Re-Expression of Poly/Oligo-Sialylated Adhesion Molecules on the Surface of Tumor Cells Disrupts Their Interaction with Immune-Effector Cells and Contributes to Pathophysiological Immune Escape
Abstract
:Simple Summary
Abstract
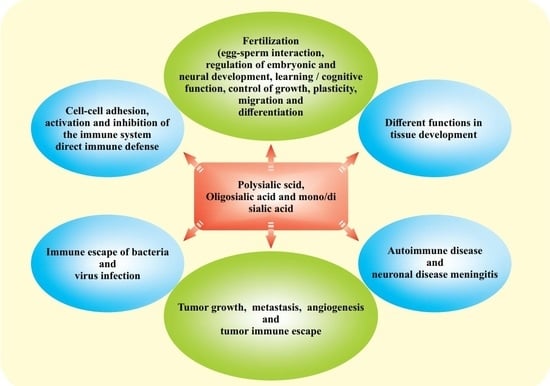
1. Introduction
2. Materials and Methods
2.1. Literature Search
2.2. Data Selection
- (1)
- The physiological and pathological roles of membrane adhesion molecules linked with oligo/poly-sialic acid glycosylation in tumor progression, apoptosis, metastasis, angiogenesis, migration, proliferation, and growth of tumors;
- (2)
- The biological roles of heterophilic and homophilic membrane adhesion molecules in neuronal and embryonic development and the development of certain neuronal diseases (such as Alzheimer's disease, Parkinson's disease, multiple sclerosis, and schizophrenia);
- (3)
- The role of sialic acid in the immune escape of tumors and pathogens (bacteria, viruses);
- (4)
- The role of sialic acid in electrostatic repulsion between immune effector and target cells (tumor or pathogens), which re-express membrane adhesion molecules linked with oligo/polysialic acid;
- (5)
- The role of sialic acid in differentiation of immune cells (T-cells), and in virus infection;
- (6)
- The role of sialic acid receptors/copartners (lectins such as siglecs or other adhesion molecules) in relation to the function of the immune cells;
- (7)
- The role of galectins, selectins, and kinases in cooperation with sialylated glycoproteins in or outside of cells.
2.3. Legend to Prisma Flow Diagram
3. Results and Discussion
3.1. Structure and Regulation of Sialic (N-Acetylneuraminic)-Acid

3.2. Physiologic Role of Poly/Oligo-Sialylated Adhesion Molecules and Their Interaction with Growth Factors and Their Receptors
3.3. Re-Expression of Polysialylated Adhesion Molecules in Cancer Progression
3.4. Lectins Are Potential Co-Partners of Sialylated Glycoproteins
3.5. I-Type Lectins or Siglecs
3.6. Sialylation of Check Point Receptors
3.7. Sialylated Glycans in Tumor Cells Prevent Galectin Induced Apoptosis, Autophagy, and Cluster Formation


3.8. Polysialylation of Glycoproteins Generates Diverse Functions
3.9. Proteins with the HNK-1 Epitope Serve a Function Similar to Poly/Oligo-Sialylated Glycoproteins
3.10. Polysialylated Glycoproteins Are Co-Receptors for Growth Factors and Their Receptors
3.11. Polysialylated NCAM-1, NRP-2, and CADM-1 Potentiate Cell Growth Signaling in Tumor Cells and Increase Tumor Progression
3.12. Characterization of NCAMPSA
3.13. NCAM-1PSA, NRP-2PSA or SynCAM-1PSA Downregulate the Expression of Adhesion Molecules
3.14. Dual Role of NCAM-1PSA
3.15. Perspectives and Therapeutic Potential
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lanitis, E.; Dangaj, D.; Irving, M.; Coukos, G. Mechanisms regulating T-cell infiltration and activity in solid tumors. Ann. Oncol. 2017, 28, xii18–xii32. [Google Scholar] [CrossRef]
- Gajewski, T.F.; Schreiber, H.; Fu, Y.X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013, 14, 1014–1022. [Google Scholar] [CrossRef] [Green Version]
