Proline Metabolism in WHO G4 Gliomas Is Altered as Compared to Unaffected Brain Tissue
Abstract
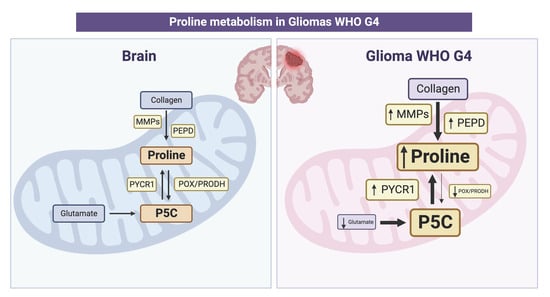
:Simple Summary
Abstract
1. Introduction
1.1. The Unsolved Problem of Glial Tumors
1.2. Proline in CellularMetabolism
1.3. Proline Pivots Rewired Tumor Metabolism
2. Materials and Methods
2.1. Study Design
2.2. Material
2.3. Real-Time PCR (RT-PCR)
2.4. Mining The Cancer Genome Atlas (TCGA) and Genotype–Tissue Expression (GTEx) Databases for Gene Expression
2.5. Expression of Selected Proteins by Western Immunoblot (WB)
2.6. Immunohistochemistry (IHC)
2.7. Gelatin Zymography Assay
2.8. Determination of PEPD Activity
2.9. Determination of Amino Acids and Organic Acids in Brain Homogenates
2.10. Statistical Analysis
3. Results
3.1. mRNA Expression of Genes Involved in the Proline Anabolic Axis and ECM Degradation Enzymes Is Enhanced
3.2. Expression of Proline Cycle Enzymes Favors Proline Biosynthesis Rather Than Utilization. Extracellular Matrix Catabolism Is Enhanced via Upregulation of Metalloproteinases and PEPD
3.3. Increased Activity of PEPD and Metalloproteinases Indicates Remodeling of Extracellular Matrix
3.4. Enzymes Responsible for Proline Biosynthesis Are Highly Upregulated in GG4 Tissues, as Opposed to the Proline Catabolic Enzyme
3.5. Concentration of Proline and Proline-Related Metabolites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Torp, S.H.; Solheim, O.; Skjulsvik, A.J. The WHO 2021 Classification of Central Nervous System tumours: A practical update on what neurosurgeons need to know-a minireview. Acta Neurochir. 2022, 164, 2453–2464. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.C.; Ashley, D.M.; López, G.Y.; Malinzak, M.; Friedman, H.S.; Khasraw, M. Management of glioblastoma: State of the art and future directions. CA Cancer J. Clin. 2020, 70, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Liu, Y. Metabolomics of Glioma. Adv. Exp. Med. Biol. 2021, 1280, 261–276. [Google Scholar] [CrossRef] [PubMed]
- McNamara, C.; Mankad, K.; Thust, S.; Dixon, L.; Limback-Stanic, C.; D’Arco, F.; Jacques, T.S.; Löbel, U. 2021 WHO classification of tumours of the central nervous system: A review for the neuroradiologist. Neuroradiology 2022, 64, 1919–1950. [Google Scholar] [CrossRef] [PubMed]
- Phang, J.M. The regulatory functions of proline and pyrroline-5-carboxylic acid. Curr. Top. Cell Regul. 1985, 25, 91–132. [Google Scholar] [CrossRef] [PubMed]
- Phang, J.M. Proline Metabolism in Cell Regulation and Cancer Biology: Recent Advances and Hypotheses. Antioxid. Redox Signal 2019, 30, 635–649. [Google Scholar] [CrossRef]
- Phang, J.M.; Yeh, G.C.; Hagedorn, C.H. The intercellular proline cycle. Life Sci. 1981, 28, 53–58. [Google Scholar] [CrossRef]
- Tanner, J.J.; Fendt, S.M.; Becker, D.F. The Proline Cycle As a Potential Cancer Therapy Target. Biochemistry 2018, 57, 3433–3444. [Google Scholar] [CrossRef]
- Phang, J.M.; Liu, W.; Hancock, C.; Christian, K.J. The proline regulatory axis and cancer. Front. Oncol. 2012, 2, 60. [Google Scholar] [CrossRef]
- Vettore, L.A.; Westbrook, R.L.; Tennant, D.A. Proline metabolism and redox; maintaining a balance in health and disease. Amino Acids 2021, 53, 1779–1788. [Google Scholar] [CrossRef]
- Phang, J.M. Perspectives, past, present and future: The proline cycle/proline-collagen regulatory axis. Amino Acids 2021, 53, 1967–1975. [Google Scholar] [CrossRef]
