Direct Interaction of miRNA and circRNA with the Oncosuppressor p53: An Intriguing Perspective in Cancer Research
Abstract
:Simple Summary
Abstract
1. Introduction
2. General Properties of miRNAs, circRNAs, and p53
2.1. Overview of miRNAs
2.2. Overview of circRNAs
2.3. Overview of p53
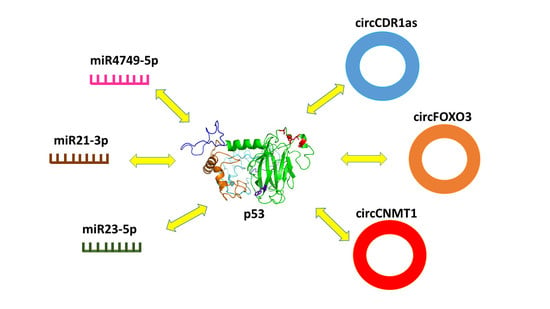
3. Interaction between p53 and miRNA
3.1. Overview of the Interplay between p53 and miRNAs
3.2. Direct Interaction between p53 and miRNA
3.2.1. Interaction between p53 and miR4749-5p
3.2.2. Interaction between p53 and miR21-3p
3.2.3. Interaction between p53 and mir23-5p
4. Interaction between p53 and circRNA
4.1. Overview of the Interplay between p53 and circRNA
4.2. Direct Interaction between p53 and circRNAs
4.2.1. Interaction between p53 and circCDR1as
4.2.2. Interaction between p53 and circFOXO3
4.2.3. Interaction between p53 and circDNMT1
5. Future Perspectives and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, L.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015, 87, 3–14. [Google Scholar] [CrossRef] [Green Version]
- Weiss, C.N.; Ito, K. Chapter Three—A Macro View of MicroRNAs: The Discovery of MicroRNAs and Their Role in Hematopoiesis and Hematologic Disease. In MiRNAs in Aging and Cancer; Galluzzi, L., Vitale, I., Eds.; Academic Press: Cambridge, MA, USA, 2017; Volume 334, pp. 99–175. ISBN 1937-6448. [Google Scholar]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, T.; Liu, Z.; Han, Z.; Ge, Q. MicroRNA Detection Specificity: Recent Advances and Future Perspective. Anal. Chem. 2019, 91, 3179–3186. [Google Scholar] [CrossRef] [Green Version]
- Shah, V.; Shah, J. Recent trends in targeting miRNAs for cancer therapy. J. Pharm. Pharmacol. 2020, 72, 1732–1749. [Google Scholar] [CrossRef]
- Vo, J.N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Zhang, Y.; Wu, Y.-M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; et al. The Landscape of Circular RNA in Cancer. Cell 2019, 176, 869–881.e13. [Google Scholar] [CrossRef] [Green Version]
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef] [PubMed]
- Panda, A.C. Circular RNAs Act as miRNA Sponges. In Circular RNAs. Advances in Experimental Medicine and Biology; Xiao, J., Ed.; Springer Singapore: Singapore, 2018; Volume 1087, pp. 67–79. ISBN 978-981-13-1426-1. [Google Scholar]
- Verduci, L.; Strano, S.; Yarden, Y.; Blandino, G. The circRNA–microRNA code: Emerging implications for cancer diagnosis and treatment. Mol. Oncol. 2019, 13, 669–680. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.R.; Bhattacharya, M.; Bhakta, S.; Saha, A.; Lee, S.-S.; Chakraborty, C. Recent research progress on circular RNAs: Biogenesis, properties, functions, and therapeutic potential. Mol. Ther. Nucleic Acids 2021, 25, 355–371. [Google Scholar] [CrossRef]
- Vogelstein, B.; Lane, D.; Levine, A.J. Surfing the p53 network. Nature 2000, 408, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Giaccia, A.J.; Kastan, M.B. The complexity of p53 modulation: Emerging patterns from divergent signals. Genes Dev. 1998, 12, 2973–2983. [Google Scholar] [CrossRef] [Green Version]
- Fields, S.; Jang, S.K. Presence of a potent transcription activating sequence in the p53 protein. Science 1990, 249, 1046–1049. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, F.; Collavin, L.; Del Sal, G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 2019, 26, 199–212. [Google Scholar] [CrossRef] [PubMed]