- Bull, C.; Boltje, T.J.; Balneger, N.; Weischer, S.M.; Wassink, M.; van Gemst, J.J.; Bloemendal, V.R.; Boon, L.; van der Vlag, J.; Heise, T.; et al. Sialic Acid Blockade Suppresses Tumor Growth by Enhancing T-cell-Mediated Tumor Immunity. Cancer Res. 2018, 78, 3574–3588. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Middleton, D.R.; Wantuch, P.L.; Ozdilek, A.; Avci, F.Y. Carbohydrates as T-cell antigens with implications in health and disease. Glycobiology 2016, 26, 1029–1040. [Google Scholar] [CrossRef]
- Rabinovich, G.A.; van Kooyk, Y.; Cobb, B.A. Glycobiology of immune responses. Ann. N. Y. Acad. Sci. 2012, 1253, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in health and disease. Nat. Rev. Nephrol. 2019, 15, 346–366. [Google Scholar] [CrossRef]
- Tian, Y.; Esteva, F.J.; Song, J.; Zhang, H. Altered expression of sialylated glycoproteins in breast cancer using hydrazide chemistry and mass spectrometry. Mol. Cell. Proteom. 2012, 11, M111.011403. [Google Scholar] [CrossRef] [Green Version]
- Colley, K.J.; Kitajima, K.; Sato, C. Polysialic acid: Biosynthesis, novel functions and applications. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 498–532. [Google Scholar] [CrossRef]
- Rodrigues, E.; Macauley, M.S. Hypersialylation in Cancer: Modulation of Inflammation and Therapeutic Opportunities. Cancers 2018, 10, 207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Freitas Junior, J.C.; Carvalho, S.; Dias, A.M.; Oliveira, P.; Cabral, J.; Seruca, R.; Oliveira, C.; Morgado-Diaz, J.A.; Reis, C.A.; Pinho, S.S. Insulin/IGF-I signaling pathways enhances tumor cell invasion through bisecting GlcNAc N-glycans modulation. an interplay with E-cadherin. PLoS ONE 2013, 8, e81579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munkley, J.; Scott, E. Targeting Aberrant Sialylation to Treat Cancer. Medicines 2019, 6, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swindall, A.F.; Bellis, S.L. Sialylation of the Fas death receptor by ST6Gal-I provides protection against Fas-mediated apoptosis in colon carcinoma cells. J. Biol. Chem. 2011, 286, 22982–22990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varki, A. Multiple changes in sialic acid biology during human evolution. Glycoconj. J. 2009, 26, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Varki, A. Biological roles of glycans. Glycobiology 2017, 27, 3–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavallaro, U.; Dejana, E. Adhesion molecule signalling: Not always a sticky business. Nat. Rev. Mol. Cell Biol. 2011, 12, 189–197. [Google Scholar] [CrossRef]
- Pinho, S.S.; Figueiredo, J.; Cabral, J.; Carvalho, S.; Dourado, J.; Magalhaes, A.; Gartner, F.; Mendonfa, A.M.; Isaji, T.; Gu, J.; et al. E-cadherin and adherens-junctions stability in gastric carcinoma: Functional implications of glycosyltransferases involving N-glycan branching biosynthesis, N-acetylglucosaminyltransferases III and V. BBA-Gen. Subj. 2013, 1830, 2690–2700. [Google Scholar] [CrossRef]
- Mallard, B.W.; Tiralongo, J. Cancer stem cell marker glycosylation: Nature, function and significance. Glycoconj. J. 2017, 34, 441–452. [Google Scholar] [CrossRef]
- Ferreira, I.G.; Pucci, M.; Venturi, G.; Malagolini, N.; Chiricolo, M.; Dall’Olio, F. Glycosylation as a Main Regulator of Growth and Death Factor Receptors Signaling. Int. J. Mol. Sci. 2018, 19, 580. [Google Scholar] [CrossRef] [Green Version]
