Production of Resveratrol by Piceid Deglycosylation Using Cellulase
Abstract
:1. Introduction
2. Results and Discussion
2.1. Model Fitting
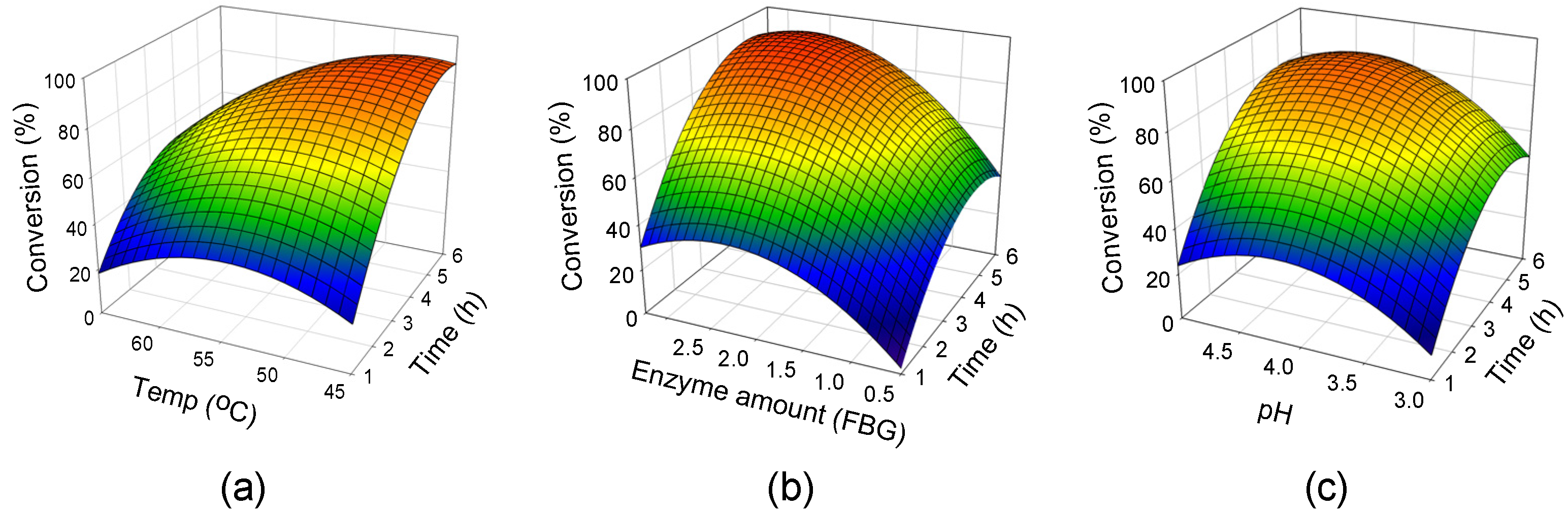
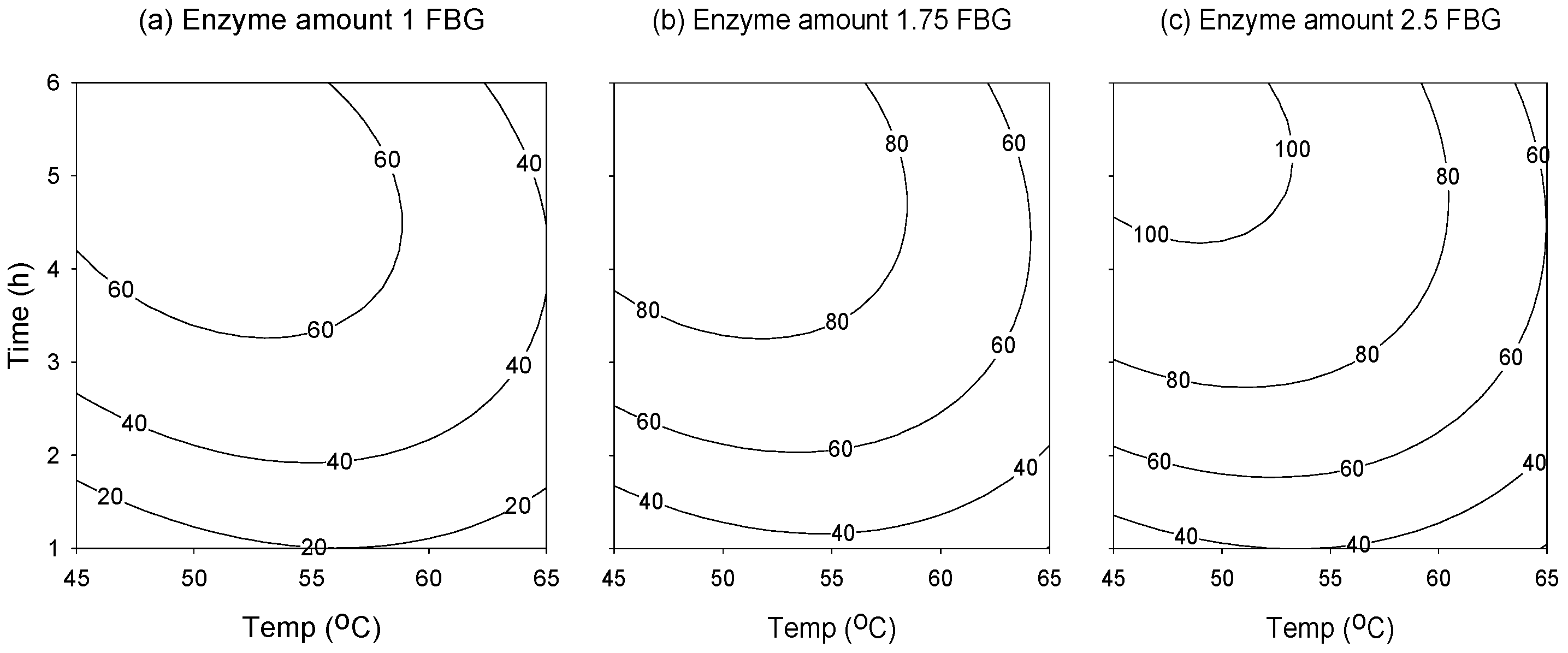
2.2. Response Surface Analysis
2.3. Attaining the Optimum Condition
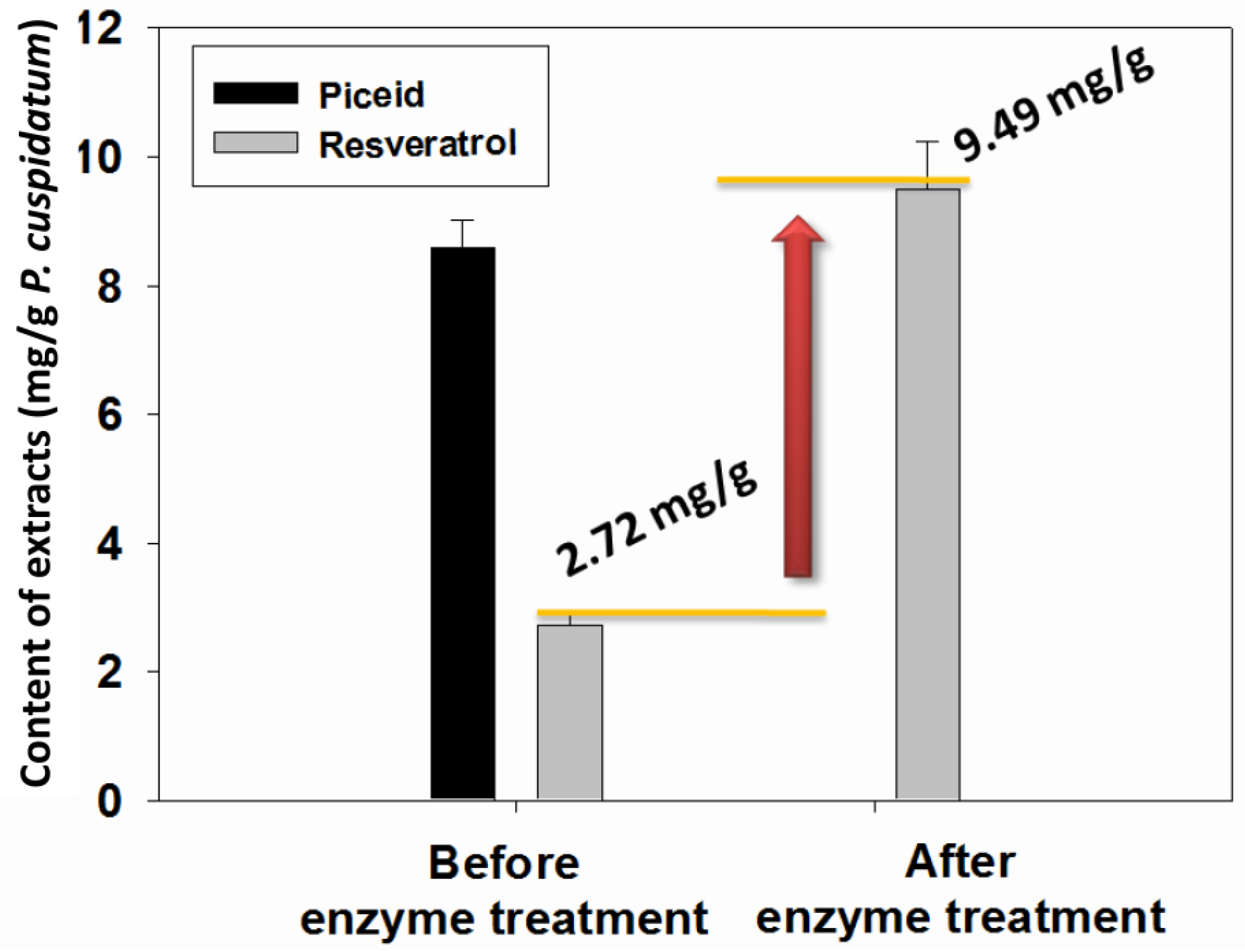
2.4. Transformation of Piceid in the Extracts of P. cuspidatum Root
3. Experimental Section
3.1. Materials
3.2. Transformation of Piceid into Resveratrol
3.3. Quantitation of Piceid and Resveratrol by HPLC
3.4. Experimental Design and Statistical Analysis
3.5. Transformation of Piceid into Resveratrol in the Extracts of P. cuspidatum Root
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Zernova, O.; Lygin, A.; Pawlowski, M.; Hill, C.; Hartman, G.; Widholm, J.; Lozovaya, V. Regulation of plant immunity through modulation of phytoalexin synthesis. Molecules 2014, 19, 7480–7496. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Fang, Y.; Li, X.; Meng, J.; Wang, H.; Li, H.; Zhang, Z.; Guo, Z. Occurrence and estimation of trans-resveratrol in one-year-old canes from seven major chinese grape producing regions. Molecules 2011, 16, 2846–2861. [Google Scholar] [CrossRef] [PubMed]
- Kurin, E.; Mučaji, P.; Nagy, M. In vitro antioxidant activities of three red wine polyphenols and their mixtures: An interaction study. Molecules 2012, 17, 14336–14348. [Google Scholar] [CrossRef] [PubMed]
- Baur, J.A.; Sinclair, D.A. Therapeutic potential of resveratrol: The in vivo evidence. Nat. Rev. Drug Discov. 2006, 5, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Jiang, A.; Qi, B.; Ma, Z.; Xiong, Y.; Dou, J.; Wang, J. Resveratrol protects against helicobacter pylori-associated gastritis by combating oxidative stress. Int. J. Mol. Sci. 2015, 16, 26061–27769. [Google Scholar] [CrossRef] [PubMed]
- Jang, M.; Cai, L.; Udeani, G.O.; Slowing, K.V.; Thomas, C.F.; Beecher, C.W.; Fong, H.H.; Farnsworth, N.R.; Kinghorn, A.D.; Mehta, R.G. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science 1997, 275, 218–220. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.Y.; Kuo, C.H.; Liu, Y.C.; Ye, L.Y.; Chen, J.H.; Shieh, C.J. Ultrasonic-assisted extraction of the botanical dietary supplement resveratrol and other constituents of Polygonum cuspidatum. J. Nat. Prod. 2012, 75, 1810–1813. [Google Scholar] [CrossRef] [PubMed]
