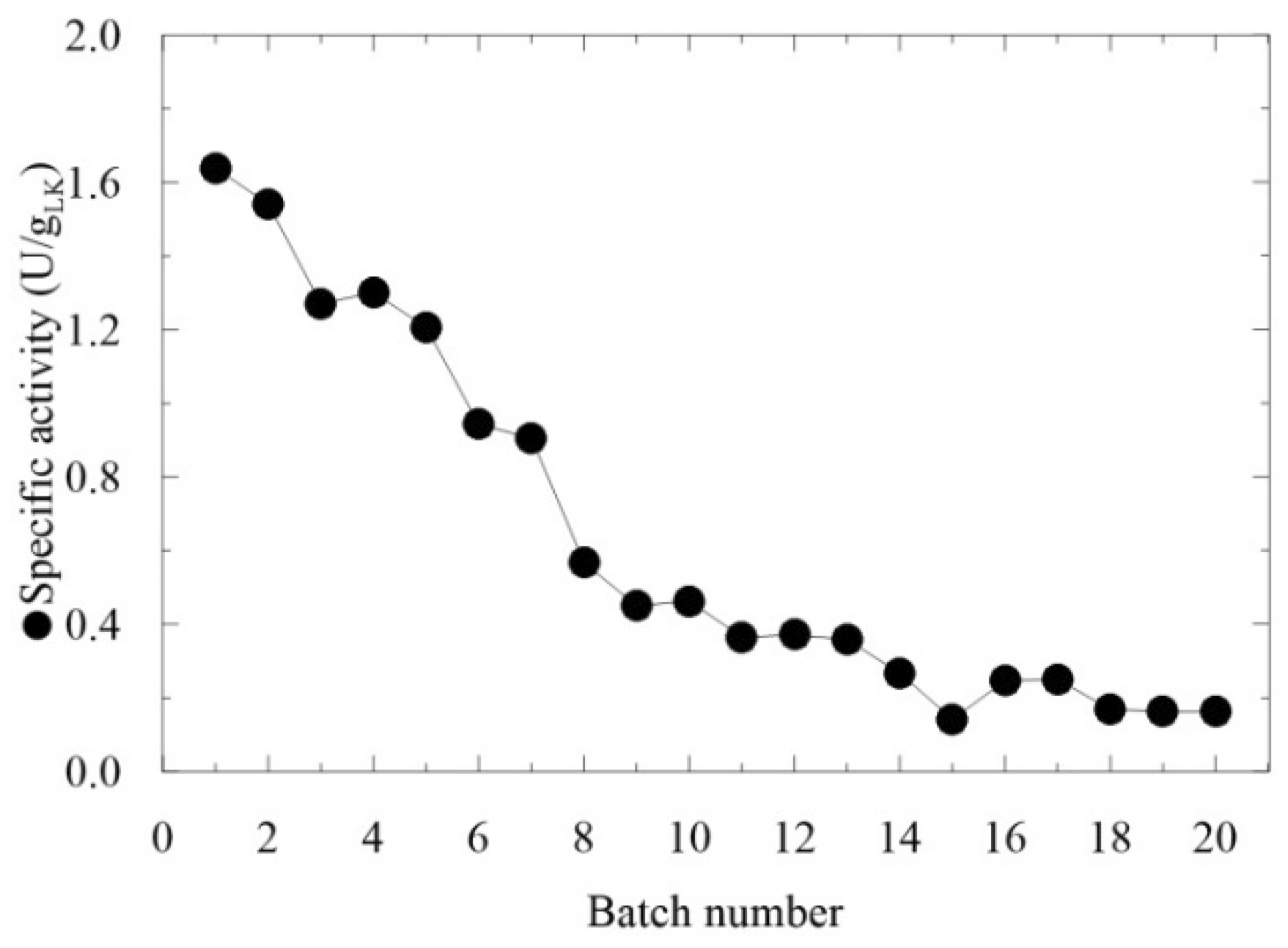
2.3. Repeated Use of LentiKats® -Immobilized AMDase CEE
2.4. Immobilization of AMDase on MMP and MMP-LentiKats®
3. Materials and Methods
3.1. Microorganism and Media
3.2. Cultivation, Induction and Crude Enzyme Preparation
3.3. Immobilization in LentiKats®
3.4. Double-Immobilization on Microparticles and Entrapment in LentiKats®
3.5. Biocatalytic Reactions with AMDase
3.6. Activity Calculations
3.7. Analytical Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Miyamoto, K.; Kourist, R. Arylmalonate decarboxylase- A highly selective bacterial biocatalyst with unknown function. Appl. Microbiol. Biotechnol. 2016, 100, 8621–8631. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, K.; Ohta, H. Purification and properties of a novel arylmalonate decarboxylase from Alcaligenes bronchisepticus KU 1201. Eur. J. Biochem. 1992, 210, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, K.; Yatake, Y.; Tamura, K.; Terao, Y.; Ohta, H. Purification and Characterization of Arylmalonate Decarboxylase from Achromobacter sp. KU1311. J. Biosci. Bioeng. 2007, 104, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.-S.; Yoon, Ch.W.; Osipov, A.; Moghavem, N.; Nwachokor, D.; Amatya, R.; Na, E.; Pantoja, J.L.; Pham, M.D.; Black, K.L.; Yu, J.S. Nanoprodrugs of NSAIDs: Preparation and characterization of flufenamic acid nanoprodrugs. J. Drug. Deliv. 2011, 2011, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Gaßmeyer, S.K.; Wetzig, J.; Mügge, C.; Assmann, M.; Enoki, J.; Hilterhaus, L.; Zuhse, R.; Miyamoto, K.; Liese, A.; Kourist, R. Arylmalonate decarboxylase catalyzed asymmetric synthesis of both enantiomers of optically pure flurbiprofen. Chem. Cat. Chem. 2016, 8, 916–921. [Google Scholar] [CrossRef]
- Terao, Y.; Ijima, Y.; Miyamoto, K.; Ohta, H. Inversion of enantioselectivity of arylmalonate decarboxylase via site-directed mutation based on the proposed reaction mechanism. J. Mol. Catal. B-Enzym. 2007, 45, 15–20. [Google Scholar] [CrossRef]
- Terao, Y.; Miyamoto, K.; Ohta, H. Improvement of the activity of arylmalonate decarboxylase by random mutagenesis. Appl. Microbiol. Biotechnol. 2006, 73, 647–653. [Google Scholar] [CrossRef]
- Kourist, R.; Miyauchi, Y.; Uemura, D.; Miyamoto, K. Engineering the promiscuous racemase activity of an arylmalonate decarboxylase. Chem. Eur. J. 2011, 17, 557–563. [Google Scholar] [CrossRef]