- Yang, L.; Wei, M.; Xing, B.; Zhang, C. Extracellular matrix and synapse formation. Biosci. Rep. 2023, 43, BSR20212411. [Google Scholar] [CrossRef]
- Eni-Aganga, I.; Lanaghan, Z.M.; Balasubramaniam, M.; Dash, C.; Pandhare, J. PROLIDASE: A Review from Discovery to its Role in Health and Disease. Front. Mol. Biosci. 2021, 8, 723003. [Google Scholar] [CrossRef]
- Li, Y.; Bie, J.; Song, C.; Liu, M.; Luo, J. PYCR, a key enzyme in proline metabolism, functions in tumorigenesis. Amino Acids 2021, 53, 1841–1850. [Google Scholar] [CrossRef]
- Liu, Y.; Mao, C.; Liu, S.; Xiao, D.; Shi, Y.; Tao, Y. Proline dehydrogenase in cancer: Apoptosis, autophagy, nutrient dependency and cancer therapy. Amino Acids 2021, 53, 1891–1902. [Google Scholar] [CrossRef]
- Phang, J.M.; Liu, W. Proline metabolism and cancer. Front. Biosci. 2012, 17, 1835–1845. [Google Scholar] [CrossRef]
- Misiura, M.; Miltyk, W. Current Understanding of the Emerging Role of Prolidase in Cellular Metabolism. Int. J. Mol. Sci. 2020, 21, 5906. [Google Scholar] [CrossRef]
- Sawicka, M.M.; Sawicki, K.; Łysoń, T.; Polityńska, B.; Miltyk, W. Proline Metabolism in Malignant Gliomas: A Systematic Literature Review. Cancers 2022, 14, 2030. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Wechselberger, C.; Doppler, C.; Bernhard, D. An Inexpensive Staining Alternative for Gelatin Zymography Gels. Methods Protoc. 2019, 2, 61. [Google Scholar] [CrossRef]
- Myara, I.; Charpentier, C.; Lemonnier, A. Optimal conditions for prolidase assay by proline colorimetric determination: Application to iminodipeptiduria. Clin. Chim. Acta 1982, 125, 193–205. [Google Scholar] [CrossRef]
- Wang, X.; Han, W.; Yang, J.; Westaway, D.; Li, L. Development of chemical isotope labeling LC-MS for tissue metabolomics and its application for brain and liver metabolome profiling in Alzheimer’s disease mouse model. Anal. Chim. Acta 2019, 1050, 95–104. [Google Scholar] [CrossRef]
- Pičmanová, M.; Moses, T.; Cortada-Garcia, J.; Barrett, G.; Florance, H.; Pandor, S.; Burgess, K. Rapid HILIC-Z ion mobility mass spectrometry (RHIMMS) method for untargeted metabolomics of complex biological samples. Metabolomics 2022, 18, 16. [Google Scholar] [CrossRef]
- Ibáñez, A.B.; Bauer, S. Analytical method for the determination of organic acids in dilute acid pretreated biomass hydrolysate by liquid chromatography-time-of-flight mass spectrometry. Biotechnol. Biofuels 2014, 7, 145. [Google Scholar] [CrossRef]
- Loreck, D.J.; Galarraga, J.; Van der Feen, J.; Phang, J.M.; Smith, B.H.; Cummins, C.J. Regulation of the pentose phosphate pathway in human astrocytes and gliomas. Metab. Brain Dis. 1987, 2, 31–46. [Google Scholar] [CrossRef]
- Shao, W.; Hao, Z.Y.; Chen, Y.F.; Du, J.; He, Q.; Ren, L.L.; Gao, Y.; Song, N.; Song, Y.; He, H.; et al. OIP5-AS1 specifies p53-driven POX transcription regulated by TRPC6 in glioma. J. Mol. Cell Biol. 2021, 13, 409–421. [Google Scholar] [CrossRef]
- Panosyan, E.H.; Lin, H.J.; Koster, J.; Lasky, J.L. In search of druggable targets for GBM amino acid metabolism. BMC Cancer 2017, 17, 162. [Google Scholar] [CrossRef]
- Cappelletti, P.; Tallarita, E.; Rabattoni, V.; Campomenosi, P.; Sacchi, S.; Pollegioni, L. Proline oxidase controls proline, glutamate, and glutamine cellular concentrations in a U87 glioblastoma cell line. PLoS ONE 2018, 13, e0196283. [Google Scholar] [CrossRef]
- Polyak, K.; Xia, Y.; Zweier, J.L.; Kinzler, K.W.; Vogelstein, B. A model for p53-induced apoptosis. Nature 1997, 389, 300–305. [Google Scholar] [CrossRef]
