Hotspots of Somatic Genetic Variation in Pituitary Neuroendocrine Tumors
Abstract
:Simple Summary
Abstract
1. Introduction
2. BRAF

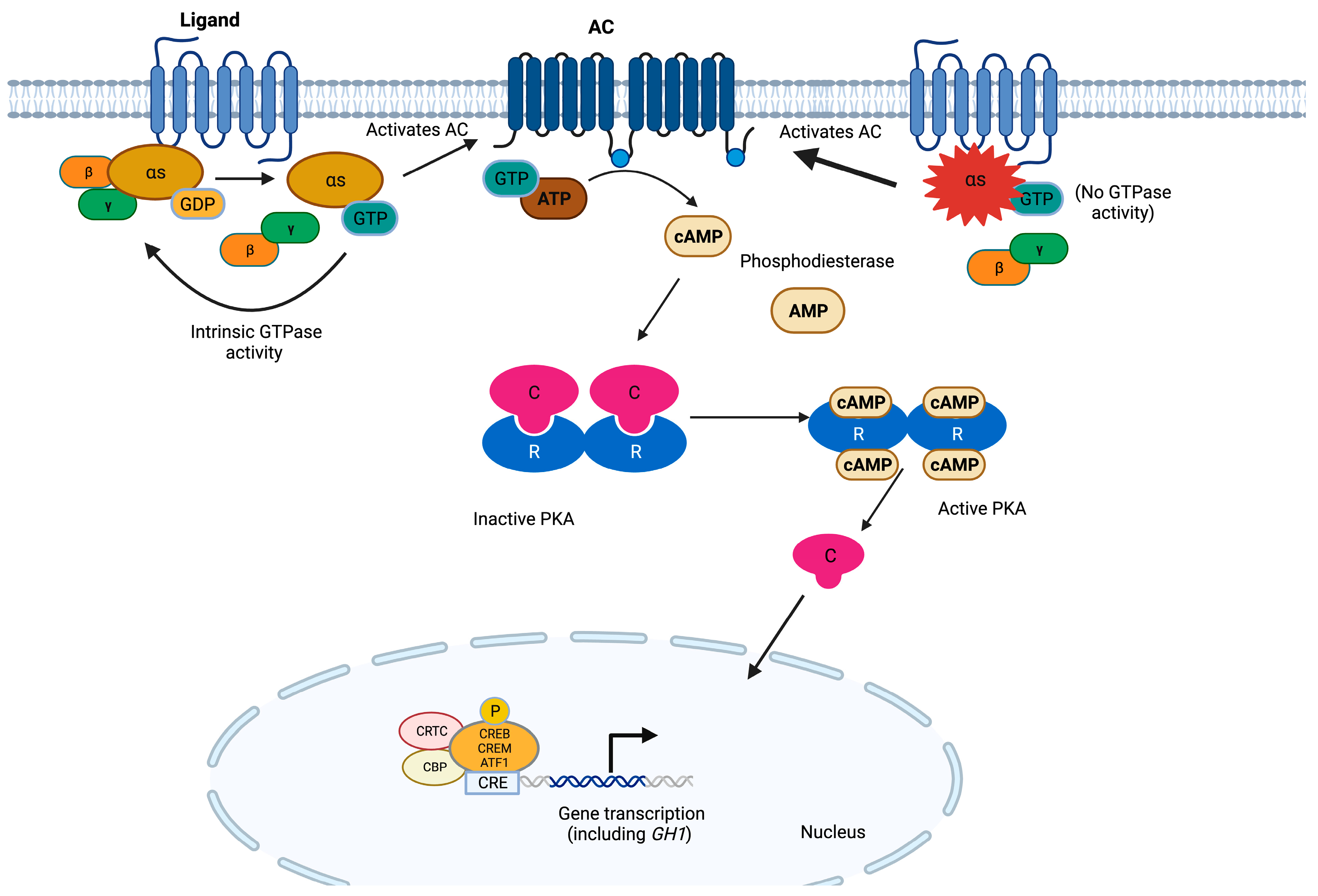
3. GNAS
4. DICER1

5. SF3B1
6. USP8
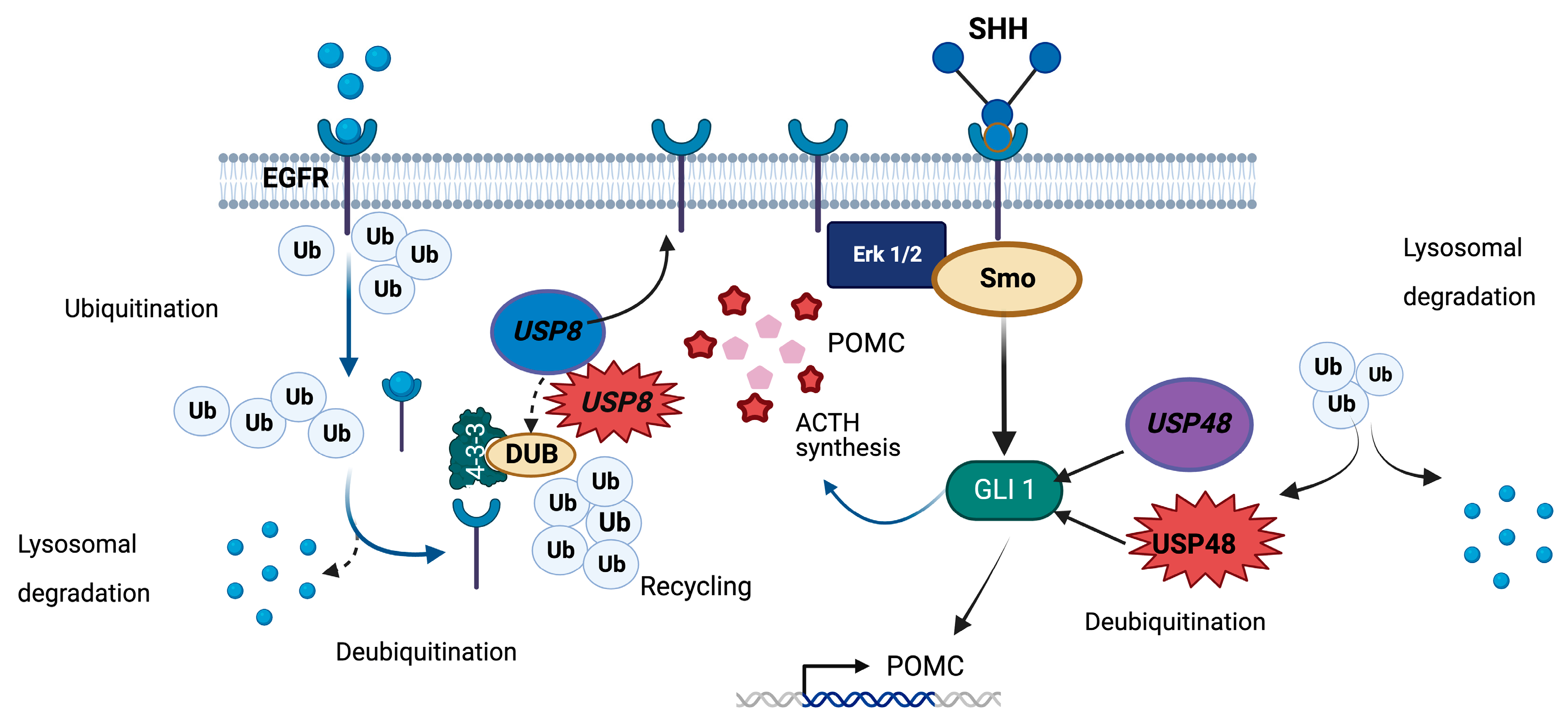
7. USP48
8. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Juul, R.I.; Nielsen, M.M.; Juul, M.; Feuerbach, L.; Pedersen, J.S. The landscape and driver potential of site-specific hotspots across cancer genomes. NPJ Genom. Med. 2021, 6, 33. [Google Scholar] [CrossRef] [PubMed]
- Cooper, D.N.; Mort, M.; Stenson, P.D.; Ball, E.V.; Chuzhanova, N.A. Methylation-mediated deamination of 5-methylcytosine appears to give rise to mutations causing human inherited disease in CpNpG trinucleotides, as well as in CpG dinucleotides. Hum. Genom. 2010, 4, 406–410. [Google Scholar] [CrossRef] [PubMed]
- Marais, G. Biased gene conversion: Implications for genome and sex evolution. Trends Genet. 2003, 19, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Nesta, A.V.; Tafur, D.; Beck, C.R. Hotspots of human mutation. Trends Genet. 2021, 37, 717–729. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Kurahashi, H.; Emanuel, B.S. Chromosomal translocations and palindromic AT-rich repeats. Curr. Opin. Genet. Dev. 2012, 22, 221–228. [Google Scholar] [CrossRef] [PubMed]
- ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Martincorena, I.; Campbell, P.J. Somatic mutation in cancer and normal cells. Science 2015, 349, 1483–1489. [Google Scholar] [CrossRef]
- Zhang, F.; Zhang, Q.; Zhu, J.; Yao, B.; Ma, C.; Qiao, N.; He, S.; Ye, Z.; Wang, Y.; Han, R.; et al. Integrated proteogenomic characterization across major histological types of pituitary neuroendocrine tumors. Cell Res. 2022, 32, 1047–1067. [Google Scholar] [CrossRef]
- Villa, C.; Baussart, B.; Assie, G.; Raverot, G.; Roncaroli, F. The World Health Organization classifications of pituitary neuroendocrine tumours: A clinico-pathological appraisal. Endocr. Relat. Cancer 2023, 30, e230021. [Google Scholar] [CrossRef]
- Landis, C.A.; Masters, S.B.; Spada, A.; Pace, A.M.; Bourne, H.R.; Vallar, L. GTPase inhibiting mutations activate the alpha chain of Gs and stimulate adenylyl cyclase in human pituitary tumours. Nature 1989, 340, 692–696. [Google Scholar] [CrossRef]
- Ma, Z.Y.; Song, Z.J.; Chen, J.H.; Wang, Y.F.; Li, S.Q.; Zhou, L.F.; Mao, Y.; Li, Y.M.; Hu, R.G.; Zhang, Z.Y.; et al. Recurrent gain-of-function USP8 mutations in Cushing’s disease. Cell Res. 2015, 25, 306–317. [Google Scholar] [CrossRef] [PubMed]
- Reincke, M.; Sbiera, S.; Hayakawa, A.; Theodoropoulou, M.; Osswald, A.; Beuschlein, F.; Meitinger, T.; Mizuno-Yamasaki, E.; Kawaguchi, K.; Saeki, Y.; et al. Mutations in the deubiquitinase gene USP8 cause Cushing’s disease. Nat. Genet. 2015, 47, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Greenman, C.; Stephens, P.; Smith, R.; Dalgliesh, G.L.; Hunter, C.; Bignell, G.; Davies, H.; Teague, J.; Butler, A.; Stevens, C.; et al. Patterns of somatic mutation in human cancer genomes. Nature 2007, 446, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Bonner, T.I.; Kerby, S.B.; Sutrave, P.; Gunnell, M.A.; Mark, G.; Rapp, U.R. Structure and biological activity of human homologs of the raf/mil oncogene. Mol. Cell Biol. 1985, 5, 1400–1407. [Google Scholar] [CrossRef] [PubMed]
- Huebner, K.; ar-Rushdi, A.; Griffin, C.A.; Isobe, M.; Kozak, C.; Emanuel, B.S.; Nagarajan, L.; Cleveland, J.L.; Bonner, T.I.; Goldsborough, M.D.; et al. Actively transcribed genes in the raf oncogene group, located on the X chromosome in mouse and human. Proc. Natl. Acad. Sci. USA 1986, 83, 3934–3938. [Google Scholar] [CrossRef] [PubMed]
- Ikawa, S.; Fukui, M.; Ueyama, Y.; Tamaoki, N.; Yamamoto, T.; Toyoshima, K. B-raf, a new member of the raf family, is activated by DNA rearrangement. Mol. Cell Biol. 1988, 8, 2651–2654. [Google Scholar] [CrossRef]
- Sithanandam, G.; Druck, T.; Cannizzaro, L.A.; Leuzzi, G.; Huebner, K.; Rapp, U.R. B-raf and a B-raf pseudogene are located on 7q in man. Oncogene 1992, 7, 795–799. [Google Scholar]
- Wellbrock, C.; Karasarides, M.; Marais, R. The RAF proteins take centre stage. Nat. Rev. Mol. Cell Biol. 2004, 5, 875–885. [Google Scholar] [CrossRef]
- Gualtieri, A.; Kyprianou, N.; Gregory, L.C.; Vignola, M.L.; Nicholson, J.G.; Tan, R.; Inoue, S.I.; Scagliotti, V.; Casado, P.; Blackburn, J.; et al. Activating mutations in BRAF disrupt the hypothalamo-pituitary axis leading to hypopituitarism in mice and humans. Nat. Commun. 2021, 12, 2028. [Google Scholar] [CrossRef]
- Hebron, K.E.; Hernandez, E.R.; Yohe, M.E. The RASopathies: From pathogenetics to therapeutics. Dis. Model Mech. 2022, 15, dmm049107. [Google Scholar] [CrossRef]
- Hoshino, R.; Chatani, Y.; Yamori, T.; Tsuruo, T.; Oka, H.; Yoshida, O.; Shimada, Y.; Ari-i, S.; Wada, H.; Fujimoto, J.; et al. Constitutive activation of the 41-/43-kDa mitogen-activated protein kinase signaling pathway in human tumors. Oncogene 1999, 18, 813–822. [Google Scholar] [CrossRef]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF gene in human cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef] [PubMed]
- Garnett, M.J.; Marais, R. Guilty as charged: B-RAF is a human oncogene. Cancer Cell 2004, 6, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Xing, M. BRAF mutation in thyroid cancer. Endocr. Relat. Cancer 2005, 12, 245–262. [Google Scholar] [CrossRef] [PubMed]
- Wan, P.T.; Garnett, M.J.; Roe, S.M.; Lee, S.; Niculescu-Duvaz, D.; Good, V.M.; Jones, C.M.; Marshall, C.J.; Springer, C.J.; Barford, D.; et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 2004, 116, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Ikenoue, T.; Hikiba, Y.; Kanai, F.; Tanaka, Y.; Imamura, J.; Imamura, T.; Ohta, M.; Ijichi, H.; Tateishi, K.; Kawakami, T.; et al. Functional analysis of mutations within the kinase activation segment of B-Raf in human colorectal tumors. Cancer Res. 2003, 63, 8132–8137. [Google Scholar]
- Wellbrock, C.; Ogilvie, L.; Hedley, D.; Karasarides, M.; Martin, J.; Niculescu-Duvaz, D.; Springer, C.J.; Marais, R. V599EB-RAF is an oncogene in melanocytes. Cancer Res. 2004, 64, 2338–2342. [Google Scholar] [CrossRef]
- Hingorani, S.R.; Jacobetz, M.A.; Robertson, G.P.; Herlyn, M.; Tuveson, D.A. Suppression of BRAF(V599E) in human melanoma abrogates transformation. Cancer Res. 2003, 63, 5198–5202. [Google Scholar]
- Grothey, A.; Fakih, M.; Tabernero, J. Management of BRAF-mutant metastatic colorectal cancer: A review of treatment options and evidence-based guidelines. Ann. Oncol. 2021, 32, 959–967. [Google Scholar] [CrossRef]
- Broccoli, A.; Terragna, C.; Nanni, L.; Martello, M.; Armuzzi, S.; Agostinelli, C.; Morigi, A.; Casadei, B.; Pellegrini, C.; Stefoni, V.; et al. Droplet digital polymerase chain reaction for the assessment of disease burden in hairy cell leukemia. Hematol. Oncol. 2022, 40, 57–62. [Google Scholar] [CrossRef]
- Haston, S.; Pozzi, S.; Carreno, G.; Manshaei, S.; Panousopoulos, L.; Gonzalez-Meljem, J.M.; Apps, J.R.; Virasami, A.; Thavaraj, S.; Gutteridge, A.; et al. MAPK pathway control of stem cell proliferation and differentiation in the embryonic pituitary provides insights into the pathogenesis of papillary craniopharyngioma. Development 2017, 144, 2141–2152. [Google Scholar] [CrossRef] [PubMed]