- Very, N.; Lefebvre, T.; El Yazidi-Belkoura, I. Drug resistance related to aberrant glycosylation in colorectal cancer. Oncotarget 2018, 9, 1380–1402. [Google Scholar] [CrossRef] [Green Version]
- Fossella, F.; McCann, J.; Tolcher, A.; Xie, H.; Hwang, L.L.; Carr, C.; Berg, K.; Fram, R. Phase II trial of BB-10901 (huN901-DM1) given weekly for four consecutive weeks every 6 weeks in patients with relapsed SCLC and CD56-positive small cell carcinoma. J. Clin. Oncol. 2005, 23 (Suppl. 16), 7159. [Google Scholar] [CrossRef]
- Laubli, H.; Borsig, L. Altered Cell Adhesion and Glycosylation Promote Cancer Immune Suppression and Metastasis. Front. Immunol. 2019, 10, 2120. [Google Scholar] [CrossRef] [Green Version]
- Bassaganas, S.; Carvalho, S.; Dias, A.M.; Perez-Garay, M.; Ortiz, M.R.; Figueras, J.; Reis, C.A.; Pinho, S.S.; Peracaula, R. Pancreatic cancer cell glycosylation regulates cell adhesion and invasion through the modulation of alpha2beta1 integrin and E-cadherin function. PLoS ONE 2014, 9, e98595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohtsubo, K.; Marth, J.D. Glycosylation in cellular mechanisms of health and disease. Cell 2006, 126, 855–867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varki, A. Glycan-based interactions involving vertebrate sialic-acid-recognizing proteins. Nature 2007, 446, 1023–1029. [Google Scholar] [CrossRef] [PubMed]
- Scott, E.; Munkley, J. Glycans as Biomarkers in Prostate Cancer. Int. J. Mol. Sci. 2019, 20, 1389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sola, R.J.; Griebenow, K. Effects of glycosylation on the stability of protein pharmaceuticals. J. Pharm. Sci. 2009, 98, 1223–1245. [Google Scholar] [CrossRef] [Green Version]
- Pearce, O.M.; Laubli, H. Sialic acids in cancer biology and immunity. Glycobiology 2016, 26, 111–128. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Li, Z.; Wang, Y.; Zheng, Q.; Li, J. Mass spectrometry for protein sialoglycosylation. Mass Spectrom. Rev. 2018, 37, 652–680. [Google Scholar] [CrossRef]
- Liu, Y.C.; Yen, H.Y.; Chen, C.Y.; Chen, C.H.; Cheng, P.F.; Juan, Y.H.; Chen, C.H.; Khoo, K.H.; Yu, C.J.; Yang, P.C.; et al. Sialylation and fucosylation of epidermal growth factor receptor suppress its dimerization and activation in lung cancer cells. Proc. Natl. Acad. Sci. USA 2011, 108, 11332–11337. [Google Scholar] [CrossRef] [Green Version]
- Lenman, A.; Liaci, A.M.; Liu, Y.; Frangsmyr, L.; Frank, M.; Blaum, B.S.; Chai, W.; Podgorski, I.I.; Harrach, B.; Benko, M.; et al. Polysialic acid is a cellular receptor for human adenovirus 52. Proc. Natl. Acad. Sci. USA 2018, 115, E4264–E4273. [Google Scholar] [CrossRef] [Green Version]
- Tong, J.; Fu, Y.; Meng, F.; Kruger, N.; Valentin-Weigand, P.; Herrler, G. The Sialic Acid Binding Activity of Human Parainfluenza Virus 3 and Mumps Virus Glycoproteins Enhances the Adherence of Group B Streptococci to HEp-2 Cells. Front. Cell Infect. Mi. 2018, 8, 280. [Google Scholar] [CrossRef] [Green Version]
- Zlatina, K.; Galuska, S.P. Polysialic Acid Modulates Only the Antimicrobial Properties of Distinct Histones. ACS Omega 2019, 4, 1601–1610. [Google Scholar] [CrossRef]