- Liu, W.; Phang, J.M. Proline dehydrogenase (oxidase) in cancer. Biofactors 2012, 38, 398–406. [Google Scholar] [CrossRef]
- Burke, L.; Guterman, I.; Palacios Gallego, R.; Britton, R.G.; Burschowsky, D.; Tufarelli, C.; Rufini, A. The Janus-like role of proline metabolism in cancer. Cell Death Discov. 2020, 6, 104. [Google Scholar] [CrossRef]
- Liu, Y.; Borchert, G.L.; Surazynski, A.; Phang, J.M. Proline oxidase, a p53-induced gene, targets COX-2/PGE2 signaling to induce apoptosis and inhibit tumor growth in colorectal cancers. Oncogene 2008, 27, 6729–6737. [Google Scholar] [CrossRef]
- Tołoczko-Iwaniuk, N.; Dziemiańczyk-Pakieła, D.; Celińska-Janowicz, K.; Zaręba, I.; Klupczyńska, A.; Kokot, Z.J.; Nowaszewska, B.K.; Reszeć, J.; Borys, J.; Miltyk, W. Proline-Dependent Induction of Apoptosis in Oral Squamous Cell Carcinoma (OSCC)-The Effect of Celecoxib. Cancers 2020, 12, 136. [Google Scholar] [CrossRef]
- Liu, Y.; Borchert, G.L.; Donald, S.P.; Diwan, B.A.; Anver, M.; Phang, J.M. Proline oxidase functions as a mitochondrial tumor suppressor in human cancers. Cancer Res. 2009, 69, 6414–6422. [Google Scholar] [CrossRef]
- Hollinshead, K.E.R.; Munford, H.; Eales, K.L.; Bardella, C.; Li, C.; Escribano-Gonzalez, C.; Thakker, A.; Nonnenmacher, Y.; Kluckova, K.; Jeeves, M.; et al. Oncogenic IDH1 Mutations Promote Enhanced Proline Synthesis through PYCR1 to Support the Maintenance of Mitochondrial Redox Homeostasis. Cell Rep. 2018, 22, 3107–3114. [Google Scholar] [CrossRef]
- De Ingeniis, J.; Ratnikov, B.; Richardson, A.D.; Scott, D.A.; Aza-Blanc, P.; De, S.K.; Kazanov, M.; Pellecchia, M.; Ronai, Z.; Osterman, A.L.; et al. Functional specialization in proline biosynthesis of melanoma. PLoS ONE 2012, 7, e45190. [Google Scholar] [CrossRef]
- Obara-Michlewska, M.; Szeliga, M. Targeting Glutamine Addiction in Gliomas. Cancers 2020, 12, 310. [Google Scholar] [CrossRef]
- Bogner, A.N.; Stiers, K.M.; Tanner, J.J. Structure, biochemistry, and gene expression patterns of the proline biosynthetic enzyme pyrroline-5-carboxylate reductase (PYCR), an emerging cancer therapy target. Amino Acids 2021, 53, 1817–1834. [Google Scholar] [CrossRef]
- Verma, A.a.S.K.a.P.S.a.S.R. Prolidase Activity and Oxidative Stress in Patients with Glioma. J. Clin. Diagn. Res. 2018, 12, BC07–BC10. [Google Scholar] [CrossRef]
- Gonullu, E.; Silav, G.; Kaya, M.; Arslan, M.; Gonullu, H.; Arslan, H.; Cebi, A.; Demir, H. Paraoxonase and Prolidase Activity in Patietns With Malignant Gliomas. J. Neurol. Sci.-Turk. 2012, 29, 778–782. [Google Scholar]
- Payne, L.S.; Huang, P.H. The pathobiology of collagens in glioma. Mol. Cancer Res. 2013, 11, 1129–1140. [Google Scholar] [CrossRef] [PubMed]
- Xue, Q.; Cao, L.; Chen, X.Y.; Zhao, J.; Gao, L.; Li, S.Z.; Fei, Z. High expression of MMP9 in glioma affects cell proliferation and is associated with patient survival rates. Oncol. Lett. 2017, 13, 1325–1330. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, R.K.; Sørensen, M.D.; Aaberg-Jessen, C.; Hermansen, S.K.; Kristensen, B.W. Expression and prognostic impact of matrix metalloproteinase-2 (MMP-2) in astrocytomas. PLoS ONE 2017, 12, e0172234. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Yu, X.; Sun, S.; Zhang, X.; Yang, W.; Zhang, J.; Jiang, Z. Increased expression of MMP-2 and MMP-9 indicates poor prognosis in glioma recurrence. Biomed. Pharmacother. 2019, 118, 109369. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, G.; Zhao, Z.; Wang, J.; Gao, G.; He, S. Matrix metalloproteinase-9 expression is increased in astrocytic glioma and associated with prognosis of patients. Jpn. J. Clin. Oncol. 2012, 42, 1060–1065. [Google Scholar] [CrossRef]