- Prieto, R.; Pascual, J.M. Craniopharyngiomas with a mixed histological pattern: The missing link to the intriguing pathogenesis of adamantinomatous and squamous-papillary varieties? Neuropathology 2013, 33, 682–686. [Google Scholar] [CrossRef]
- Brastianos, P.K.; Taylor-Weiner, A.; Manley, P.E.; Jones, R.T.; Dias-Santagata, D.; Thorner, A.R.; Lawrence, M.S.; Rodriguez, F.J.; Bernardo, L.A.; Schubert, L.; et al. Exome sequencing identifies BRAF mutations in papillary craniopharyngiomas. Nat. Genet. 2014, 46, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Larkin, S.J.; Preda, V.; Karavitaki, N.; Grossman, A.; Ansorge, O. BRAF V600E mutations are characteristic for papillary craniopharyngioma and may coexist with CTNNB1-mutated adamantinomatous craniopharyngioma. Acta Neuropathol. 2014, 127, 927–929. [Google Scholar] [CrossRef] [PubMed]
- Holsken, A.; Sill, M.; Merkle, J.; Schweizer, L.; Buchfelder, M.; Flitsch, J.; Fahlbusch, R.; Metzler, M.; Kool, M.; Pfister, S.M.; et al. Adamantinomatous and papillary craniopharyngiomas are characterized by distinct epigenomic as well as mutational and transcriptomic profiles. Acta Neuropathol. Commun. 2016, 4, 20. [Google Scholar] [CrossRef] [PubMed]
- Goschzik, T.; Gessi, M.; Dreschmann, V.; Gebhardt, U.; Wang, L.; Yamaguchi, S.; Wheeler, D.A.; Lauriola, L.; Lau, C.C.; Muller, H.L.; et al. Genomic alterations of adamantinomatous and papillary craniopharyngioma. J. Neuropathol. Exp. Neurol. 2017, 76, 126–134. [Google Scholar] [CrossRef]
- Yoshimoto, K.; Hatae, R.; Suzuki, S.O.; Hata, N.; Kuga, D.; Akagi, Y.; Amemiya, T.; Sangatsuda, Y.; Mukae, N.; Mizoguchi, M.; et al. High-resolution melting and immunohistochemical analysis efficiently detects mutually exclusive genetic alterations of adamantinomatous and papillary craniopharyngiomas. Neuropathology 2018, 38, 3–10. [Google Scholar] [CrossRef]
- Omay, S.B.; Chen, Y.N.; Almeida, J.P.; Ruiz-Trevino, A.S.; Boockvar, J.A.; Stieg, P.E.; Greenfield, J.P.; Souweidane, M.M.; Kacker, A.; Pisapia, D.J.; et al. Do craniopharyngioma molecular signatures correlate with clinical characteristics? J. Neurosurg. 2018, 128, 1473–1478. [Google Scholar] [CrossRef]
- La Corte, E.; Younus, I.; Pivari, F.; Selimi, A.; Ottenhausen, M.; Forbes, J.A.; Pisapia, D.J.; Dobri, G.A.; Anand, V.K.; Schwartz, T.H. BRAF V600E mutant papillary craniopharyngiomas: A single-institutional case series. Pituitary 2018, 21, 571–583. [Google Scholar] [CrossRef]
- Chen, J.; Jian, X.; Deng, S.; Ma, Z.; Shou, X.; Shen, Y.; Zhang, Q.; Song, Z.; Li, Z.; Peng, H.; et al. Identification of recurrent USP48 and BRAF mutations in Cushing’s disease. Nat. Commun. 2018, 9, 3171. [Google Scholar] [CrossRef]
- Sbiera, S.; Pérez-Rivas, L.G.; Taranets, L.; Weigand, I.; Flitsch, J.; Graf, E.; Monoranu, C.M.; Saeger, W.; Hagel, C.; Honegger, J.; et al. Driver mutations in USP8 wild-type Cushing’s disease. Neuro Oncol. 2019, 21, 1273–1283. [Google Scholar] [CrossRef] [PubMed]
- Uzilov, A.V.; Taik, P.; Cheesman, K.C.; Javanmard, P.; Ying, K.; Roehnelt, A.; Wang, H.; Fink, M.Y.; Lau, C.Y.; Moe, A.S.; et al. USP8 and TP53 drivers are associated with CNV in a corticotroph adenoma cohort enriched for aggressive tumors. J. Clin. Endocrinol. Metab. 2021, 106, 826–842. [Google Scholar] [CrossRef] [PubMed]
- Abraham, A.P.; Pai, R.; Beno, D.L.; Chacko, G.; Asha, H.S.; Rajaratnam, S.; Kapoor, N.; Thomas, N.; Chacko, A.G. USP8, USP48, and BRAF mutations differ in their genotype-phenotype correlation in Asian Indian patients with Cushing’s disease. Endocrine 2022, 75, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Ramírez, L.C.; Pankratz, N.; Lane, J.; Faucz, F.R.; Chittiboina, P.; Kay, D.M.; Beethem, Z.; Mills, J.L.; Stratakis, C.A. Genetic drivers of Cushing’s disease: Frequency and associated phenotypes. Genet. Med. 2022, 24, 2516–2525. [Google Scholar] [CrossRef] [PubMed]
- Schmid, S.; Solomon, D.A.; Perez, E.; Thieme, A.; Kleinschmidt-DeMasters, B.K.; Giannini, C.; Reinhardt, A.; Asa, S.L.; Mete, O.; Stichel, D.; et al. Genetic and epigenetic characterization of posterior pituitary tumors. Acta Neuropathol. 2021, 142, 1025–1043. [Google Scholar] [CrossRef] [PubMed]
- Hatzivassiliou, G.; Song, K.; Yen, I.; Brandhuber, B.J.; Anderson, D.J.; Alvarado, R.; Ludlam, M.J.; Stokoe, D.; Gloor, S.L.; Vigers, G.; et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature 2010, 464, 431–435. [Google Scholar] [CrossRef]
- Su, F.; Viros, A.; Milagre, C.; Trunzer, K.; Bollag, G.; Spleiss, O.; Reis-Filho, J.S.; Kong, X.; Koya, R.C.; Flaherty, K.T.; et al. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N. Engl. J. Med. 2012, 366, 207–215. [Google Scholar] [CrossRef]
- Aziz, S.N.; Proano, L.; Cruz, C.; Tenemaza, M.G.; Monteros, G.; Hassen, G.; Baskar, A.; Argudo, J.M.; Duenas, J.B.; Fabara, S.P. Vemurafenib in the Treatment of Erdheim Chester Disease: A Systematic Review. Cureus 2022, 14, e25935. [Google Scholar] [CrossRef]
- Maitre, E.; Paillassa, J.; Troussard, X. Novel targeted treatments in hairy cell leukemia and other hairy cell-like disorders. Front. Oncol. 2022, 12, 1068981. [Google Scholar] [CrossRef]
- Fernandez, M.F.; Choi, J.; Sosman, J. New approaches to targeted therapy in melanoma. Cancers 2023, 15, 3224. [Google Scholar] [CrossRef]
- Guaitoli, G.; Zullo, L.; Tiseo, M.; Dankner, M.; Rose, A.A.; Facchinetti, F. Non-small-cell lung cancer: How to manage. Drugs Context 2023, 12, 2022-11-3. [Google Scholar] [CrossRef] [PubMed]
- Hamidi, S.; Hofmann, M.C.; Iyer, P.C.; Cabanillas, M.E.; Hu, M.I.; Busaidy, N.L.; Dadu, R. Review article: New treatments for advanced differentiated thyroid cancers and potential mechanisms of drug resistance. Front. Endocrinol. 2023, 14, 1176731. [Google Scholar] [CrossRef] [PubMed]
- Juratli, T.A.; Jones, P.S.; Wang, N.; Subramanian, M.; Aylwin, S.J.B.; Odia, Y.; Rostami, E.; Gudjonsson, O.; Shaw, B.L.; Cahill, D.P.; et al. Targeted treatment of papillary craniopharyngiomas harboring BRAF V600E mutations. Cancer 2019, 125, 2910–2914. [Google Scholar] [CrossRef] [PubMed]
- Brastianos, P.K.; Twohy, E.; Geyer, S.; Gerstner, E.R.; Kaufmann, T.J.; Tabrizi, S.; Kabat, B.; Thierauf, J.; Ruff, M.W.; Bota, D.A.; et al. BRAF-MEK inhibition in newly diagnosed papillary craniopharyngiomas. N. Engl. J. Med. 2023, 389, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Grenier-Chartrand, F.; Barrit, S.; Racu, M.L.; Luce, S.; Spitaels, J.; Sadeghi-Meibodi, N.; Lebrun, L.; Salmon, I.; Lefranc, F.; De Witte, O. Dabrafenib monotherapy for a recurrent BRAFV600E-mutated TTF-1-positive posterior pituitary tumor. Acta. Neurochir. 2022, 164, 737–742. [Google Scholar] [CrossRef] [PubMed]
- Sollfrank, L.; Lettmaier, S.; Erdmann, M.; Uslu, U. Panniculitis under successful targeted inhibition of the MAPK/ERK signaling pathway in a patient with BRAF V600E-mutated spindle cell oncocytoma of the pituitary gland. Anticancer Res. 2019, 39, 3955–3959. [Google Scholar] [CrossRef] [PubMed]
- Dawoud, F.M.; Naylor, R.M.; Giannini, C.; Swanson, A.A.; Meyer, F.B.; Uhm, J.H. TTF-1 positive posterior pituitary tumor: Limitations of current treatment and potential new hope in BRAF V600E mutation variants. Clin. Neurol. Neurosurg. 2020, 196, 106059. [Google Scholar] [CrossRef]