- Freiberger, F.; Claus, H.; Gunzel, A.; Oltmann-Norden, I.; Vionnet, J.; Muhlenhoff, M.; Vogel, U.; Vann, W.F.; Gerardy-Schahn, R.; Stummeyer, K. Biochemical characterization of a Neisseria meningitidis polysialyltransferase reveals novel functional motifs in bacterial sialyltransferases. Mol. Microbiol. 2007, 65, 1258–1275. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.; Chen, X. Aldolase-catalyzed synthesis of beta-D-galp-(1-->9)-D-KDN: A novel acceptor for sialyltransferases. Org. Lett. 2006, 8, 2393–2396. [Google Scholar] [CrossRef]
- Ghosh, S. Sialic acid and biology of life: An introduction. In Sialic Acids and Sialoglycoconjugates in the Biology of Life, Health and Disease; Academic Press: Cambridge, MA, USA, 2020; Volume 1. [Google Scholar]
- Kooner, A.S.; Yu, H.; Chen, X. Synthesis of N-Glycolylneuraminic Acid (Neu5Gc) and Its Glycosides. Front. Immunol. 2019, 10, 2004. [Google Scholar] [CrossRef]
- Schauer, R. Sialic acids as regulators of molecular and cellular interactions. Curr. Opin. Struc. Biol. 2009, 19, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Schauer, R.; Kamerling, J.P. Exploration of the Sialic Acid World. Adv. Carbohydr. Chem. Biochem. 2018, 75, 1–213. [Google Scholar] [CrossRef] [PubMed]
- Hawsawi, M. Exploring the Scope and Limitations of the Oxidative Deamination of N-Acetyl Neuraminic Acid; Wayne State University: Detroit, MI, USA, 2020. [Google Scholar]
- Zhang, R.; Loers, G.; Schachner, M.; Boelens, R.; Wienk, H.; Siebert, S.; Eckert, T.; Kraan, S.; Rojas-Macias, M.A.; Lutteke, T.; et al. Molecular Basis of the Receptor Interactions of Polysialic Acid (polySia), polySia Mimetics, and Sulfated Polysaccharides. ChemMedChem 2016, 11, 990–1002. [Google Scholar] [CrossRef]
- Zhou, G.P.; Huang, R.B.; Troy, F.A., 2nd. 3D structural conformation and functional domains of polysialyltransferase ST8Sia IV required for polysialylation of neural cell adhesion molecules. Protein Pept. Lett. 2015, 22, 137–148. [Google Scholar] [CrossRef]
- Teoh, S.T.; Ogrodzinski, M.P.; Ross, C.; Hunter, K.W.; Lunt, S.Y. Sialic Acid Metabolism: A Key Player in Breast Cancer Metastasis Revealed by Metabolomics. Front. Oncol. 2018, 8, 174. [Google Scholar] [CrossRef]
- Muhlenhoff, M.; Rollenhagen, M.; Werneburg, S.; Gerardy-Schahn, R.; Hildebrandt, H. Polysialic acid: Versatile modification of NCAM, SynCAM 1 and neuropilin-2. Neurochem. Res. 2013, 38, 1134–1143. [Google Scholar] [CrossRef] [PubMed]
- Elkashef, S.M.; Allison, S.J.; Sadiq, M.; Basheer, H.A.; Ribeiro Morais, G.; Loadman, P.M.; Pors, K.; Falconer, R.A. Polysialic acid sustains cancer cell survival and migratory capacity in a hypoxic environment. Sci. Rep. 2016, 6, 33026. [Google Scholar] [CrossRef] [Green Version]
- Falconer, R.A.; Errington, R.J.; Shnyder, S.D.; Smith, P.J.; Patterson, L.H. Polysialyltransferase: A new target in metastatic cancer. Curr. Cancer Drug. Tar. 2012, 12, 925–939. [Google Scholar] [CrossRef]
- Kronewitter, S.R.; Marginean, I.; Cox, J.T.; Zhao, R.; Hagler, C.D.; Shukla, A.K.; Carlson, T.S.; Adkins, J.N.; Camp, D.G., 2nd; Moore, R.J.; et al. Polysialylated N-glycans identified in human serum through combined developments in sample preparation, separations, and electrospray ionization-mass spectrometry. Anal. Chem. 2014, 86, 8700–8710. [Google Scholar] [CrossRef] [Green Version]