- Matoishi, K.; Ueda, M.; Miyamoto, K.; Ohta, H. Mechanism of asymmetric decarboxylation of α-aryl-α-methylmalonate catalyzed by arylmalonate decarboxylase originated from Alcaligenes bronchisepticus. J. Mol. Catal. B-Enzym. 2004, 27, 161–168. [Google Scholar] [CrossRef]
- Bornscheuer, U.T. Immobilizing enzymes: How to create more suitable biocatalysts. Angew. Chem. Int. Ed. 2003, 42, 3336–3337. [Google Scholar] [CrossRef]
- Brady, D.; Jordaan, J. Advances in enzyme immobilization. Biotechnol. Lett. 2009, 31, 1639–1650. [Google Scholar] [CrossRef] [PubMed]
- Bommarius, A.S.; Pave, M.F. Stabilizing biocatalysts. Chem. Soc. Rev. 2013, 42, 6534–6565. [Google Scholar] [CrossRef] [PubMed]
- Mohamad, N.R.; Marzuki, N.H.C.; Buang, N.A.; Huyop, F.; Wahab, R.A.; Mohamad. An overview of technologies for immobilization of enzymes and surface analysis techniques for immobilized enzymes. Biotechnol. Biotechnol. Equip. 2015, 29, 205–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krasňan, V.; Stloukal, R.; Rosenberg, M.; Rebroš, M. Immobilization of cells and enzymes to LentiKats®. Appl. Microbiol. Biotechnol. 2016, 100, 2535–2553. [Google Scholar] [CrossRef] [PubMed]
- Jesionowski, T.; Zdarta, J.; Krajewska, B. Enzyme immobilization by adsorption: a review. Adsorption 2014, 20, 801–821. [Google Scholar] [CrossRef] [Green Version]
- Zdarta, J.; Meyer, A.S.; Jesionowski, T.; Pinelo, M. A General Overview of Support Materials for Enzyme Immobilization: Characteristics, Properties, Practical Utility. Catalysts 2018, 8, 92. [Google Scholar] [CrossRef]
- Shinkai, M. Functional magnetic particles for medical application. J. Biosci. Bioeng. 2002, 94, 606–613. [Google Scholar] [CrossRef]
- Ngomsik, A.-F.; Bee, A.; Draye, M.; Cote, G.; Cabuil, V. Magnetic Nano- and Microparticles for Metal Removal and Environmental Applications: A. Review. C. R. Chim. 2005, 8, 963–970. [Google Scholar] [CrossRef]
- Mehta, R.V.; Upadhyay, R.V.; Charles, S.W.; Ramchand, C.N. Direct binding of protein to magnetic particles. Biotechnol. Tech. 1997, 11, 493–496. [Google Scholar] [CrossRef]
- Koneracká, M.; Kopčanský, P.; Timko, M.; Ramchand, C.N.; de Sequeira, A.; Trevan, M. Direct binding procedure of proteins and enzymes to fine magnetic particles. J. Mol. Catal. B- Enzym. 2002, 18, 13–18. [Google Scholar] [CrossRef]