- Chang, F.; Steelman, L.S.; Lee, J.T.; Shelton, J.G.; Navolanic, P.M.; Blalock, W.L.; Franklin, R.A.; McCubrey, J.A. Signal transduction mediated by the Ras/Raf/MEK/ERK pathway from cytokine receptors to transcription factors: Potential targeting for therapeutic intervention. Leukemia 2003, 17, 1263–1293. [Google Scholar] [CrossRef]
- Derwich, A.; Sykutera, M.; Brominska, B.; Rubis, B.; Ruchala, M.; Sawicka-Gutaj, N. The role of activation of PI3K/AKT/mTOR and RAF/MEK/ERK pathways in aggressive pituitary adenomas-new potential therapeutic approach—A systematic review. Int. J. Mol. Sci. 2023, 24, 10952. [Google Scholar] [CrossRef]
- Ishida, M.; Moore, G.E. The role of imprinted genes in humans. Mol. Aspects Med. 2013, 34, 826–840. [Google Scholar] [CrossRef]
- Hayward, B.E.; Moran, V.; Strain, L.; Bonthron, D.T. Bidirectional imprinting of a single gene: GNAS1 encodes maternally, paternally, and biallelically derived proteins. Proc. Natl. Acad. Sci. USA 1998, 95, 15475–15480. [Google Scholar] [CrossRef] [PubMed]
- Hayward, B.E.; Bonthron, D.T. An imprinted antisense transcript at the human GNAS1 locus. Hum. Mol. Genet 2000, 9, 835–841. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2023. Nucleic. Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, G.; Ballare, E.; Giammona, E.; Beck-Peccoz, P.; Spada, A. The gsalpha gene: Predominant maternal origin of transcription in human thyroid gland and gonads. J. Clin. Endocrinol. Metab. 2002, 87, 4736–4740. [Google Scholar] [CrossRef]
- Syrovatkina, V.; Alegre, K.O.; Dey, R.; Huang, X.Y. Regulation, signaling, and physiological functions of G-proteins. J. Mol. Biol. 2016, 428, 3850–3868. [Google Scholar] [CrossRef]
- Peverelli, E.; Mantovani, G.; Lania, A.G.; Spada, A. cAMP in the pituitary: An old messenger for multiple signals. J. Mol. Endocrinol. 2014, 52, R67–R77. [Google Scholar] [CrossRef]
- Duc, N.M.; Kim, H.R.; Chung, K.Y. Structural mechanism of G protein activation by G protein-coupled receptor. Eur. J. Pharmacol. 2015, 763, 214–222. [Google Scholar] [CrossRef]
- Lambright, D.G.; Noel, J.P.; Hamm, H.E.; Sigler, P.B. Structural determinants for activation of the alpha-subunit of a heterotrimeric G protein. Nature 1994, 369, 621–628. [Google Scholar] [CrossRef]
- Pierce, K.L.; Premont, R.T.; Lefkowitz, R.J. Seven-transmembrane receptors. Nat. Rev. Mol. Cell. Biol. 2002, 3, 639–650. [Google Scholar] [CrossRef]
- Hernandez-Ramirez, L.C.; Trivellin, G.; Stratakis, C.A. Cyclic 3’,5’-adenosine monophosphate (cAMP) signaling in the anterior pituitary gland in health and disease. Mol. Cell Endocrinol. 2018, 463, 72–86. [Google Scholar] [CrossRef]
- Landis, C.A.; Harsh, G.; Lyons, J.; Davis, R.L.; McCormick, F.; Bourne, H.R. Clinical characteristics of acromegalic patients whose pituitary tumors contain mutant Gs protein. J. Clin. Endocrinol. Metab. 1990, 71, 1416–1420. [Google Scholar] [CrossRef] [PubMed]
- Lyons, J.; Landis, C.A.; Harsh, G.; Vallar, L.; Grunewald, K.; Feichtinger, H.; Duh, Q.Y.; Clark, O.H.; Kawasaki, E.; Bourne, H.R. Two G protein oncogenes in human endocrine tumors. Science 1990, 249, 655–659. [Google Scholar] [CrossRef] [PubMed]
- Suarez, H.G.; du Villard, J.A.; Caillou, B.; Schlumberger, M.; Parmentier, C.; Monier, R. gsp mutations in human thyroid tumours. Oncogene 1991, 6, 677–679. [Google Scholar] [PubMed]
- Spada, A.; Vallar, L. G-protein oncogenes in acromegaly. Horm. Res. 1992, 38, 90–93. [Google Scholar] [CrossRef] [PubMed]
- Hosoi, E.; Yokogoshi, Y.; Hosoi, E.; Horie, H.; Sano, T.; Yamada, S.; Saito, S. Analysis of the Gs alpha gene in growth hormone-secreting pituitary adenomas by the polymerase chain reaction-direct sequencing method using paraffin-embedded tissues. Acta Endocrinol. 1993, 129, 301–306. [Google Scholar] [CrossRef]
- Parma, J.; Duprez, L.; Van Sande, J.; Cochaux, P.; Gervy, C.; Mockel, J.; Dumont, J.; Vassart, G. Somatic mutations in the thyrotropin receptor gene cause hyperfunctioning thyroid adenomas. Nature 1993, 365, 649–651. [Google Scholar] [CrossRef]
- Tordjman, K.; Stern, N.; Ouaknine, G.; Yossiphov, Y.; Razon, N.; Nordenskjold, M.; Friedman, E. Activating mutations of the Gs alpha-gene in nonfunctioning pituitary tumors. J. Clin. Endocrinol. Metab. 1993, 77, 765–769. [Google Scholar] [CrossRef]
- Yoshimoto, K.; Iwahana, H.; Fukuda, A.; Sano, T.; Itakura, M. Rare mutations of the Gs alpha subunit gene in human endocrine tumors. Mutation detection by polymerase chain reaction-primer-introduced restriction analysis. Cancer 1993, 72, 1386–1393. [Google Scholar] [CrossRef]
- Michiels, F.M.; Caillou, B.; Talbot, M.; Dessarps-Freichey, F.; Maunoury, M.T.; Schlumberger, M.; Mercken, L.; Monier, R.; Feunteun, J. Oncogenic potential of guanine nucleotide stimulatory factor alpha subunit in thyroid glands of transgenic mice. Proc. Natl. Acad. Sci. USA 1994, 91, 10488–10492. [Google Scholar] [CrossRef]
- Williamson, E.A.; Ince, P.G.; Harrison, D.; Kendall-Taylor, P.; Harris, P.E. G-protein mutations in human pituitary adrenocorticotrophic hormone-secreting adenomas. Eur. J. Clin. Invest 1995, 25, 128–131. [Google Scholar] [CrossRef]
- Williamson, E.A.; Johnson, S.J.; Foster, S.; Kendall-Taylor, P.; Harris, P.E. G protein gene mutations in patients with multiple endocrinopathies. J. Clin. Endocrinol. Metab. 1995, 80, 1702–1705. [Google Scholar] [CrossRef] [PubMed]
- Yang, I.M.; Woo, J.T.; Kim, S.W.; Kim, J.W.; Kim, Y.S.; Choi, Y.K. Characteristics of acromegalic patients with a good response to octreotide, a somatostatin analogue. Clin. Endocrinol. 1995, 42, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Fragoso, M.C.B.V.; Latronico, C.; Carvalho, F.M.; Zerbini, M.C.N.; Marcondes, J.A.M.; Araujo, L.M.B.; Lando, V.S.; Frazzatto, E.T.; Mendonca, B.B.; Villares, S.M.F. Activating mutation of the stimulatory G protein (gsp) as a putative cause of ovarian and testicular human stromal Leydig cell tumors. J. Clin. Endocr. Metab. 1998, 83, 2074–2078. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Tang, D.; Deng, J.; Su, C. Detection of gsp oncogene in growth hormone-secreting pituitary adenomas and the study of clinical characteristics of acromegalic patients with gsp-positive pituitary tumors. Chin. Med. J. 1998, 111, 891–894. [Google Scholar] [PubMed]
- Buchfelder, M.; Fahlbusch, R.; Merz, T.; Symowski, H.; Adams, E.F. Clinical correlates in acromegalic patients with pituitary tumors expressing GSP oncogenes. Pituitary 1999, 1, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Persani, L.; Lania, A.; Alberti, L.; Romoli, R.; Mantovani, G.; Filetti, S.; Spada, A.; Conti, M. Induction of specific phosphodiesterase isoforms by constitutive activation of the cAMP pathway in autonomous thyroid adenomas. J. Clin. Endocrinol. Metab. 2000, 85, 2872–2878. [Google Scholar] [CrossRef] [PubMed]
- Krohn, K.; Paschke, R. Somatic mutations in thyroid nodular disease. Mol. Genet. Metab. 2002, 75, 202–208. [Google Scholar] [CrossRef]
- Mendoza, V.; Sosa, E.; Espinosa-de-los-Monteros, A.L.; Salcedo, M.; Guinto, G.; Cheng, S.; Sandoval, C.; Mercado, M. GSPalpha mutations in Mexican patients with acromegaly: Potential impact on long term prognosis. Growth Horm. IGF. Res. 2005, 15, 28–32. [Google Scholar] [CrossRef]