- RodrIguez, E.; Schetters, S.T.T.; van Kooyk, Y. The tumour glyco-code as a novel immune checkpoint for immunotherapy. Nat. Rev. Immunol. 2018, 18, 204–211. [Google Scholar] [CrossRef] [PubMed]
- Stowell, S.R.; Ju, T.; Cummings, R.D. Protein glycosylation in cancer. Annu. Rev. Pathol. 2015, 10, 473–510. [Google Scholar] [CrossRef] [Green Version]
- Vajaria, B.N.; Begum, R.; Patel, P.S. Clinical Significance of Glycosylation Changes in Oral Cancer. Glycobiology 2015, 25, 1300–1301. [Google Scholar]
- Munkley, J.; Elliott, D.J. Hallmarks of glycosylation in cancer. Oncotarget 2016, 7, 35478–35489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanas, A.; Sahasrabudhe, N.M.; Rodriguez, E.; van Kooyk, Y.; van Vliet, S.J. Fucosylated Antigens in Cancer: An Alliance toward Tumor Progression, Metastasis, and Resistance to Chemotherapy. Front. Oncol. 2018, 8, 39. [Google Scholar] [CrossRef]
- Gong, L.; Zhou, X.; Yang, J.; Jiang, Y.; Yang, H. Effects of the regulation of polysialyltransferase ST8SiaII on the invasiveness and metastasis of small cell lung cancer cells. Oncol. Rep. 2017, 37, 131–138. [Google Scholar] [CrossRef] [Green Version]
- Sato, C.; Kitajima, K. Sialic Acids in Neurology. In Advances in Carbohydrate Chemistry and Biochemistry; Academic Press: Cambridge, MA, USA, 2019; Volume 76, pp. 1–64. [Google Scholar]
- Bruses, J.L.; Rutishauser, U. Roles, regulation, and mechanism of polysialic acid function during neural development. Biochimie 2001, 83, 635–643. [Google Scholar] [CrossRef]
- Bonfanti, L. PSA-NCAM in mammalian structural plasticity and neurogenesis. Prog. Neurobiol. 2006, 80, 129–164. [Google Scholar] [CrossRef]
- Burgess, A.; Wainwright, S.R.; Shihabuddin, L.S.; Rutishauser, U.; Seki, T.; Aubert, I. Polysialic acid regulates the clustering, migration, and neuronal differentiation of progenitor cells in the adult hippocampus. Dev. Neurobiol. 2008, 68, 1580–1590. [Google Scholar] [CrossRef] [PubMed]
- Rutishauser, U. Polysialic acid in the plasticity of the developing and adult vertebrate nervous system. Nat. Rev. Neurosci. 2008, 9, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Rutishauser, U.; El Maarouf, A. Polysialic Acid in the CNS: Plasticity and Repair. Glycobiology 2008, 18, 944. [Google Scholar]
- Mehrabian, M.; Hildebrandt, H.; Schmitt-Ulms, G. NCAM1 Polysialylation: The Prion Protein’s Elusive Reason for Being? ASN Neuro 2016, 8, 1759091416679074. [Google Scholar] [CrossRef] [Green Version]
- Boutin, C.; Schmitz, B.; Cremer, H.; Diestel, S. NCAM expression induces neurogenesis in vivo. Eur. J. Neurosci. 2009, 30, 1209–1218. [Google Scholar] [CrossRef] [PubMed]
- Gibson, N.J. Cell adhesion molecules in context: CAM function depends on the neighborhood. Cell Adh. Migr. 2011, 5, 48–51. [Google Scholar] [CrossRef] [Green Version]