- Vousden, K.H.; Lane, D.P. p53 in health and disease. Nat. Rev. Mol. Cell Biol. 2007, 8, 275–283. [Google Scholar]
- Crunkhorn, S. Stapled peptide reactivates p53. Nat. Rev. Drug Discov. 2013, 12, 741. [Google Scholar] [CrossRef]
- Yamada, T.; Christov, K.; Shilkaitis, A.; Bratescu, L.; Green, A.; Santini, S.; Bizzarri, A.R.; Cannistraro, S.; Beattie, C.W. p28, A first in class peptide inhibitor of cop1 binding to p53. Br. J. Cancer 2013, 108, 2495–2504. [Google Scholar] [CrossRef] [Green Version]
- Navarro, F.; Lieberman, J. miR-34 and p53: New insights into a complex functional relationship. PLoS ONE 2015, 10, e0132767. [Google Scholar] [CrossRef]
- Tseng, H.-H.; Tseng, Y.-K.; You, J.-J.; Kang, B.-H.; Wang, T.-H.; Yang, C.-M.; Chen, H.-C.; Liou, H.-H.; Liu, P.-F.; Ger, L.-P.; et al. Next-generation Sequencing for microRNA Profiling: MicroRNA-21-3p Promotes Oral Cancer Metastasis. Anticancer Res. 2017, 37, 1059–1066. [Google Scholar] [PubMed] [Green Version]
- Chen, S.; Thorne, R.F.; Zhang, X.D.; Wu, M.; Liu, L. Non-coding RNAs, guardians of the p53 galaxy. Semin. Cancer Biol. 2020, 75, 72–83. [Google Scholar] [CrossRef]
- Singh, D.; Khan, M.A.; Siddique, H.R. Role of p53-miRNAs circuitry in immune surveillance and cancer development: A potential avenue for therapeutic intervention. In Seminars in Cell & Developmental Biology; Academic Press: Cambridge, MA, USA, 2021. [Google Scholar] [CrossRef]
- Alles, J.; Fehlmann, T.; Fischer, U.; Backes, C.; Galata, V.; Minet, M.; Hart, M.; Abu-Halima, M.; Grässer, F.A.; Lenhof, H.P.; et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019, 47, 3353–3364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, H.C.; Wani, S.; Steptoe, A.L.; Krishnan, K.; Nones, K.; Nourbakhsh, E.; Vlassov, A.; Grimmond, S.M.; Cloonan, N. Imperfect centered miRNA binding sites are common and can mediate repression of target mRNAs. Genome Biol. 2014, 15, R51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szabo, L.; Salzman, J. Detecting circular RNAs: Bioinformatic and experimental challenges. Nat. Rev. Genet. 2016, 17, 679–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, K.V.; Mattick, J.S. The rise of regulatory RNA. Nat. Rev. Genet. 2014, 15, 423–437. [Google Scholar] [CrossRef] [Green Version]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs Are the Predominant Transcript Isoform from Hundreds of Human Genes in Diverse Cell Types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knupp, D.; Miura, P. CircRNA accumulation: A new hallmark of aging? Mech. Ageing Dev. 2018, 173, 71–79. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Bach, D.-H.; Lee, S.K.; Sood, A.K. Circular RNAs in Cancer. Mol. Ther. Nucleic Acids 2019, 16, 118–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, W.Y.; Cai, Z.R.; Liu, J.; Wang, D.S.; Ju, H.Q.; Xu, R.H. Circular RNA: Metabolism, functions and interactions with proteins. Mol. Cancer 2020, 19, 172. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 2020, 21, 475–490. [Google Scholar] [CrossRef]
- Joerger, A.C.; Fersht, A.R. The tumor suppressor p53: From structures to drug discovery. Cold Spring Harb. Perspect. Biol. 2010, 2, a000919. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.J.; Gorina, S.; Jeffrey, P.D.; Pavletich, N.P. Crystal-structure of a p53 tumor-suppressor DNA complex: Understanding tumorigenic mutations. Science 1994, 265, 346–355. [Google Scholar] [CrossRef]