- Freda, P.U.; Chung, W.K.; Matsuoka, N.; Walsh, J.E.; Kanibir, M.N.; Kleinman, G.; Wang, Y.; Bruce, J.N.; Post, K.D. Analysis of GNAS mutations in 60 growth hormone secreting pituitary tumors: Correlation with clinical and pathological characteristics and surgical outcome based on highly sensitive GH and IGF-I criteria for remission. Pituitary 2007, 10, 275–282. [Google Scholar] [CrossRef]
- Taboada, G.F.; Tabet, A.L.; Naves, L.A.; de Carvalho, D.P.; Gadelha, M.R. Prevalence of gsp oncogene in somatotropinomas and clinically non-functioning pituitary adenomas: Our experience. Pituitary 2009, 12, 165–169. [Google Scholar] [CrossRef]
- Lu, J.Y.; Hung, P.J.; Chen, P.L.; Yen, R.F.; Kuo, K.T.; Yang, T.L.; Wang, C.Y.; Chang, T.C.; Huang, T.S.; Chang, C.C. Follicular thyroid carcinoma with NRAS Q61K and GNAS R201H mutations that had a good (131)I treatment response. Endocrinol. Diabetes Metab. Case Rep. 2016, 2016, 150067. [Google Scholar] [CrossRef] [PubMed]
- Ronchi, C.L.; Peverelli, E.; Herterich, S.; Weigand, I.; Mantovani, G.; Schwarzmayr, T.; Sbiera, S.; Allolio, B.; Honegger, J.; Appenzeller, S.; et al. Landscape of somatic mutations in sporadic GH-secreting pituitary adenomas. Eur. J. Endocrinol. 2016, 174, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Ritvonen, E.; Pitkanen, E.; Karppinen, A.; Vehkavaara, S.; Demir, H.; Paetau, A.; Schalin-Jantti, C.; Karhu, A. Impact of AIP and inhibitory G protein alpha 2 proteins on clinical features of sporadic GH-secreting pituitary adenomas. Eur. J. Endocrinol. 2017, 176, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Puig-Domingo, M.; Gil, J.; Sampedro-Nunez, M.; Jorda, M.; Webb, S.M.; Serra, G.; Pons, L.; Salinas, I.; Blanco, A.; Marques-Pamies, M.; et al. Molecular profiling for acromegaly treatment: A validation study. Endocr. Relat. Cancer 2020, 27, 375–389. [Google Scholar] [CrossRef] [PubMed]
- Tirosh, A.; Jin, D.X.; De Marco, L.; Laitman, Y.; Friedman, E. Activating genomic alterations in the Gs alpha gene (GNAS) in 274 694 tumors. Genes Chromosomes Cancer 2020, 59, 503–516. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.; Kim, K.; Kim, D.; Moon, J.H.; Kim, E.H.; Kim, S.H.; Ku, C.R.; Lee, E.J. Associations of GNAS mutations with surgical outcomes in patients with growth hormone-secreting pituitary adenoma. Endocrinol. Metab. 2021, 36, 342–350. [Google Scholar] [CrossRef] [PubMed]
- Romanet, P.; Galluso, J.; Kamenicky, P.; Hage, M.; Theodoropoulou, M.; Roche, C.; Graillon, T.; Etchevers, H.C.; De Murat, D.; Mougel, G.; et al. Somatotroph tumors and the epigenetic status of the GNAS locus. Int. J. Mol. Sci. 2021, 22, 7570. [Google Scholar] [CrossRef] [PubMed]
- Yamato, A.; Nagano, H.; Gao, Y.; Matsuda, T.; Hashimoto, N.; Nakayama, A.; Yamagata, K.; Yokoyama, M.; Gong, Y.; Shi, X.; et al. Proteogenomic landscape and clinical characterization of GH-producing pituitary adenomas/somatotroph pituitary neuroendocrine tumors. Commun. Biol. 2022, 5, 1304. [Google Scholar] [CrossRef]
- O’Hayre, M.; Vazquez-Prado, J.; Kufareva, I.; Stawiski, E.W.; Handel, T.M.; Seshagiri, S.; Gutkind, J.S. The emerging mutational landscape of G proteins and G-protein-coupled receptors in cancer. Nat. Rev. Cancer 2013, 13, 412–424. [Google Scholar] [CrossRef]
- Turan, S.; Bastepe, M. GNAS Spectrum of Disorders. Curr. Osteoporos. Rep. 2015, 13, 146–158. [Google Scholar] [CrossRef]
- Ritterhouse, L.L.; Vivero, M.; Mino-Kenudson, M.; Sholl, L.M.; Iafrate, A.J.; Nardi, V.; Dong, F. GNAS mutations in primary mucinous and non-mucinous lung adenocarcinomas. Mod. Pathol. 2017, 30, 1720–1727. [Google Scholar] [CrossRef] [PubMed]
- Faias, S.; Duarte, M.; Pereira, L.; Chaves, P.; Cravo, M.; Dias Pereira, A.; Albuquerque, C. Methylation changes at the GNAS imprinted locus in pancreatic cystic neoplasms are important for the diagnosis of malignant cysts. World J. Gastrointest. Oncol. 2020, 12, 1056–1064. [Google Scholar] [CrossRef] [PubMed]
- McCune, D.J. Osteitis fibrosa cystica: The case of a nine year old girl who also exhibits precocious puberty, multiple pigmentation of the skin and hyperthyroidism. Am. J. Dis. Child 1936, 52, 743–744. [Google Scholar]
- Weinstein, L.S.; Shenker, A.; Gejman, P.V.; Merino, M.J.; Friedman, E.; Spiegel, A.M. Activating mutations of the stimulatory G protein in the McCune-Albright syndrome. N. Engl. J. Med. 1991, 325, 1688–1695. [Google Scholar] [CrossRef] [PubMed]
- Boyce, A.M.; Florenzano, P.; de Castro, L.F.; Collins, M.T. Fibrous dysplasia/McCune-Albright syndrome. In GeneReviews®; Adam, M.P., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 2019. [Google Scholar]
- Lumbroso, S.; Paris, F.; Sultan, C. Activating Gsalpha mutations: Analysis of 113 patients with signs of McCune-Albright syndrome—A European Collaborative Study. J. Clin. Endocrinol. Metab. 2004, 89, 2107–2113. [Google Scholar] [CrossRef]
- Idowu, B.D.; Al-Adnani, M.; O’Donnell, P.; Yu, L.; Odell, E.; Diss, T.; Gale, R.E.; Flanagan, A.M. A sensitive mutation-specific screening technique for GNAS1 mutations in cases of fibrous dysplasia: The first report of a codon 227 mutation in bone. Histopathology 2007, 50, 691–704. [Google Scholar] [CrossRef] [PubMed]
- Spencer, T.; Pan, K.S.; Collins, M.T.; Boyce, A.M. The clinical spectrum of McCune-Albright syndrome and its management. Horm. Res. Paediatr. 2019, 92, 347–356. [Google Scholar] [CrossRef]
- Boyce, A.M.; Collins, M.T. Fibrous dysplasia/McCune-Albright syndrome: A rare, mosaic disease of Galpha s activation. Endocr. Rev. 2020, 41, 345–370. [Google Scholar] [CrossRef]
- Spada, A.; Vallar, L.; Faglia, G. G-proteins and hormonal signalling in human pituitary tumors: Genetic mutations and functional alterations. Front. Neuroendocrinol. 1993, 14, 214–232. [Google Scholar] [CrossRef]
- Weinstein, L.S. The stimulatory G protein alpha-subunit gene: Mutations and imprinting lead to complex phenotypes. J. Clin. Endocrinol. Metab. 2001, 86, 4622–4626. [Google Scholar] [CrossRef]
- Adams, E.F.; Lei, T.; Buchfelder, M.; Petersen, B.; Fahlbusch, R. Biochemical characteristics of human pituitary somatotropinomas with and without gsp mutations: In vitro cell culture studies. J. Clin. Endocrinol. Metab. 1995, 80, 2077–2081. [Google Scholar] [CrossRef] [PubMed]
- Rymuza, J.; Kober, P.; Rusetska, N.; Mossakowska, B.J.; Maksymowicz, M.; Nyc, A.; Baluszek, S.; Zielinski, G.; Kunicki, J.; Bujko, M. Transcriptomic classification of pituitary neuroendocrine tumors causing acromegaly. Cells 2022, 11, 3846. [Google Scholar] [CrossRef] [PubMed]
- Spada, A.; Arosio, M.; Bochicchio, D.; Bazzoni, N.; Vallar, L.; Bassetti, M.; Faglia, G. Clinical, biochemical, and morphological correlates in patients bearing growth hormone-secreting pituitary-tumors with or without constitutively active adenylyl cyclase. J. Clin. Endocr. Metab. 1990, 71, 1421–1426. [Google Scholar] [CrossRef] [PubMed]
- Larkin, S.; Reddy, R.; Karavitaki, N.; Cudlip, S.; Wass, J.; Ansorge, O. Granulation pattern, but not GSP or GHR mutation, is associated with clinical characteristics in somatostatin-naive patients with somatotroph adenomas. Eur. J. Endocrinol. 2013, 168, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Lasolle, H.; Elsensohn, M.H.; Wierinckx, A.; Alix, E.; Bonnefille, C.; Vasiljevic, A.; Cortet, C.; Decoudier, B.; Sturm, N.; Gaillard, S.; et al. Chromosomal instability in the prediction of pituitary neuroendocrine tumors prognosis. Acta Neuropathol. Commun. 2020, 8, 190. [Google Scholar] [CrossRef] [PubMed]