- Tang, H.; Chang, H.; Dong, Y.; Guo, L.; Shi, X.; Wu, Y.; Huang, Y.; He, Y. Architecture of cell-cell adhesion mediated by sidekicks. Proc. Natl. Acad. Sci. USA 2018, 115, 9246–9251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villringer, S.; Madl, J.; Sych, T.; Manner, C.; Imberty, A.; Romer, W. Lectin-mediated protocell crosslinking to mimic cell-cell junctions and adhesion. Sci. Rep. 2018, 8, 1932. [Google Scholar] [CrossRef]
- Baldwin, K.T.; Eroglu, C. Molecular mechanisms of astrocyte-induced synaptogenesis. Curr. Opin. Neurobiol. 2017, 45, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Bhide, G.P. Biophysical and Biochemical Determinants of Protein-Specific Polysialylation. Ph.D Thesis, University of Illinois at Chicago, Chicago, IL, USA, 2017. [Google Scholar]
- Kiermaier, E.; Moussion, C.; Veldkamp, C.T.; Gerardy-Schahn, R.; de Vries, I.; Williams, L.G.; Chaffee, G.R.; Phillips, A.J.; Freiberger, F.; Imre, R.; et al. Polysialylation controls dendritic cell trafficking by regulating chemokine recognition. Science 2016, 351, 186–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Werneburg, S.; Buettner, F.F.; Erben, L.; Mathews, M.; Neumann, H.; Muhlenhoff, M.; Hildebrandt, H. Polysialylation and lipopolysaccharide-induced shedding of E-selectin ligand-1 and neuropilin-2 by microglia and THP-1 macrophages. Glia 2016, 64, 1314–1330. [Google Scholar] [CrossRef] [PubMed]
- Fujioka, Y.; Nishide, S.; Ose, T.; Suzuki, T.; Kato, I.; Fukuhara, H.; Fujioka, M.; Horiuchi, K.; Satoh, A.O.; Nepal, P. A sialylated voltage-dependent Ca2+ channel binds hemagglutinin and mediates influenza a virus entry into mammalian cells. Cell Host Microbe 2018, 23, 809–818.e805. [Google Scholar] [CrossRef] [Green Version]
- Jing, X.; Liang, H.; Hao, C.; Yang, X.; Cui, X. Overexpression of MUC1 predicts poor prognosis in patients with breast cancer. Oncol. Rep. 2019, 41, 801–810. [Google Scholar] [CrossRef]
- Yabe, U.; Sato, C.; Matsuda, T.; Kitajima, K. Polysialic acid in human milk. CD36 is a new member of mammalian polysialic acid-containing glycoprotein. J. Biol. Chem. 2003, 278, 13875–13880. [Google Scholar] [CrossRef] [Green Version]
- Angata, K.; Chan, D.; Thibault, J.; Fukuda, M. Molecular dissection of the ST8Sia IV polysialyltransferase. Distinct domains are required for neural cell adhesion molecule recognition and polysialylation. J. Biol. Chem. 2004, 279, 25883–25890. [Google Scholar] [CrossRef] [Green Version]
- Cao, L.; Wang, X.; Yang, J.; Guo, J.; Li, X.; Yang, X.; Tan, Z.; Guan, F. NCAM and attached polysialic acid affect behaviors of breast epithelial cells through differential signaling pathways. Res. Square 2020. (Under Revision). [Google Scholar]
- Rollenhagen, M.; Kuckuck, S.; Ulm, C.; Hartmann, M.; Galuska, S.P.; Geyer, R.; Geyer, H.; Muhlenhoff, M. Polysialylation of the synaptic cell adhesion molecule 1 (SynCAM 1) depends exclusively on the polysialyltransferase ST8SiaII in vivo. J. Biol. Chem. 2012, 287, 35170–35180. [Google Scholar] [CrossRef] [Green Version]
- Ellis, L.M. The role of neuropilins in cancer. Mol. Cancer Ther. 2006, 5, 1099–1107. [Google Scholar] [CrossRef] [Green Version]
- Pellet-Many, C.; Frankel, P.; Jia, H.; Zachary, I. Neuropilins: Structure, function and role in disease. Biochem. J. 2008, 411, 211–226. [Google Scholar] [CrossRef] [Green Version]