- Spier, A.P.; Bavaresco, C.S.; Wyse, Â.T.; Carvalho, D.; Freitas Sarkis, J.J. Effects of resveratrol and purple grape juice on nucleotide hydrolysis by adult rat serum. Food Chem. 2007, 103, 565–571. [Google Scholar] [CrossRef]
- Djoko, B.; Chiou, R.Y.Y.; Shee, J.J.; Liu, Y.W. Characterization of immunological activities of peanut stilbenoids, arachidin-1, piceatannol, and resveratrol on lipopolysaccharide-induced inflammation of raw 264.7 macrophages. J. Agric. Food Chem. 2007, 55, 2376–2383. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.F.; Yang, J.Y.; Wang, F.; Wang, X.X. Resveratrol: Botanical origin, pharmacological activity and applications. Chin. J. Nat. Med. 2013, 11, 1–15. [Google Scholar] [CrossRef]
- Kuo, C.H.; Chen, B.Y.; Liu, Y.C.; Chang, C.M.; Deng, T.S.; Chen, J.H.; Shieh, C.J. Optimized ultrasound-assisted extraction of phenolic compounds from Polygonum cuspidatum. Molecules 2013, 19, 67–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Counet, C.; Callemien, D.; Collin, S. Chocolate and cocoa: New sources of trans-resveratrol and trans-piceid. Food Chem. 2006, 98, 649–657. [Google Scholar] [CrossRef]
- Wang, L.; Xu, M.; Liu, C.; Wang, J.; Xi, H.; Wu, B.; Loescher, W.; Duan, W.; Fan, P.; Li, S. Resveratrols in grape berry skins and leaves in Vitis germplasm. PLoS ONE 2013, 8, e61642. [Google Scholar] [CrossRef] [PubMed]
- Lepak, A.; Gutmann, A.; Kulmer, S.T.; Nidetzky, B. Creating a water-soluble resveratrol-based antioxidant by site-selective enzymatic glucosylation. ChemBioChem 2015, 16, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.H.; Hsiao, F.W.; Dai, S.M.; Chang, C.M.J.; Lee, C.C.; Liu, Y.C.; Shieh, C.J. Lipase catalyzed acetylation of 3, 5, 4′-trihydroxystilbene: Optimization and kinetics study. Bioprocess Biosyst. Eng. 2012, 35, 1137–1145. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.H.; Hsiao, F.W.; Chen, J.H.; Hsieh, C.W.; Liu, Y.C.; Shieh, C.J. Kinetic aspects of ultrasound-accelerated lipase catalyzed acetylation and optimal synthesis of 4′-acetoxyresveratrol. Ultrason. Sonochem. 2013, 20, 546–552. [Google Scholar] [CrossRef] [PubMed]