- Neou, M.; Villa, C.; Armignacco, R.; Jouinot, A.; Raffin-Sanson, M.L.; Septier, A.; Letourneur, F.; Diry, S.; Diedisheim, M.; Izac, B.; et al. Pangenomic classification of pituitary neuroendocrine tumors. Cancer Cell 2020, 37, 123–134.e125. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, G.; Bondioni, S.; Lania, A.G.; Corbetta, S.; de Sanctis, L.; Cappa, M.; Di Battista, E.; Chanson, P.; Beck-Peccoz, P.; Spada, A. Parental origin of Gsalpha mutations in the McCune-Albright syndrome and in isolated endocrine tumors. J. Clin. Endocrinol. Metab. 2004, 89, 3007–3009. [Google Scholar] [CrossRef]
- Hayward, B.E.; Barlier, A.; Korbonits, M.; Grossman, A.B.; Jacquet, P.; Enjalbert, A.; Bonthron, D.T. Imprinting of the G(s)alpha gene GNAS1 in the pathogenesis of acromegaly. J. Clin. Investig. 2001, 107, R31–R36. [Google Scholar] [CrossRef]
- Picard, C.; Silvy, M.; Gerard, C.; Buffat, C.; Lavaque, E.; Figarella-Branger, D.; Dufour, H.; Gabert, J.; Beckers, A.; Brue, T.; et al. Gs alpha overexpression and loss of Gs alpha imprinting in human somatotroph adenomas: Association with tumor size and response to pharmacologic treatment. Int. J. Cancer 2007, 121, 1245–1252. [Google Scholar] [CrossRef]
- Altarejos, J.Y.; Montminy, M. CREB and the CRTC co-activators: Sensors for hormonal and metabolic signals. Nat. Rev. Mol. Cell Biol. 2011, 12, 141–151. [Google Scholar] [CrossRef]
- Schultz, K.A.P.; Stewart, D.R.; Kamihara, J.; Bauer, A.J.; Merideth, M.A.; Stratton, P.; Huryn, L.A.; Harris, A.K.; Doros, L.; Field, A.; et al. DICER1 tumor predisposition. In GeneReviews®; Adam, M.P., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 2020. [Google Scholar]
- Hill, D.A.; Ivanovich, J.; Priest, J.R.; Gurnett, C.A.; Dehner, L.P.; Desruisseau, D.; Jarzembowski, J.A.; Wikenheiser-Brokamp, K.A.; Suarez, B.K.; Whelan, A.J.; et al. DICER1 mutations in familial pleuropulmonary blastoma. Science 2009, 325, 965. [Google Scholar] [CrossRef] [PubMed]
- Slade, I.; Bacchelli, C.; Davies, H.; Murray, A.; Abbaszadeh, F.; Hanks, S.; Barfoot, R.; Burke, A.; Chisholm, J.; Hewitt, M.; et al. DICER1 syndrome: Clarifying the diagnosis, clinical features and management implications of a pleiotropic tumour predisposition syndrome. J. Med. Genet. 2011, 48, 273–278. [Google Scholar] [CrossRef] [PubMed]
- de Kock, L.; Wang, Y.C.; Revil, T.; Badescu, D.; Rivera, B.; Sabbaghian, N.; Wu, M.; Weber, E.; Sandoval, C.; Hopman, S.M.; et al. High-sensitivity sequencing reveals multi-organ somatic mosaicism causing DICER1 syndrome. J. Med. Genet. 2016, 53, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Schultz, K.A.P.; Williams, G.M.; Kamihara, J.; Stewart, D.R.; Harris, A.K.; Bauer, A.J.; Turner, J.; Shah, R.; Schneider, K.; Schneider, K.W.; et al. DICER1 and associated conditions: Identification of at-risk individuals and recommended surveillance strategies. Clin. Cancer Res. 2018, 24, 2251–2261. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.; Lee, H.; Ghahremani, S.; Kempert, P.; Ischander, M.; Teitell, M.A.; Nelson, S.F.; Martinez-Agosto, J.A. Expanding the phenotype of mutations in DICER1: Mosaic missense mutations in the RNase IIIb domain of DICER1 cause GLOW syndrome. J. Med. Genet. 2014, 51, 294–302. [Google Scholar] [CrossRef] [PubMed]
- Brenneman, M.; Field, A.; Yang, J.; Williams, G.; Doros, L.; Rossi, C.; Schultz, K.A.; Rosenberg, A.; Ivanovich, J.; Turner, J.; et al. Temporal order of RNase IIIb and loss-of-function mutations during development determines phenotype in pleuropulmonary blastoma/DICER1 syndrome: A unique variant of the two-hit tumor suppression model. F1000Res 2015, 4, 214. [Google Scholar] [CrossRef] [PubMed]
- MacRae, I.J.; Zhou, K.; Li, F.; Repic, A.; Brooks, A.N.; Cande, W.Z.; Adams, P.D.; Doudna, J.A. Structural basis for double-stranded RNA processing by Dicer. Science 2006, 311, 195–198. [Google Scholar] [CrossRef]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef]
- Foulkes, W.D.; Priest, J.R.; Duchaine, T.F. DICER1: Mutations, microRNAs and mechanisms. Nat. Rev. Cancer 2014, 14, 662–672. [Google Scholar] [CrossRef]
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744. [Google Scholar] [CrossRef]
- Svoboda, P. Key mechanistic principles and considerations concerning RNA interference. Front. Plant Sci. 2020, 11, 1237. [Google Scholar] [CrossRef] [PubMed]
- de Boer, C.M.; Eini, R.; Gillis, A.M.; Stoop, H.; Looijenga, L.H.; White, S.J. DICER1 RNase IIIb domain mutations are infrequent in testicular germ cell tumours. BMC. Res. Notes 2012, 5, 569. [Google Scholar] [CrossRef] [PubMed]
- Heravi-Moussavi, A.; Anglesio, M.S.; Cheng, S.W.; Senz, J.; Yang, W.; Prentice, L.; Fejes, A.P.; Chow, C.; Tone, A.; Kalloger, S.E.; et al. Recurrent somatic DICER1 mutations in nonepithelial ovarian cancers. N. Engl. J. Med. 2012, 366, 234–242. [Google Scholar] [CrossRef] [PubMed]
- de Kock, L.; Plourde, F.; Carter, M.T.; Hamel, N.; Srivastava, A.; Meyn, M.S.; Arseneau, J.; Bouron-Dal, S.D.; Foulkes, W.D. Germ-line and somatic DICER1 mutations in a pleuropulmonary blastoma. Pediatr. Blood Cancer 2013, 60, 2091–2092. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.K.; Sabbaghian, N.; Xu, B.; Addidou-Kalucki, S.; Bernard, C.; Zou, D.; Reeve, A.E.; Eccles, M.R.; Cole, C.; Choong, C.S.; et al. Biallelic DICER1 mutations occur in Wilms tumours. J. Pathol. 2013, 230, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Tomiak, E.; de, K.L.; Grynspan, D.; Ramphal, R.; Foulkes, W.D. DICER1 mutations in an adolescent with cervical embryonal rhabdomyosarcoma (cERMS). Pediatr. Blood Cancer 2014, 61, 568–569. [Google Scholar] [CrossRef] [PubMed]
- de Kock, L.; Sabbaghian, N.; Soglio, D.B.D.; Guillerman, R.P.; Park, B.K.; Chami, R.; Deal, C.L.; Priest, J.R.; Foulkes, W.D. Exploring the association between DICER1 mutations and differentiated thyroid carcinoma. J. Clin. Endocr. Metab. 2014, 99, E1072–E1077. [Google Scholar] [CrossRef]
- de Kock, L.; Sabbaghian, N.; Druker, H.; Weber, E.; Hamel, N.; Miller, S.; Choong, C.S.; Gottardo, N.G.; Kees, U.R.; Rednam, S.P.; et al. Germ-line and somatic DICER1 mutations in pineoblastoma. Acta. Neuropathol. 2014, 128, 583–595. [Google Scholar] [CrossRef]
- de Kock, L.; Sabbaghian, N.; Plourde, F.; Srivastava, A.; Weber, E.; Bouron-Dal Soglio, D.; Hamel, N.; Choi, J.H.; Park, S.H.; Deal, C.L.; et al. Pituitary blastoma: A pathognomonic feature of germ-line DICER1 mutations. Acta. Neuropathol. 2014, 128, 111–122. [Google Scholar] [CrossRef]
- Doros, L.A.; Rossi, C.T.; Yang, J.; Field, A.; Williams, G.M.; Messinger, Y.; Cajaiba, M.M.; Perlman, E.J.; Schultz, A.; Cathro, H.P.; et al. DICER1 mutations in childhood cystic nephroma and its relationship to DICER1-renal sarcoma. Mod. Pathol. 2014, 27, 1267–1280. [Google Scholar] [CrossRef]
- Murray, M.J.; Bailey, S.; Raby, K.L.; Saini, H.K.; de, K.L.; Burke, G.A.; Foulkes, W.D.; Enright, A.J.; Coleman, N.; Tischkowitz, M. Serum levels of mature microRNAs in DICER1-mutated pleuropulmonary blastoma. Oncogenesis 2014, 3, e87. [Google Scholar] [CrossRef] [PubMed]
- Pugh, T.J.; Yu, W.; Yang, J.; Field, A.L.; Ambrogio, L.; Carter, S.L.; Cibulskis, K.; Giannikopoulos, P.; Kiezun, A.; Kim, J.; et al. Exome sequencing of pleuropulmonary blastoma reveals frequent biallelic loss of TP53 and two hits in DICER1 resulting in retention of 5p-derived miRNA hairpin loop sequences. Oncogene 2014, 33, 5295–5302. [Google Scholar] [CrossRef] [PubMed]