- Bhide, G.P.; Fernandes, N.R.; Colley, K.J. Sequence Requirements for Neuropilin-2 Recognition by ST8SiaIV and Polysialylation of Its O-Glycans. J. Biol. Chem. 2016, 291, 9444–9457. [Google Scholar] [CrossRef] [Green Version]
- Sulpice, E.; Plouet, J.; Berge, M.; Allanic, D.; Tobelem, G.; Merkulova-Rainon, T. Neuropilin-1 and neuropilin-2 act as coreceptors, potentiating proangiogenic activity. Blood 2008, 111, 2036–2045. [Google Scholar] [CrossRef]
- Grun, D.; Adhikary, G.; Eckert, R.L. VEGF-A acts via neuropilin-1 to enhance epidermal cancer stem cell survival and formation of aggressive and highly vascularized tumors. Oncogene 2016, 35, 4379–4387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winther, M.; Berezin, V.; Walmod, P.S. NCAM2/OCAM/RNCAM: Cell adhesion molecule with a role in neuronal compartmentalization. Int. J. Biochem. Cell Biol. 2012, 44, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Leshchyns’ka, I.; Liew, H.T.; Shepherd, C.; Halliday, G.M.; Stevens, C.H.; Ke, Y.D.; Ittner, L.M.; Sytnyk, V. Abeta-dependent reduction of NCAM2-mediated synaptic adhesion contributes to synapse loss in Alzheimer’s disease. Nat. Commun. 2015, 6, 8836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parcerisas, A.; Pujadas, L.; Ortega-Gasco, A.; Perello-Amoros, B.; Viais, R.; Hino, K.; Figueiro-Silva, J.; La Torre, A.; Trullas, R.; Simo, S.; et al. NCAM2 Regulates Dendritic and Axonal Differentiation through the Cytoskeletal Proteins MAP2 and 14-3-3. Cereb. Cortex 2020, 30, 3781–3799. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, K.K.; Falkesgaard, M.H.; Winther, M.; Roed, N.K.; Quistgaard, C.L.; Teisen, M.N.; Edslev, S.M.; Petersen, D.L.; Aljubouri, A.; Christensen, C.; et al. NCAM2 Fibronectin type-III domains form a rigid structure that binds and activates the Fibroblast Growth Factor Receptor. Sci. Rep. 2018, 8, 8957. [Google Scholar] [CrossRef] [PubMed]
- Sheng, L.; Leshchyns’ka, I.; Sytnyk, V. Neural Cell Adhesion Molecule 2 (NCAM2)-Induced c-Src-Dependent Propagation of Submembrane Ca2+ Spikes Along Dendrites Inhibits Synapse Maturation. Cereb. Cortex 2019, 29, 1439–1459. [Google Scholar] [CrossRef]
- Kulahin, N.; Walmod, P.S. The neural cell adhesion molecule NCAM2/OCAM/RNCAM, a close relative to NCAM. Adv. Exp. Med. Biol. 2010, 663, 403–420. [Google Scholar] [CrossRef]
- Kim, W.; Watanabe, H.; Lomoio, S.; Tesco, G. Spatiotemporal processing of neural cell adhesion molecules 1 and 2 by BACE1 in vivo. J. Biol. Chem. 2021, 296, 100372. [Google Scholar] [CrossRef]
- Niland, S.; Eble, J.A. Neuropilins in the context of tumor vasculature. Int. J. Mol. Sci. 2019, 20, 639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neufeld, G.; Kessler, O. The semaphorins: Versatile regulators of tumour progression and tumour angiogenesis. Nat. Rev. Cancer 2008, 8, 632–645. [Google Scholar] [CrossRef] [PubMed]
- Gaur, P.; Bielenberg, D.R.; Samuel, S.; Bose, D.; Zhou, Y.; Gray, M.J.; Dallas, N.A.; Fan, F.; Xia, L.; Lu, J.; et al. Role of class 3 semaphorins and their receptors in tumor growth and angiogenesis. Clin. Cancer Res. 2009, 15, 6763–6770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khare, N.; Fascetti, N.; DaRocha, S.; Chiquet-Ehrismann, R.; Baumgartner, S. Expression patterns of two new members of the Semaphorin family in Drosophila suggest early functions during embryogenesis. Mech. Dev. 2000, 91, 393–397. [Google Scholar] [CrossRef]