- Signorelli, P.; Ghidoni, R. Resveratrol as an anticancer nutrient: Molecular basis, open questions and promises. J. Nutr. Biochem. 2005, 16, 449–466. [Google Scholar] [CrossRef] [PubMed]
- Henry, C.; Vitrac, X.; Decendit, A.; Ennamany, R.; Krisa, S.; Mérillon, J.M. Cellular uptake and efflux of trans-piceid and its aglycone trans-resveratrol on the apical membrane of human intestinal caco-2 cells. J. Agric. Food Chem. 2005, 53, 798–803. [Google Scholar] [CrossRef] [PubMed]
- Ladurner, A.; Schachner, D.; Schueller, K.; Pignitter, M.; Heiss, E.; Somoza, V.; Dirsch, V. Impact of trans-resveratrol-sulfates and -glucuronides on endothelial nitric oxide synthase activity, nitric oxide release and intracellular reactive oxygen species. Molecules 2014, 19, 16724–16736. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, L.; Guo, Y.X.; Dong, Y.S.; Zhang, D.J.; Xiu, Z.L. Biotransformation of piceid in Polygonum cuspidatum to resveratrol by Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2007, 75, 763–768. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.H.; Lee, C.K. Enhanced enzymatic hydrolysis of sugarcane bagasse by n-methylmorpholine-n-oxide pretreatment. Bioresour. Technol. 2009, 100, 866–871. [Google Scholar] [CrossRef] [PubMed]
- Shafiee, A.; Tsipouras, A.; Bouffard, A.; Onishi, J.C.; Guan, Z.; Motamedi, H. Enzymatic deglycosylation of enfumafungin, a triterpene glycoside natural product, and its chemically synthesized analogues. J. Mol. Catal. B 2001, 16, 27–32. [Google Scholar] [CrossRef]
- Horii, K.; Adachi, T.; Matsuda, T.; Tanaka, T.; Sahara, H.; Shibasaki, S.; Ogino, C.; Hata, Y.; Ueda, M.; Kondo, A. Improvement of isoflavone aglycones production using β-glucosidase secretory produced in recombinant Aspergillus oryzae. J. Mol. Catal. B 2009, 59, 297–301. [Google Scholar] [CrossRef]