- Anglesio, M.S.; Wang, Y.; Yang, W.; Senz, J.; Wan, A.; Heravi-Moussavi, A.; Salamanca, C.; Maines-Bandiera, S.; Huntsman, D.G.; Morin, G.B. Cancer-associated somatic DICER1 hotspot mutations cause defective miRNA processing and reverse-strand expression bias to predominantly mature 3p strands through loss of 5p strand cleavage. J. Pathol. 2013, 229, 400–409. [Google Scholar] [CrossRef]
- Chong, A.S.; Han, H.; Albrecht, S.; Weon, Y.C.; Park, S.K.; Foulkes, W.D. DICER1 syndrome in a young adult with pituitary blastoma. Acta. Neuropathol. 2021, 142, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.P.Y.; Kelsey, M.M.; Sabbaghian, N.; Park, S.H.; Deal, C.L.; Esbenshade, A.J.; Ploner, O.; Peet, A.; Traunecker, H.; Ahmed, Y.H.E.; et al. Clinical outcomes and complications of pituitary blastoma. J. Clin. Endocrinol. Metab. 2021, 106, 351–363. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.P.; Li, K.K.; Chow, C.; Chan, S.; Leung, A.W.; Shing, M.M.; To, K.F.; Chan, D.T.; Chan, G.C.; Ng, H.K. Expanding the clinical and molecular spectrum of pituitary blastoma. Acta. Neuropathol. 2022, 143, 415–417. [Google Scholar] [CrossRef] [PubMed]
- Scheithauer, B.W.; Kovacs, K.; Horvath, E.; Kim, D.S.; Osamura, R.Y.; Ketterling, R.P.; Lloyd, R.V.; Kim, O.L. Pituitary blastoma. Acta. Neuropathol. 2008, 116, 657–666. [Google Scholar] [CrossRef]
- Nadaf, J.; de Kock, L.; Chong, A.S.; Korbonits, M.; Thorner, P.; Benlimame, N.; Fu, L.; Peet, A.; Warner, J.; Ploner, O.; et al. Molecular characterization of DICER1-mutated pituitary blastoma. Acta. Neuropathol. 2021, 141, 929–944. [Google Scholar] [CrossRef]
- Thorner, P.S.; Chong, A.S.; Nadaf, J.; Benlimame, N.; Marrano, P.; Chami, R.; Fu, L.; Foulkes, W.D. PRAME protein expression in DICER1-related tumours. J. Pathol. Clin. Res. 2022, 8, 579–581. [Google Scholar] [CrossRef]
- Xu, Y.; Zou, R.; Wang, J.; Wang, Z.W.; Zhu, X. The role of the cancer testis antigen PRAME in tumorigenesis and immunotherapy in human cancer. Cell Prolif. 2020, 53, e12770. [Google Scholar] [CrossRef]
- Asada, K.; Canestrari, E.; Fu, X.; Li, Z.; Makowski, E.; Wu, Y.C.; Mito, J.K.; Kirsch, D.G.; Baraban, J.; Paroo, Z. Rescuing dicer defects via inhibition of an anti-dicing nuclease. Cell Rep. 2014, 9, 1471–1481. [Google Scholar] [CrossRef] [PubMed]
- Asada, K.; Canestrari, E.; Paroo, Z. A druggable target for rescuing microRNA defects. Bioorg Med. Chem. Lett. 2016, 26, 4942–4946. [Google Scholar] [CrossRef] [PubMed]
- Muller, M.; Fazi, F.; Ciaudo, C. Argonaute proteins: From structure to function in development and pathological cell fate determination. Front Cell Dev. Biol. 2019, 7, 360. [Google Scholar] [CrossRef] [PubMed]
- Thunders, M.; Delahunt, B. Gene of the month: DICER1: Ruler and controller. J. Clin. Pathol. 2021, 74, 69–72. [Google Scholar] [CrossRef]
- Meiklejohn, K.M.; Darbinyan, A.; Barbieri, A.L. A review of DICER1: Structure, function and contribution to disease. Diagnostic Histopathology 2022, 28, 329–336. [Google Scholar] [CrossRef]
- Cretu, C.; Schmitzova, J.; Ponce-Salvatierra, A.; Dybkov, O.; De Laurentiis, E.I.; Sharma, K.; Will, C.L.; Urlaub, H.; Luhrmann, R.; Pena, V. Molecular architecture of SF3b and structural consequences of its cancer-related mutations. Mol. Cell 2016, 64, 307–319. [Google Scholar] [CrossRef]
- Alsafadi, S.; Houy, A.; Battistella, A.; Popova, T.; Wassef, M.; Henry, E.; Tirode, F.; Constantinou, A.; Piperno-Neumann, S.; Roman-Roman, S.; et al. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat. Commun. 2016, 7, 10615. [Google Scholar] [CrossRef]
- Darman, R.B.; Seiler, M.; Agrawal, A.A.; Lim, K.H.; Peng, S.; Aird, D.; Bailey, S.L.; Bhavsar, E.B.; Chan, B.; Colla, S.; et al. Cancer-associated SF3B1 hotspot mutations induce cryptic 3’ splice site selection through use of a different branch point. Cell Rep. 2015, 13, 1033–1045. [Google Scholar] [CrossRef]
- DeBoever, C.; Ghia, E.M.; Shepard, P.J.; Rassenti, L.; Barrett, C.L.; Jepsen, K.; Jamieson, C.H.; Carson, D.; Kipps, T.J.; Frazer, K.A. Transcriptome sequencing reveals potential mechanism of cryptic 3’ splice site selection in SF3B1-mutated cancers. PLoS Comput. Biol. 2015, 11, e1004105. [Google Scholar] [CrossRef]
- Will, C.L.; Lührmann, R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011, 3, a003707. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Gong, Q.; Wang, Y.; Li, M.; Wang, L.; Ding, H.; Li, P. The biological function and clinical significance of SF3B1 mutations in cancer. Biomark Res. 2020, 8, 38. [Google Scholar] [CrossRef]
- Li, C.; Xie, W.; Rosenblum, J.S.; Zhou, J.; Guo, J.; Miao, Y.; Shen, Y.; Wang, H.; Gong, L.; Li, M.; et al. Somatic SF3B1 hotspot mutation in prolactinomas. Nat. Commun. 2020, 11, 2506. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.; Perez-Rivas, L.G.; Zhao, Y.; Chasseloup, F.; Lasolle, H.; Cortet, C.; Descotes, F.; Villa, C.; Baussart, B.; Burman, P.; et al. Prevalence and clinical correlations of SF3B1 variants in lactotroph tumours. Eur. J. Endocrinol. 2023, 189, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Cass, D.M.; Berglund, J.A. The SF3b155 N-terminal domain is a scaffold important for splicing. Biochemistry 2006, 45, 10092–10101. [Google Scholar] [CrossRef] [PubMed]
- Papaemmanuil, E.; Cazzola, M.; Boultwood, J.; Malcovati, L.; Vyas, P.; Bowen, D.; Pellagatti, A.; Wainscoat, J.S.; Hellstrom-Lindberg, E.; Gambacorti-Passerini, C.; et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med. 2011, 365, 1384–1395. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lawrence, M.S.; Wan, Y.; Stojanov, P.; Sougnez, C.; Stevenson, K.; Werner, L.; Sivachenko, A.; DeLuca, D.S.; Zhang, L.; et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N. Engl. J. Med. 2011, 365, 2497–2506. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Sanada, M.; Shiraishi, Y.; Nowak, D.; Nagata, Y.; Yamamoto, R.; Sato, Y.; Sato-Otsubo, A.; Kon, A.; Nagasaki, M.; et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011, 478, 64–69. [Google Scholar] [CrossRef]
- Biankin, A.V.; Waddell, N.; Kassahn, K.S.; Gingras, M.C.; Muthuswamy, L.B.; Johns, A.L.; Miller, D.K.; Wilson, P.J.; Patch, A.M.; Wu, J.; et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012, 491, 399–405. [Google Scholar] [CrossRef]
- Stephens, P.J.; Tarpey, P.S.; Davies, H.; Van Loo, P.; Greenman, C.; Wedge, D.C.; Nik-Zainal, S.; Martin, S.; Varela, I.; Bignell, G.R.; et al. The landscape of cancer genes and mutational processes in breast cancer. Nature 2012, 486, 400–404. [Google Scholar] [CrossRef]
- Harbour, J.W.; Roberson, E.D.; Anbunathan, H.; Onken, M.D.; Worley, L.A.; Bowcock, A.M. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat. Genet. 2013, 45, 133–135. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Krauthammer, M.; Halaban, R. Rare SF3B1 R625 mutations in cutaneous melanoma. Melanoma Res. 2014, 24, 332–334. [Google Scholar] [CrossRef]