- Duan, J.; Nilsson, L. Effect of Zn2+ on DNA recognition and stability of the p53 DNA-binding domain. Biochemistry 2006, 45, 7483–7492. [Google Scholar] [CrossRef] [PubMed]
- Kruse, J.-P.P.; Gu, W. Modes of p53 Regulation. Cell 2009, 137, 609–622. [Google Scholar] [CrossRef] [Green Version]
- Chillemi, G.; Davidovich, P.; Abramo, M.D.; Mametnabiev, T.; Garabadzhiu, A.V.; Desideri, A.; Melino, G. Molecular dynamics of the full-length p53 monomer. Cell Cycle 2013, 12, 3098–3108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vousden, K.H.; Prives, C. Blinded by the Light: The Growing Complexity of p53. Cell 2009, 137, 413–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hafner, A.; Bulyk, M.L.; Jambhekar, A.; Lahav, G. The multiple mechanisms that regulate p53 activity and cell fate. Nat. Rev. Mol. Cell Biol. 2019, 20, 199–210. [Google Scholar] [CrossRef]
- Sullivan, K.D.; Galbraith, M.D.; Andrysik, Z.; Espinosa, J.M. Mechanisms of transcriptional regulation by p53. Cell Death Differ. 2018, 25, 133–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Momand, J.; Zambetti, G.P.; Olson, D.C.; George, D.; Levine, A.J. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 1992, 69, 1237–1245. [Google Scholar] [CrossRef]
- Kussie, P.H.; Gorina, S.; Marechal, V.; Elenbaas, B.; Moreau, J.; Levine, A.J.; Pavletich, N.P. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 1996, 274, 948–953. [Google Scholar] [CrossRef]
- Momand, J.; Wu, H.; Dasgupta, G. MDM2—Master regulator of the p53 tumor suppressor protein. Gene 2000, 242, 15–29. [Google Scholar] [CrossRef]
- Wade, M.; Li, Y.C.; Wahl, G.M. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat. Rev. Cancer 2013, 13, 83–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schon, O.; Friedler, A.; Bycroft, M.; Freund, S.M.V.; Fersht, A.R. Molecular mechanism of the interaction between Mdm2 and p53. J. Mol. Biol. 2002, 323, 491–501. [Google Scholar] [CrossRef]
- Chéne, P.; Chène, P. Inhibition of the p53-MDM2 interaction: Targeting a protein-protein interface. Mol. Can. Res. 2004, 2, 20–28. [Google Scholar]
- Moll, U.M.; Petrenko, O. The MDM2-p53 Interaction. Mol. Cancer Res. 2003, 1, 1001–1008. [Google Scholar] [PubMed]
- Wei, X.; Wu, S.; Song, T.; Chen, L.; Gao, M.; Borcherds, W.; Daughdrill, G.W.; Chen, J. Secondary interaction between MDMX and p53 core domain inhibits p53 DNA binding. Proc. Natl. Acad. Sci. USA 2016, 113, E2558–E2563. [Google Scholar] [CrossRef] [Green Version]
- Dornan, D.; Wertz, I.; Shimizu, H.; Arnott, D.; Frantz, G.D.; Dowd, P.; O’Rourke, K.; Koeppen, H.; Dixit, V.M. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 2004, 429, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Oren, M.; Rotter, V. Mutant p53 gain-of-function in cancer. Cold Spring Harb. Perspect. Biol. 2010, 2, a001107. [Google Scholar] [CrossRef] [PubMed]
- Levrero, M.; De Laurenzi, V.; Costanzo, A.; Gong, J.; Wang, J.Y.; Melino, G. The p53/p63/p73 family of transcription factors: Overlapping and distinct functions. J. Cell Sci. 2000, 113, 1661–1670. [Google Scholar] [CrossRef] [PubMed]
- Di Como, C.J.; Gaiddon, C.; Prives, C. p73 function is inhibited by tumor-derived p53 mutants in mammalian cells. Mol. Cell. Biol. 1999, 19, 1438–1449. [Google Scholar] [CrossRef] [Green Version]