- La Torre, G.L.; Laganà, G.; Bellocco, E.; Vilasi, F.; Salvo, F.; Dugo, G. Improvement on enzymatic hydrolysis of resveratrol glucosides in wine. Food Chem. 2004, 85, 259–266. [Google Scholar] [CrossRef]
- Matsakas, L.; Christakopoulos, P. Ethanol production from enzymatically treated dried food waste using enzymes produced on-site. Sustainability 2015, 7, 1446–1458. [Google Scholar] [CrossRef]
- Decker, C.H.; Visser, J.; Schreier, P. β-glucosidases from five black Aspergillus species: Study of their physico-chemical and biocatalytic properties. J. Agric. Food Chem. 2000, 48, 4929–4936. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Liao, W.; Chen, S. Production of cellulase/β-glucosidase by the mixed fungi culture Trichoderma reesei and Aspergillus phoenicis on dairy manure. Process Biochem. 2005, 40, 3087–3094. [Google Scholar] [CrossRef]
- Kuo, C.H.; Lee, C.K. Enhancement of enzymatic saccharification of cellulose by cellulose dissolution pretreatments. Carbohydr. Polym. 2009, 77, 41–46. [Google Scholar] [CrossRef]
- Naika, G.S.; Kaul, P.; Prakash, V. Purification and characterization of a new endoglucanase from Aspergillus aculeatus. J. Agric. Food Chem. 2007, 55, 7566–7572. [Google Scholar] [CrossRef] [PubMed]
- Humbird, D.; Davis, R.; Tao, L.; Kinchin, C.; Hsu, D.; Aden, A.; Schoen, P.; Lukas, J.; Olthof, B.; Worley, M. Process Design and Economics for Biochemical Conversion of Lignocellulosic Biomass to Ethanol: Dilute-Acid Pretreatment and Enzymatic Hydrolysis of Corn Stover; National Renewable Energy Laboratory (NREL): Golden, CO, USA, 2011. [Google Scholar]
- Lau, M.W.; Bals, B.D.; Chundawat, S.P.; Jin, M.; Gunawan, C.; Balan, V.; Jones, A.D.; Dale, B.E. An integrated paradigm for cellulosic biorefineries: Utilization of lignocellulosic biomass as self-sufficient feedstocks for fuel, food precursors and saccharolytic enzyme production. Energy Environ. Sci. 2012, 5, 7100–7110. [Google Scholar] [CrossRef]