- Tamai, S.; Ichinose, T.; Tsutsui, T.; Tanaka, S.; Garaeva, F.; Sabit, H.; Nakada, M. Tumor Microenvironment in Glioma Invasion. Brain Sci. 2022, 12, 505. [Google Scholar] [CrossRef]
- Karimi, N.; Kheiri, H.; Zarrinpour, V.; Forghanifard, M.M. Bioinformatic analysis of MMP family members in GBM. Inform. Med. Unlocked 2023, 39, 101240. [Google Scholar] [CrossRef]
- Jin, Y.; Xiao, W.; Song, T.; Feng, G.; Dai, Z. Expression and Prognostic Significance of p53 in Glioma Patients: A Meta-analysis. Neurochem. Res. 2016, 41, 1723–1731. [Google Scholar] [CrossRef]
- Ludwig, K.; Kornblum, H.I. Molecular markers in glioma. J. Neurooncol. 2017, 134, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Saadeh, F.S.; Mahfouz, R.; Assi, H.I. EGFR as a clinical marker in glioblastomas and other gliomas. Int. J. Biol. Markers 2018, 33, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Maus, A.; Peters, G.J. Glutamate and α-ketoglutarate: Key players in glioma metabolism. Amino Acids 2017, 49, 21–32. [Google Scholar] [CrossRef] [PubMed]
- de Groot, J.; Sontheimer, H. Glutamate and the biology of gliomas. Glia 2011, 59, 1181–1189. [Google Scholar] [CrossRef]
- Ferreira, A.G.K.; Biasibetti-Brendler, H.; Sidegum, D.S.V.; Loureiro, S.O.; Figueiró, F.; Wyse, A.T.S. Effect of Proline on Cell Death, Cell Cycle, and Oxidative Stress in C6 Glioma Cell Line. Neurotox. Res. 2021, 39, 327–334. [Google Scholar] [CrossRef]
- Ding, J.; Kuo, M.L.; Su, L.; Xue, L.; Luh, F.; Zhang, H.; Wang, J.; Lin, T.G.; Zhang, K.; Chu, P.; et al. Human mitochondrial pyrroline-5-carboxylate reductase 1 promotes invasiveness and impacts survival in breast cancers. Carcinogenesis 2017, 38, 519–531. [Google Scholar] [CrossRef]
- She, Y.; Mao, A.; Li, F.; Wei, X. P5CR1 protein expression and the effect of gene-silencing on lung adenocarcinoma. PeerJ 2019, 7, e6934. [Google Scholar] [CrossRef]
- Oscilowska, I.; Rolkowski, K.; Baszanowska, W.; Huynh, T.Y.L.; Lewoniewska, S.; Nizioł, M.; Sawicka, M.; Bielawska, K.; Szoka, P.; Miltyk, W.; et al. Proline Dehydrogenase/Proline Oxidase (PRODH/POX) Is Involved in the Mechanism of Metformin-Induced Apoptosis in C32 Melanoma Cell Line. Int. J. Mol. Sci. 2022, 23, 2354. [Google Scholar] [CrossRef]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sawicka, M.M.; Sawicki, K.; Jadeszko, M.; Bielawska, K.; Supruniuk, E.; Reszeć, J.; Prokop-Bielenia, I.; Polityńska, B.; Jadeszko, M.; Rybaczek, M.; et al. Proline Metabolism in WHO G4 Gliomas Is Altered as Compared to Unaffected Brain Tissue. Cancers 2024, 16, 456. https://doi.org/10.3390/cancers16020456
Sawicka MM, Sawicki K, Jadeszko M, Bielawska K, Supruniuk E, Reszeć J, Prokop-Bielenia I, Polityńska B, Jadeszko M, Rybaczek M, et al. Proline Metabolism in WHO G4 Gliomas Is Altered as Compared to Unaffected Brain Tissue. Cancers. 2024; 16(2):456. https://doi.org/10.3390/cancers16020456
Chicago/Turabian StyleSawicka, Magdalena M., Karol Sawicki, Marek Jadeszko, Katarzyna Bielawska, Elżbieta Supruniuk, Joanna Reszeć, Izabela Prokop-Bielenia, Barbara Polityńska, Mateusz Jadeszko, Magdalena Rybaczek, and et al. 2024. "Proline Metabolism in WHO G4 Gliomas Is Altered as Compared to Unaffected Brain Tissue" Cancers 16, no. 2: 456. https://doi.org/10.3390/cancers16020456