- Moscetti, I.; Bizzarri, A.R.; Cannistraro, S. Binding kinetics of mutant p53R175H with wild type p53 and p63: A Surface Plasmon Resonance and Atomic Force Spectroscopy study. Biophys. Chem. 2017, 228, 55–61. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, C.; Zhao, Y.; Feng, Z. MicroRNA Control of p53. J. Cell Biochem. 2017, 14, 7–14. [Google Scholar] [CrossRef]
- Suzuki, H.I.; Yamagata, K.; Sugimoto, K.; Iwamoto, T.; Kato, S.; Miyazono, K. Modulation of microRNA processing by p53. Nature 2009, 460, 529. [Google Scholar] [CrossRef]
- Yan, H.; Huang, W.; Rao, J.; Yuan, J. miR-21 regulates ischemic neuronal injury via the p53/Bcl-2/Bax signaling pathway. Aging 2021, 13, 22242–22255. [Google Scholar] [CrossRef]
- Cortez, M.A.; Ivan, C.; Valdecanas, D.; Wang, X.; Peltier, H.J.; Ye, Y.; Araujo, L.; Carbone, D.P.; Shilo, K.; Giri, D.K.; et al. PDL1 Regulation by p53 via miR-34. JNCI J. Natl. Cancer Inst. 2016, 108, djv303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pu, X.; Huang, G.; Guo, H.; Guo, C.; Li, H.; Ye, S.; Ling, S.; Jiang, L.; Tian, Y.; Lin, T. Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J. Gastroenterol. Hepatol. 2010, 25, 1674–1680. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, Y.; Fang, J.-Y.; Xu, J. Gain-of-function miRNA signature by mutant p53 associates with poor cancer outcome. Oncotarget 2016, 7, 11056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Aung, L.H.H.; Long, B.; Qin, D.; An, S.; Li, P. miR-23a binds to p53 and enhances its association with miR-128 promoter. Sci. Rep. 2015, 5, 16422. [Google Scholar] [CrossRef] [Green Version]
- Moscetti, I.; Cannistraro, S.; Bizzarri, A.R. Probing direct interaction of oncomiR-21-3p with the tumor suppressor p53 by fluorescence, FRET and atomic force spectroscopy. Arch. Biochem. Biophys. 2019, 671, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Bizzarri, A.R.; Cannistraro, S. Investigation of a Direct Interaction between miR4749 and the Tumor Suppressor p53 by Fluorescence, FRET and Molecular Modeling. Biomolecules 2020, 10, 346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quan, W.; Yao, Y.; Xianhua, C.; Xiaodong, P.; Qi, H.; Dong, W.; Youcai, D.; Xiaohui, L.; Jun, Y.; Jihong, Z. Competing endogenous RNA screening based on long noncoding RNA-messenger RNA co-expression profile in Hepatitis B virus-associated hepatocarcinogenesis. J. Tradit. Chinese Med. 2017, 37, 510–521. [Google Scholar] [CrossRef]
- Ho, K.-H.; Kuo, T.-C.; Lee, Y.-T.; Chen, P.-H.; Shih, C.-M.; Cheng, C.-H.; Liu, A.-J.; Lee, C.-C.; Chen, K.-C. Xanthohumol regulates miR-4749-5p-inhibited RFC2 signaling in enhancing temozolomide cytotoxicity to glioblastoma. Life Sci. 2020, 254, 117807. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer US: New York, NY, USA, 2006; ISBN 978-0-387-31278-1. [Google Scholar]
- Funari, G.; Domenici, F.; Nardinocchi, L.; Puca, R.; D’Orazi, G.; Bizzarri, A.R.; Cannistraro, S. Interaction of p53 with Mdm2 and azurin as studied by atomic force spectroscopy. J. Mol. Recognit. 2010, 23, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Moscetti, I.; Cannistraro, S.; Bizzarri, A.R. Surface plasmon resonance sensing of biorecognition interactions within the tumor suppressor P53 network. Sensors 2017, 17, 2680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medintz, N.H.I. FRET—Förster Resonance Energy Transfer from Theory to Applications; Wiley-VCHVerlag: Weinheim, Germany, 2014. [Google Scholar]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuböck, R.; Hofacker, I.L. The Vienna RNA websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef] [Green Version]