- Klein-Marcuschamer, D.; Oleskowicz-Popiel, P.; Simmons, B.A.; Blanch, H.W. The challenge of enzyme cost in the production of lignocellulosic biofuels. Biotechnol. Bioeng. 2012, 109, 1083–1087. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.H.; Chen, H.H.; Chen, J.H.; Liu, Y.C.; Shieh, C.J. High yield of wax ester synthesized from cetyl alcohol and octanoic acid by lipozyme rmim and novozym 435. Int. J. Mol. Sci. 2012, 13, 11694–11704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taherzadeh, M.J.; Karimi, K. Enzymatic-based hydrolysis processes for ethanol. BioResources 2007, 2, 707–738. [Google Scholar]
- Chen, J.; Liu, D.; Shi, B.; Wang, H.; Cheng, Y.; Zhang, W. Optimization of hydrolysis conditions for the production of glucomanno-oligosaccharides from konjac using β-mannanase by response surface methodology. Carbohydr. Polym. 2013, 93, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Takada, G.; Kawaguchi, T.; Sumitani, J.I.; Arai, M. Expression of Aspergillus aculeatus No. F-50 cellobiohydrolase I (cbh1) and Beta.-glucosidase 1 (bgll) genes by Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 1998, 62, 1615–1618. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhao, L.C.; Li, W.; Zhang, L.X.; Zhang, J.; Liang, J. Highly efficient biotransformation of polydatin to resveratrol by snailase hydrolysis using response surface methodology optimization. Molecules 2013, 18, 9717–9726. [Google Scholar] [CrossRef] [PubMed]

| Run | Independent Variable | Conversion(%) | |||
|---|---|---|---|---|---|
| X1 (°C) | X2 (h) | X3 (FBG) | X4 | Actual Values a | |
| Temperature | Time | Enzyme Amount | pH | ||
| 1 | 45 | 4.75 | 2.5 | 3 | 68.31 ± 3.71 |
| 2 | 35 | 3.5 | 1.75 | 4 | 39.04 ± 3.18 |
| 3 | 55 | 3.5 | 1.75 | 6 | 19.01 ± 0.04 |
| 4 | 55 | 3.5 | 1.75 | 4 | 80.91 ± 1.95 |
| 5 | 65 | 4.75 | 2.5 | 5 | 49.04 ± 7.55 |
| 6 | 45 | 2.25 | 2.5 | 5 | 52.25 ± 5.41 |
| 7 | 65 | 2.25 | 2.5 | 5 | 39.88 ± 1.78 |
| 8 | 45 | 2.25 | 1 | 5 | 30.34 ± 2.03 |
| 9 | 55 | 3.5 | 1.75 | 4 | 82.26 ± 2.70 |
| 10 | 45 | 4.75 | 1 | 3 | 27.13 ± 0.17 |
| 11 | 55 | 6 | 1.75 | 4 | 98.39 ± 0.33 |
| 12 | 55 | 1 | 1.75 | 4 | 25.49 ± 0.48 |
| 13 | 65 | 2.25 | 2.5 | 3 | 2.17 ± 0.09 |
| 14 | 65 | 4.75 | 1 | 3 | 1.23 ± 0.04 |
| 15 | 65 | 4.75 | 2.5 | 3 | 2.50 ± 0.18 |