- Yang, H.M.; Hsiao, S.J.; Schaeffer, D.F.; Lai, C.; Remotti, H.E.; Horst, D.; Mansukhani, M.M.; Horst, B.A. Identification of recurrent mutational events in anorectal melanoma. Mod. Pathol. 2017, 30, 286–296. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Li, C.; Fang, Q.; Liu, Y.; Wang, D.; Chen, Y.; Xie, W.; Zhang, Y. The SF3B1(R625H) mutation promotes prolactinoma tumor progression through aberrant splicing of DLG1. J. Exp. Clin. Cancer Res. 2022, 41, 26. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Borrego, M.C.; Fuentes-Fayos, A.C.; Venegas-Moreno, E.; Rivero-Cortes, E.; Dios, E.; Moreno-Moreno, P.; Madrazo-Atutxa, A.; Remon, P.; Solivera, J.; Wildemberg, L.E.; et al. Splicing machinery is dysregulated in pituitary neuroendocrine tumors and is associated with aggressiveness features. Cancers 2019, 11, 1439. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, X.; Zhang, H.; Liu, F.; Bi, Y.; Zhang, Y.; Cheng, C.; Liu, J. Knockdown of SF3B1 inhibits cell proliferation, invasion and migration triggering apoptosis in breast cancer via aberrant splicing. Breast Cancer 2020, 27, 464–476. [Google Scholar] [CrossRef] [PubMed]
- Effenberger, K.A.; Urabe, V.K.; Jurica, M.S. Modulating splicing with small molecular inhibitors of the spliceosome. Wiley Interdiscip. Rev. RNA 2017, 8, e1381. [Google Scholar] [CrossRef]
- Zhang, D.; Meng, F. A comprehensive overview of structure-activity relationships of small-molecule splicing modulators targeting SF3B1 as anticancer agents. ChemMedChem 2020, 15, 2098–2120. [Google Scholar] [CrossRef]
- Steensma, D.P.; Wermke, M.; Klimek, V.M.; Greenberg, P.L.; Font, P.; Komrokji, R.S.; Yang, J.; Brunner, A.M.; Carraway, H.E.; Ades, L.; et al. Phase I first-in-human dose escalation study of the oral SF3B1 modulator H3B-8800 in myeloid neoplasms. Leukemia 2021, 35, 3542–3550. [Google Scholar] [CrossRef]
- Pérez-Rivas, L.G.; Theodoropoulou, M.; Ferrau, F.; Nusser, C.; Kawaguchi, K.; Stratakis, C.A.; Faucz, F.R.; Wildemberg, L.E.; Assie, G.; Beschorner, R.; et al. The gene of the ubiquitin-specific protease 8 is frequently mutated in adenomas causing Cushing’s disease. J. Clin. Endocrinol. Metab. 2015, 100, E997–E1004. [Google Scholar] [CrossRef]
- Hayashi, K.; Inoshita, N.; Kawaguchi, K.; Ibrahim Ardisasmita, A.; Suzuki, H.; Fukuhara, N.; Okada, M.; Nishioka, H.; Takeuchi, Y.; Komada, M.; et al. The USP8 mutational status may predict drug susceptibility in corticotroph adenomas of Cushing’s disease. Eur. J. Endocrinol. 2016, 174, 213–226. [Google Scholar] [CrossRef] [PubMed]
- de Araujo, L.J.; Lerario, A.M.; de Castro, M.; Martins, C.S.; Bronstein, M.D.; Machado, M.C.; Trarbach, E.B.; Villares Fragoso, M.C. Transcriptome Analysis Showed a Differential Signature between Invasive and Non-invasive Corticotrophinomas. Front. Endocrinol. 2017, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Faucz, F.R.; Tirosh, A.; Tatsi, C.; Berthon, A.; Hernández-Ramírez, L.C.; Settas, N.; Angelousi, A.; Correa, R.; Papadakis, G.Z.; Chittiboina, P.; et al. Somatic USP8 gene mutations are a common cause of pediatric Cushing disease. J. Clin. Endocrinol. Metab. 2017, 102, 2836–2843. [Google Scholar] [CrossRef] [PubMed]
- Albani, A.; Pérez-Rivas, L.G.; Dimopoulou, C.; Zopp, S.; Colon-Bolea, P.; Roeber, S.; Honegger, J.; Flitsch, J.; Rachinger, W.; Buchfelder, M.; et al. The USP8 mutational status may predict long-term remission in patients with Cushing’s disease. Clin. Endocrinol. 2018, 89, 454–458. [Google Scholar] [CrossRef]
- Ballmann, C.; Thiel, A.; Korah, H.E.; Reis, A.C.; Saeger, W.; Stepanow, S.; Kohrer, K.; Reifenberger, G.; Knobbe-Thomsen, C.B.; Knappe, U.J.; et al. USP8 mutations in pituitary Cushing adenomas-targeted analysis by next-generation sequencing. J. Endocr. Soc. 2018, 2, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Bujko, M.; Kober, P.; Boresowicz, J.; Rusetska, N.; Paziewska, A.; Dabrowska, M.; Piascik, A.; Pekul, M.; Zielinski, G.; Kunicki, J.; et al. USP8 mutations in corticotroph adenomas determine a distinct gene expression profile irrespective of functional tumour status. Eur. J. Endocrinol. 2019, 181, 615–627. [Google Scholar] [CrossRef] [PubMed]
- Losa, M.; Mortini, P.; Pagnano, A.; Detomas, M.; Cassarino, M.F.; Pecori Giraldi, F. Clinical characteristics and surgical outcome in USP8-mutated human adrenocorticotropic hormone-secreting pituitary adenomas. Endocrine 2019, 63, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Weigand, I.; Knobloch, L.; Flitsch, J.; Saeger, W.; Monoranu, C.M.; Hofner, K.; Herterich, S.; Rotermund, R.; Ronchi, C.L.; Buchfelder, M.; et al. Impact of USP8 gene mutations on protein deregulation in Cushing disease. J. Clin. Endocrinol. Metab. 2019, 104, 2535–2546. [Google Scholar] [CrossRef]
- Castellnou, S.; Vasiljevic, A.; Lapras, V.; Raverot, V.; Alix, E.; Borson-Chazot, F.; Jouanneau, E.; Raverot, G.; Lasolle, H. SST5 expression and USP8 mutation in functioning and silent corticotroph pituitary tumors. Endocr. Connect. 2020, 9, 243–253. [Google Scholar] [CrossRef]
- Martins, C.S.; Camargo, R.C.; Coeli-Lacchini, F.B.; Saggioro, F.P.; Moreira, A.C.; de Castro, M. USP8 mutations and cell cycle regulation in corticotroph adenomas. Horm. Metab. Res. 2020, 52, 117–123. [Google Scholar] [CrossRef]
- Sesta, A.; Cassarino, M.F.; Terreni, M.; Ambrogio, A.G.; Libera, L.; Bardelli, D.; Lasio, G.; Losa, M.; Pecori Giraldi, F. Ubiquitin-specific protease 8 mutant corticotrope adenomas present unique secretory and molecular features and shed light on the role of ubiquitylation on ACTH processing. Neuroendocrinology 2020, 110, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Pasternak-Pietrzak, K.; Faucz, F.R.; Stratakis, C.A.; Moszczynska, E.; Roszkowski, M.; Grajkowska, W.; Pronicki, M.; Szalecki, M. Is there a common cause for paediatric Cushing’s disease? Endokrynol. Pol. 2021, 72, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Treppiedi, D.; Barbieri, A.M.; Di Muro, G.; Marra, G.; Mangili, F.; Catalano, R.; Esposito, E.; Ferrante, E.; Serban, A.L.; Locatelli, M.; et al. Genetic Profiling of a Cohort of Italian Patients with ACTH-Secreting Pituitary Tumors and Characterization of a Novel USP8 Gene Variant. Cancers 2021, 13, 4022. [Google Scholar] [CrossRef] [PubMed]
- Rebollar-Vega, R.G.; Zuarth-Vázquez, J.M.; Hernández-Ramírez, L.C. Clinical spectrum of USP8 pathogenic variants in Cushing’s disease. Arch. Med. Res. 2023, 102899. [Google Scholar] [CrossRef] [PubMed]
- Clague, M.J.; Urbe, S.; Komander, D. Breaking the chains: Deubiquitylating enzyme specificity begets function. Nat. Rev. Mol. Cell Biol. 2019, 20, 338–352. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, E.; Iura, T.; Mukai, A.; Yoshimori, T.; Kitamura, N.; Komada, M. Regulation of epidermal growth factor receptor down-regulation by UBPY-mediated deubiquitination at endosomes. Mol. Biol. Cell 2005, 16, 5163–5174. [Google Scholar] [CrossRef]
- Alwan, H.A.; van Leeuwen, J.E. UBPY-mediated epidermal growth factor receptor (EGFR) de-ubiquitination promotes EGFR degradation. J. Biol. Chem. 2007, 282, 1658–1669. [Google Scholar] [CrossRef]
- Berlin, I.; Schwartz, H.; Nash, P.D. Regulation of epidermal growth factor receptor ubiquitination and trafficking by the USP8.STAM complex. J. Biol. Chem. 2010, 285, 34909–34921. [Google Scholar] [CrossRef]
- Crespo-Yàñez, X.; Aguilar-Gurrieri, C.; Jacomin, A.C.; Journet, A.; Mortier, M.; Taillebourg, E.; Soleilhac, E.; Weissenhorn, W.; Fauvarque, M.O. CHMP1B is a target of USP8/UBPY regulated by ubiquitin during endocytosis. PLoS Genet. 2018, 14, e1007456. [Google Scholar] [CrossRef]