- Popenda, M.; Szachniuk, M.; Antczak, M.; Purzycka, K.J.; Lukasiak, P.; Bartol, N.; Blazewicz, J.; Adamiak, R.W. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012, 40, e112. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Zhang, D.; Zhou, P.; Li, B.; Huang, S.-Y. HDOCK: A web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017, 45, W365–W373. [Google Scholar] [CrossRef]
- Lou, J.; Hao, Y.; Lin, K.; Lyu, Y.; Chen, M.; Wang, H.; Zou, D.; Jiang, X.; Wang, R.; Jin, D.; et al. Circular RNA CDR1as disrupts the p53/MDM2 complex to inhibit Gliomagenesis. Mol. Cancer 2020, 19, 138. [Google Scholar] [CrossRef] [PubMed]
- Du, W.W.; Fang, L.; Yang, W.; Wu, N.; Awan, F.M.; Yang, Z.; Yang, B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017, 24, 357–370. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Du, W.W.; Lyu, J.; Dong, J.; Zhang, C.; Yang, W.; He, A.; Kwok, Y.S.S.; Ma, J.; Wu, N.; et al. Enhanced breast cancer progression by mutant p53 is inhibited by the circular RNA circ-Ccnb1. Cell Death Differ. 2018, 25, 2195–2208. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Tsao, C. Emerging role of microRNA-21 in cancer. Biomed. Rep. 2016, 5, 395–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jenike, A.E.; Halushka, M.K. miR-21: A non-specific biomarker of all maladies. Biomark. Res. 2021, 9, 18. [Google Scholar] [CrossRef]
- Buller, B.; Liu, X.; Wang, X.; Zhang, R.L.; Zhang, L.; Hozeska-Solgot, A.; Chopp, M.; Zhang, Z.G. MicroRNA-21 protects neurons from ischemic death. FEBS J. 2010, 277, 4299–4307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bautista-Sánchez, D.; Arriaga-Canon, C.; Pedroza-Torres, A.; De La Rosa-Velázquez, I.A.; González-Barrios, R.; Contreras-Espinosa, L.; Montiel-Manríquez, R.; Castro-Hernández, C.; Fragoso-Ontiveros, V.; Álvarez-Gómez, R.M.; et al. The Promising Role of miR-21 as a Cancer Biomarker and Its Importance in RNA-Based Therapeutics. Mol. Ther. Nucleic Acids 2020, 20, 409–420. [Google Scholar] [CrossRef]
- Báez-Vega, P.M.; Echevarría Vargas, I.M.; Valiyeva, F.; Encarnación-Rosado, J.; Roman, A.; Flores, J.; Marcos-Martínez, M.J.; Vivas-Mejía, P.E. Targeting miR-21-3p inhibits proliferation and invasion of ovarian cancer cells. Oncotarget 2016, 7, 36321–36337. [Google Scholar] [CrossRef]
- Hasáková, K.; Bilcikova, J.; Vician, M.; Reis, R.; Michal, Z.; Herichova, I. Gender-dependent expression of leading and passenger strand of miR-21 and miR-16 in human colorectal cancer and adjacent colonic tissues. Physiol. Res. 2017, 66, S575–S582. [Google Scholar] [CrossRef]
- Peralta-Zaragoza, O.; Deas, J.; Meneses-Acosta, A.; De la O-Gómez, F.; Fernández-Tilapa, G.; Gómez-Cerón, C.; Benítez-Boijseauneau, O.; Burguete-García, A.; Torres-Poveda, K.; Bermúdez-Morales, V.H.; et al. Relevance of miR-21 in regulation of tumor suppressor gene PTEN in human cervical cancer cells. BMC Cancer 2016, 16, 215. [Google Scholar] [CrossRef] [Green Version]