- Wong, L.S.; Okrasa, K.; Micklefield, J. Site-selective immobilisation of functional enzymes on to polystyrene nanoparticles. Org. Biomol. Chem. 2010, 8, 782–787. [Google Scholar] [CrossRef] [PubMed]
- Aßmann, M.; Mügge, C.; Gaßmeyer, S.K.; Enoki, J.; Hilterhaus, L.; Kourist, R.; Liese, A.; Kara, S. Improvement of the process stability of arylmalonate decarboxylase by immobilization for biocatalytic profen synthesis. Front. Microbiol. 2017, 8, 448. [Google Scholar] [CrossRef]
- Lakshmi, G.J.; Kotra, S.R.; Peravali, J.B.; Kumar, P.P.B.S.; Rao, K.R.S.S. Molecular cloning, high level expression and activity analysis of constructed human interleukin-25 using industrially important IPTG inducible Escherichia coli BL21(DE3). J. Biosci. Biotechnol. 2014, 6, 19–30. [Google Scholar] [CrossRef]
- Torello, G.; van Midden, N.; Stloukal, R.; Hanefeld, U. Immobilized Hydroxynitrile Lyase: A Comparative Study of Recyclability. ChemCatChem 2014, 6, 1096–1102. [Google Scholar] [CrossRef]
- Reboš, M.; Rosenberg, M.; Mlichová, Z.; Krištofíková, Ľ.; Paluch, M. A simple entrapment of glucoamylase into LentiKats® as an efficient catalyst for maltodextrin hydrolysis. Enzym. Microb. Tech. 2006, 39, 800–804. [Google Scholar] [CrossRef]
- Dowd, J.E.; Riggs, D.S. A Comparison of Estimates of Michaelis-Menten Kinetic Constants from Various Linear Transformations. J. Biol. Chem. 1965, 240, 863–869. [Google Scholar]
- Kawasaki, T.; Saito, K.; Ohta, H. The Mode of Substrate- Recognition Mechanism of Arylmalonate Decarboxylase. Chem. Lett. 1997, 26, 351–352. [Google Scholar] [CrossRef]
- Okrasa, K.; Levy, C.; Hauer, B.; Baudendistel, N.; Leys, D.; Micklefield, J. Structure and Mechanism of an Unusual Malonate Decarboxylase and Related Racemases. Chem. Eur. J. 2008, 14, 6609–6613. [Google Scholar] [CrossRef] [PubMed]
- Okrasa, K.; Levy, C.; Wilding, M.; Goodall, M.; Baudendistel, N.; Hauer, B.; Leys, D.; Micklefield, J. Structure-guided directed evolution of alkenyl and arylmalonate decarboxylases. Angew. Chem. Int. Ed. 2009, 48, 7691–7694. [Google Scholar] [CrossRef]
- Rebros, M.; Rosenberg, M.; Mlichova, Z.; Kristofikova, L. Hydrolysis of sucrose by invertase entrapped in polyvinyl alcohol hydrogel capsules. Food Chem. 2007, 102, 784–787. [Google Scholar] [CrossRef]