- Theodoropoulou, M.; Reincke, M.; Fassnacht, M.; Komada, M. Decoding the genetic basis of Cushing’s disease: USP8 in the spotlight. Eur. J. Endocrinol. 2015, 173, M73–M83. [Google Scholar] [CrossRef]
- Islam, M.T.; Chen, F.; Chen, H. The oncogenic role of ubiquitin specific peptidase (USP8) and its signaling pathways targeting for cancer therapeutics. Arch. Biochem. Biophys. 2021, 701, 108811. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Rivas, L.G.; Theodoropoulou, M.; Puar, T.H.; Fazel, J.; Stieg, M.R.; Ferrau, F.; Assie, G.; Gadelha, M.R.; Deutschbein, T.; Fragoso, M.C.; et al. Somatic USP8 mutations are frequent events in corticotroph tumor progression causing Nelson’s tumor. Eur. J. Endocrinol. 2018, 178, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Treppiedi, D.; Marra, G.; Di Muro, G.; Esposito, E.; Barbieri, A.M.; Catalano, R.; Mangili, F.; Bravi, F.; Locatelli, M.; Lania, A.G.; et al. P720R USP8 Mutation Is Associated with a Better Responsiveness to Pasireotide in ACTH-Secreting PitNETs. Cancers 2022, 14, 2455. [Google Scholar] [CrossRef]
- Jian, F.F.; Li, Y.F.; Chen, Y.F.; Jiang, H.; Chen, X.; Zheng, L.L.; Zhao, Y.; Wang, W.Q.; Ning, G.; Bian, L.G.; et al. Inhibition of Ubiquitin-specific Peptidase 8 Suppresses Adrenocorticotropic Hormone Production and Tumorous Corticotroph Cell Growth in AtT20 Cells. Chin. Med. J. 2016, 129, 2102–2108. [Google Scholar] [CrossRef] [PubMed]
- Shichi, H.; Fukuoka, H.; Kanzawa, M.; Yamamoto, M.; Yamamoto, N.; Suzuki, M.; Urai, S.; Matsumoto, R.; Kanie, K.; Fujita, Y.; et al. Responsiveness to DDAVP in Cushing’s disease is associated with USP8 mutations through enhancing AVPR1B promoter activity. Pituitary 2022, 25, 496–507. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.; Persky, R.; Stegemann, R.; Hernández-Ramírez, L.C.; Zeltser, D.; Lodish, M.B.; Chen, A.; Keil, M.F.; Tatsi, C.; Faucz, F.R.; et al. Germline USP8 mutation associated with pediatric Cushing disease and other clinical features: A new syndrome. J. Clin. Endocrinol. Metab. 2019, 104, 4676–4682. [Google Scholar] [CrossRef]
- Fukuoka, H.; Cooper, O.; Ben-Shlomo, A.; Mamelak, A.; Ren, S.G.; Bruyette, D.; Melmed, S. EGFR as a therapeutic target for human, canine, and mouse ACTH-secreting pituitary adenomas. J. Clin. Investig. 2011, 121, 4712–4721. [Google Scholar] [CrossRef]
- Kageyama, K.; Asari, Y.; Sugimoto, Y.; Niioka, K.; Daimon, M. Ubiquitin-specific protease 8 inhibitor suppresses adrenocorticotropic hormone production and corticotroph tumor cell proliferation. Endocr. J. 2020, 67, 177–184. [Google Scholar] [CrossRef]
- Treppiedi, D.; Di Muro, G.; Marra, G.; Barbieri, A.M.; Mangili, F.; Catalano, R.; Serban, A.; Ferrante, E.; Locatelli, M.; Lania, A.G.; et al. USP8 inhibitor RA-9 reduces ACTH release and cell growth in tumor corticotrophs. Endocr. Relat. Cancer 2021, 28, 573–582. [Google Scholar] [CrossRef]
- Clague, M.J.; Urbe, S. Ubiquitin: Same molecule, different degradation pathways. Cell 2010, 143, 682–685. [Google Scholar] [CrossRef]
- Mossakowska, B.J.; Rusetska, N.; Konopinski, R.; Kober, P.; Maksymowicz, M.; Pekul, M.; Zielinski, G.; Styk, A.; Kunicki, J.; Bujko, M. The expression of cell cycle-related genes in USP8-mutated corticotroph neuroendocrine pituitary tumors and their possible role in cell cycle-targeting treatment. Cancers 2022, 14, 5594. [Google Scholar] [CrossRef] [PubMed]
- Xia, R.; Jia, H.; Fan, J.; Liu, Y.; Jia, J. USP8 promotes smoothened signaling by preventing its ubiquitination and changing its subcellular localization. PLoS Biol. 2012, 10, e1001238. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Vallese, S.; Peretto, I.; Jacq, X.; Rain, J.C.; Colland, F.; Guedat, P. Synthesis and biological evaluation of 9-oxo-9H-indeno[1,2-b]pyrazine-2,3-dicarbonitrile analogues as potential inhibitors of deubiquitinating enzymes. ChemMedChem 2010, 5, 552–558. [Google Scholar] [CrossRef] [PubMed]
| Gene | Hotspot Defect | Effect on Protein | PitNET Subtypes Affected | Clinical Phenotype | Pharmacological Agents Targeting the Hotspot |
|---|---|---|---|---|---|
| BRAF | Somatic p.V600E | Destabilization of the inactive conformation of BRAF, promoting its activation | Corticotropinomas | Unclear (very infrequent genetic defect in PitNETs) | Vemurafenib, dabrafenib, and encorafenib, as monotherapy or combined with MEK inhibitors (in clinical use for other tumor types) |
| GNAS | Somatic missense variants affecting residues 201 or 227 | Stabilization of Gsα in its active conformation and inhibition of its GTPase activity | Somatotropinomas. Less frequently: NF-PitNETs and corticotropinomas | Older patients. Controversial: low GH or IGF1, small tumors with slow growth rate, densely granulated somatotropinomas | None reported |
| DICER1 | Germline and (or?) * somatic variants affecting residues 1705, 1709, 1809, 1810, or 1813 | Loss of RNase IIIb activity | PitBs | CD in infants, neonates, or, less frequently, in childhood and young adulthood. Rarely: silent corticotropinomas. Large, aggressive, and poorly differentiated tumors with an oncofetal signature. Presentation as isolated tumors or as part of DICER1 syndrome | Not reported. Indirect approach: pharmacological inhibition of the endonuclease complex TSN-TSNAX (not available for clinical use) |
| SF3B1 | Somatic p.R625H and p.R625C | Impaired interaction with the BP and with other U2 snRNP components | Prolactinomas | High serum prolactin, large tumors with increased Ki67 index, increased progression-free survival and mortality, and requirement for multiple treatments. Occasionally: metastatic PitNETs | H3B-8800 and pladienolide B (not available for clinical use) |
| USP8 | Somatic variants affecting residues 718–720 | Impaired interaction with 14-3-3, proteolytic cleavage, and enhanced deubiquitinase activity | Corticotropinomas | CD in young adults and teenagers, most frequently women, increased SST5 and MGMT, rarely Crooke’s cell adenomas, not associated with Nelson’s syndrome. Controversial: effects on tumor size, hormone secretion, response to dynamic tests, and remission/recurrence rates | DUBs-IN-2 and RA-9 (not available for clinical use). Indirect: gefitinib (in clinical use for other tumor types, under research for corticotropinomas) |
| USP48 | Somatic p.M415I or p.M415V | Enhanced deubiquitinase activity | Corticotropinomas | Controversial: small tumors in one study, high rate of cavernous sinus invasion in a different study. Requires further evaluation | Potential, not yet tested: DUBs-IN (not available for clinical use) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torres-Morán, M.; Franco-Álvarez, A.L.; Rebollar-Vega, R.G.; Hernández-Ramírez, L.C. Hotspots of Somatic Genetic Variation in Pituitary Neuroendocrine Tumors. Cancers 2023, 15, 5685. https://doi.org/10.3390/cancers15235685
Torres-Morán M, Franco-Álvarez AL, Rebollar-Vega RG, Hernández-Ramírez LC. Hotspots of Somatic Genetic Variation in Pituitary Neuroendocrine Tumors. Cancers. 2023; 15(23):5685. https://doi.org/10.3390/cancers15235685
Chicago/Turabian StyleTorres-Morán, Mariana, Alexa L. Franco-Álvarez, Rosa G. Rebollar-Vega, and Laura C. Hernández-Ramírez. 2023. "Hotspots of Somatic Genetic Variation in Pituitary Neuroendocrine Tumors" Cancers 15, no. 23: 5685. https://doi.org/10.3390/cancers15235685