- Bizzarri, A.R.; Cannistraro, S. The application of atomic force spectroscopy to the study of biological complexes undergoing a biorecognition process. Chem. Soc. Rev. 2010, 39, 734–749. [Google Scholar] [CrossRef]
- Bizzarri, A.R.; Cannistraro, S. Atomic Force Spectroscopy in Biological Complex Formation: Strategies and Perspectives. J. Phys. Chem. B 2009, 113, 16449–16464. [Google Scholar] [CrossRef] [PubMed]
- Evans, E. Probing the relation between force-lifetime- and chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. Struct. 2001, 30, 5–128. [Google Scholar] [CrossRef] [Green Version]
- Nooren, I.M.; Thornton, J.M. Structural characterization and functional significance of transient protein-protein interactions. EMBO J. 2003, 22, 3486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moscetti, I.; Bizzarri, A.R.; Cannistraro, S. Imaging and kinetics of the bimolecular complex formed by the tumor suppressor p53 with ubiquitin ligase COP1 as studied by atomic force microscopy and surface plasmon resonance. Int. J. Nanomed. 2018, 13, 251–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moscetti, I.; Teveroni, E.; Moretti, F.; Bizzarri, A.R.; Cannistraro, S. MDM2–MDM4 molecular interaction investigated by atomic force spectroscopy and surface plasmon resonance. Int. J. Nanomed. 2016, 11, 4221–4229. [Google Scholar]
- Taranta, M.; Bizzarri, A.R.; Cannistraro, S. Probing the interaction between p53 and the bacterial protein azurin by single molecule force spectroscopy. J. Mol. Recognit. 2008, 21, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Fuhrmann, A.; Schoening, J.C.; Anselmetti, D.; Staiger, D.; Ros, R. Quantitative analysis of single-molecule RNA-protein interaction. Biophys. J. 2009, 96, 5030–5039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, N.; Tan, H.Y.; Feng, Y.G.; Zhang, C.; Chen, F.; Feng, Y. microRNA-23a in human cancer: Its roles, mechanisms and therapeutic relevance. Cancers 2019, 11, 7. [Google Scholar] [CrossRef] [Green Version]
- Huang, F.-Y.; Wong, D.K.-H.; Seto, W.-K.; Lai, C.-L.; Yuen, M.-F. Estradiol induces apoptosis via activation of miRNA-23a and p53: Implication for gender difference in liver cancer development. Oncotarget 2015, 6, 34941–34952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaudhary, R.; Muys, B.R.; Grammatikakis, I.; De, S.; Abdelmohsen, K.; Li, X.L.; Zhu, Y.; Daulatabad, S.V.; Tsitsipatis, D.; Meltzer, P.S.; et al. A Circular RNA from the MDM2 Locus Controls Cell Cycle Progression by Suppressing p53 Levels. Mol. Cell. Biol. 2020, 40, e00473-19. [Google Scholar] [CrossRef] [PubMed]
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.H.; Nicolaou, A.; et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016, 7, 12429. [Google Scholar] [CrossRef] [Green Version]
- Shi, Z.; Chen, T.; Yao, Q.; Zheng, L.; Zhang, Z.; Wang, J.; Hu, Z.; Cui, H.; Han, Y.; Han, X.; et al. The circular RNA ciRS-7 promotes APP and BACE1 degradation in an NF-κB-dependent manner. FEBS J. 2017, 284, 1096–1109. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Zeng, X.; Shan, R.; Wen, W.; Li, J.; Tan, J.; Li, L.; Wan, R. The Emerging Picture of the Roles of CircRNA-CDR1as in Cancer. Front. Cell Dev. Biol. 2020, 8, 1499. [Google Scholar] [CrossRef]