- Markošová, K.; Dolejš, I.; Stloukal, R.; Rios-Solis, L.; Rosenberg, M.; Micheletti, M.; Lye, G.J.; Turner, N.J.; Rebroš, M. Immobilisation and kinetics of monoamine oxidase (MAO-N-D5) enzyme in polyvinyl alcohol gels. J. Mol. Catal. B-Enzym. 2016, 129, 69–74. [Google Scholar] [CrossRef] [Green Version]
- Cárdenas-Fernández, M.; López, C.; Álvaro, G.; López-Santín, J. Immobilized L-aspartate ammonia-lyase from Bacillus sp. YM55-1 as biocatalyst for highly concentrated L-aspartate synthesis. Bioprocess. Biosyst. Eng. 2012, 35, 1437–1444. [Google Scholar] [CrossRef] [PubMed]
- Wilson, L.; Illanes, A.; Pessela, B.C.; Abian, O.; Fernández-Lafuente, R.; Guisán, J.M. Encapsulation of crosslinked penicillin G acylase aggregates in Lentikats: Evaluation of a novel biocatalyst in organic media. Biotechnol. Bioeng. 2004, 86, 558–562. [Google Scholar] [CrossRef] [PubMed]
- Sirvio, J.; Hyvakko, U.; Liimatainen, H.; Niinimaki, J.; Hormi, O. Periodate oxidation of cellulose at elevated temperatures using metal salts as cellulose activators. Cabrohyd. Polym. 2011, 83, 1293–1297. [Google Scholar] [CrossRef]
- Zhang, D.H.; Yuwen, L.X.; Peng, L.J. Parameters Affecting the Performance of Immobilized Enzyme. J. Chem.-NY 2013, 2013, 1–7. [Google Scholar] [CrossRef]
- Zajkoska, P.; Rosenberg, M.; Heath, R.; Malone, K.J.; Stloukal, R.; Turner, N.J.; Rebroš, M. Immobilised whole-cell recombinant monoamine oxidase biocatalysis. Appl. Microbiol. Biotechnol. 2015, 99, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
| Kinetic Parameters | CEE Free | CEE LentiKats® | CEE MMP-LentiKats® |
|---|---|---|---|
| Km [mM] | 25.2 | 38.6 | 67.2 |
| Vmax [U/mgCEE] | 204.5 | 80 | 1.85 |
| kcat [s−1] | 88.62 | 34.67 | 0.8 |
| kcat/Km [s−1.mM−1] | 3.52 | 0.9 | 0.01 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Markošová, K.; Husarčíková, J.; Halásová, M.; Kourist, R.; Rosenberg, M.; Stloukal, R.; Zajoncová, L.; Rebroš, M. Immobilization of Arylmalonate Decarboxylase. Catalysts 2018, 8, 603. https://doi.org/10.3390/catal8120603
Markošová K, Husarčíková J, Halásová M, Kourist R, Rosenberg M, Stloukal R, Zajoncová L, Rebroš M. Immobilization of Arylmalonate Decarboxylase. Catalysts. 2018; 8(12):603. https://doi.org/10.3390/catal8120603
Chicago/Turabian StyleMarkošová, Kristína, Jana Husarčíková, Monika Halásová, Robert Kourist, Michal Rosenberg, Radek Stloukal, Ludmila Zajoncová, and Martin Rebroš. 2018. "Immobilization of Arylmalonate Decarboxylase" Catalysts 8, no. 12: 603. https://doi.org/10.3390/catal8120603