| 16 | 65 | 2.25 | 1 | 5 | 20.03 ± 0.80 |
| 17 | 55 | 3.5 | 1.75 | 4 | 75.24 ± 1.64 |
| 18 | 45 | 4.75 | 2.5 | 5 | 84.21 ± 2.66 |
| 19 | 55 | 3.5 | 0.25 | 4 | 19.84 ± 1.25 |
| 20 | 55 | 3.5 | 3.25 | 4 | 97.24 ± 1.01 |
| 21 | 65 | 2.25 | 1 | 3 | 1.01 ± 0.08 |
| 22 | 65 | 4.75 | 1 | 5 | 31.83 ± 2.20 |
| 23 | 45 | 2.25 | 1 | 3 | 14.23 ± 0.21 |
| 24 | 45 | 4.75 | 1 | 5 | 56.85 ± 0.75 |
| 25 | 45 | 2.25 | 2.5 | 3 | 33.44 ± 1.33 |
| 26 | 75 | 3.5 | 1.75 | 4 | 1.73 ± 0.10 |
| 27 | 55 | 3.5 | 1.75 | 2 | 0.37 ± 0.04 |
| Factor a | Conversion (Y) | |||
|---|---|---|---|---|
| Sum of Squares | df | F-Value | Prob > F | |
| Model | 23,999.79 | 14 | 10.17 | 0.0001 * |
| Linear term | ||||
| X1 | 3594.42 | 1 | 21.34 | 0.0006 * |
| X2 | 3118.40 | 1 | 18.51 | 0.0010 * |
| X3 | 3849.26 | 1 | 22.85 | 0.0004 * |
| X4 | 2639.37 | 1 | 15.67 | 0.0019 * |
| Quadratic | ||||
| X12 | 5248.53 | 1 | 31.1 | 0.0001 * |
| X22 | 598.27 | 1 | 3.55 | 0.0839 |
| X32 | 806.20 | 1 | 4.78 | 0.0492 * |
| X42 | 7190.4 | 1 | 42.69 | <0.0001 * |
| Interactions | ||||
| X1X2 | 448.74 | 1 | 2.66 | 0.1285 |
| X1X3 | 307.99 | 1 | 1.82 | 0.2012 |
| X1X4 | 177.68 | 1 | 1.05 | 0.3246 |
| X2X3 | 38.74 | 1 | 0.23 | 0.6401 |
| X2X4 | 60.54 | 1 | 0.35 | 0.5599 |
| X3X4 | 34.56 | 1 | 0.20 | 0.6586 |
| Residual | 2020.83 | 12 | - | - |
| Lack of Fit | 1993.05 | 10 | 14.34 | 0.0669 |
| Pure Error | 27.78 | 2 | - | - |
| R-Squared | 0.92 | - | - | - |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuo, C.-H.; Chen, B.-Y.; Liu, Y.-C.; Chen, J.-H.; Shieh, C.-J. Production of Resveratrol by Piceid Deglycosylation Using Cellulase. Catalysts 2016, 6, 32. https://doi.org/10.3390/catal6030032
Kuo C-H, Chen B-Y, Liu Y-C, Chen J-H, Shieh C-J. Production of Resveratrol by Piceid Deglycosylation Using Cellulase. Catalysts. 2016; 6(3):32. https://doi.org/10.3390/catal6030032
Chicago/Turabian StyleKuo, Chia-Hung, Bao-Yuan Chen, Yung-Chuan Liu, Jiann-Hwa Chen, and Chwen-Jen Shieh. 2016. "Production of Resveratrol by Piceid Deglycosylation Using Cellulase" Catalysts 6, no. 3: 32. https://doi.org/10.3390/catal6030032