- Guo, D.; Li, F.; Zhao, X.; Long, B.; Zhang, S.; Wang, A.; Cao, D.; Sun, J.; Li, B. Circular RNA expression and association with the clinicopathological characteristics in papillary thyroid carcinoma. Oncol. Rep. 2020, 44, 519–532. [Google Scholar] [CrossRef]
- Yang, Q.; Du, W.W.; Wu, N.; Yang, W.; Awan, F.M.; Fang, L.; Ma, J.; Li, X.; Zeng, Y.; Yang, Z.; et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017, 24, 1609–1620. [Google Scholar] [CrossRef] [Green Version]
- Piwecka, M.; Glažar, P.; Hernandez-Miranda, L.R.; Memczak, S.; Wolf, S.A.; Rybak-Wolf, A.; Filipichyk, A.; Klironomos, F.; Plass, M.; Rajewsky, N.; et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017, 357, eaam8526. [Google Scholar]
- Rao, D.; Yu, C.; Sheng, J.; Lv, E.; Huang, W. The Emerging Roles of circFOXO3 in Cancer. Front. Cell Dev. Biol. 2021, 9, 1446. [Google Scholar] [CrossRef]
- Link, W. Introduction to FOXO Biology. In FOXO Transcription Factors: Methods and Protocols; Link, W., Ed.; Springer: New York, NY, USA, 2019; pp. 1–9. ISBN 978-1-4939-8900-3. [Google Scholar]
- Huang, W.; Huang, F.; Feng, C. CircFoxo3 Promotes Adriamycin Resistance Through Regulation of miR-199a-5p/ATP Binding Cassette Subfamily C Member 1 Axis in Hepatocellular Carcinoma. OncoTargets. Ther. 2020, 13, 5113–5122. [Google Scholar] [CrossRef] [PubMed]
- Kong, Z.; Wan, X.; Lu, Y.; Zhang, Y.; Huang, Y.; Xu, Y.; Liu, Y.; Zhao, P.; Xiang, X.; Li, L.; et al. Circular RNA circFOXO3 promotes prostate cancer progression through sponging miR-29a-3p. J. Cell. Mol. Med. 2020, 24, 799–813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ai, Y.; Wu, S.; Zou, C.; Wei, H. Circular RNA circFOXO3 regulates KDM2A by targeting miR-214 to promote tumor growth and metastasis in oral squamous cell carcinoma. J. Cell. Mol. Med. 2021, 1–11. [Google Scholar] [CrossRef]
- Du, W.W.; Yang, W.; Li, X.; Awan, F.M.; Yang, Z.; Fang, L.; Lyu, J.; Li, F.; Peng, C.; Krylov, S.N.; et al. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene 2018, 37, 5829–5842. [Google Scholar] [CrossRef] [PubMed]













| p53-RNA Interaction | Domain of p53 Involved in the Binding | Proposed Regulation Mechanism | Ref. |
|---|---|---|---|
| p53-miR4749 | DBD | Inhibition of p53-DNA interaction | [70] |
| p53-miR21 | DBD | Inhibition of p53-DNA interaction | [69] |
| p53-miR23 | CTD | Promotion of p53-miR128 interaction, favouring apoptosis due to oxidative stress | [68] |
| p53-circCDR1 | DBD | Disruption of the p53-MDM2 complex, favouring an anticancer action | [80] |
| p53-circFOXO3 | CTD | Promotion of MDM2 ubiquitination, favouring cell apoptosis | [81] |
| p53-circCNM1 | CTD | Inhibition of p53 transcription leading to tumor cell proliferation | [82] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bizzarri, A.R.; Cannistraro, S. Direct Interaction of miRNA and circRNA with the Oncosuppressor p53: An Intriguing Perspective in Cancer Research. Cancers 2021, 13, 6108. https://doi.org/10.3390/cancers13236108
Bizzarri AR, Cannistraro S. Direct Interaction of miRNA and circRNA with the Oncosuppressor p53: An Intriguing Perspective in Cancer Research. Cancers. 2021; 13(23):6108. https://doi.org/10.3390/cancers13236108
Chicago/Turabian StyleBizzarri, Anna Rita, and Salvatore Cannistraro. 2021. "Direct Interaction of miRNA and circRNA with the Oncosuppressor p53: An Intriguing Perspective in Cancer Research" Cancers 13, no. 23: 6108. https://doi.org/10.3390/cancers13236108