- Xiao, W. Class 5 Semaphorins Mediate Synapse Elimination and Activity-Dependent Synaptic Plasticity in Hippocampal Neurons. Ph.D Thesis, University of British Columbia, Vancouver, BC, Canada, 2017. [Google Scholar]
- Mucka, P.; Levonyak, N.; Geretti, E.; Zwaans, B.M.M.; Li, X.; Adini, I.; Klagsbrun, M.; Adam, R.M.; Bielenberg, D.R. Inflammation and Lymphedema Are Exacerbated and Prolonged by Neuropilin 2 Deficiency. Am. J. Pathol. 2016, 186, 2803–2812. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Chen, Q.; Yin, D.; Shi, S.; Yu, L.; Zhou, S.; Chen, E.; Zhou, Z.; Shi, Y.; Fan, J.; et al. Novel role of semaphorin 3A in the growth and progression of hepatocellular carcinoma. Oncol. Rep. 2017, 37, 3313–3320. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, F.; Kalb, R.G.; Strittmatter, S.M. Molecular basis of semaphorin-mediated axon guidance. J. Neurobiol. 2000, 44, 219–229. [Google Scholar] [CrossRef]
- Tamagnone, L.; Comoglio, P.M. Signalling by semaphorin receptors: Cell guidance and beyond. Trends Cell Biol. 2000, 10, 377–383. [Google Scholar] [CrossRef]
- Iragavarapu-Charyulu, V.; Wojcikiewicz, E.; Urdaneta, A. Semaphorins in angiogenesis and autoimmune diseases: Therapeutic targets? Front. Immunol. 2020, 11, 346. [Google Scholar] [CrossRef]
- Geretti, E.; Klagsbrun, M. Neuropilins: Novel targets for anti-angiogenesis therapies. Cell Adh. Migr. 2007, 1, 56–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christensen, C.; Ambartsumian, N.; Gilestro, G.; Thomsen, B.; Comoglio, P.; Tamagnone, L.; Guldberg, P.; Lukanidin, E. Proteolytic processing converts the repelling signal Sema3E into an inducer of invasive growth and lung metastasis. Cancer Res. 2005, 65, 6167–6177. [Google Scholar] [CrossRef] [Green Version]
- Tian, L.; Coletti, D.; Li, Z.L. Angiotensin II induces the exocytosis of galectin-3 via integrin alphav/AKT/NF-kappaB signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7183. [Google Scholar] [CrossRef] [PubMed]
- Bielenberg, D.R.; Hida, Y.; Shimizu, A.; Kaipainen, A.; Kreuter, M.; Kim, C.C.; Klagsbrun, M. Semaphorin 3F, a chemorepulsant for endothelial cells, induces a poorly vascularized, encapsulated, nonmetastatic tumor phenotype. J. Clin. Investig. 2004, 114, 1260–1271. [Google Scholar] [CrossRef] [Green Version]
- Osada, R.; Horiuchi, A.; Kikuchi, N.; Ohira, S.; Ota, M.; Katsuyama, Y.; Konishi, I. Expression of semaphorins, vascular endothelial growth factor, and their common receptor neuropilins and alleic loss of semaphorin locus in epithelial ovarian neoplasms: Increased ratio of vascular endothelial growth factor to semaphorin is a poor prognostic factor in ovarian carcinomas. Hum. Pathol. 2006, 37, 1414–1425. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Rodriguez, E.R.; Reimert, D.V.; Shu, T.; Fritzsch, B.; Richards, L.J.; Kolodkin, A.L.; Ginty, D.D. Neuropilin-1 conveys semaphorin and VEGF signaling during neural and cardiovascular development. Dev. Cell 2003, 5, 45–57. [Google Scholar] [CrossRef] [Green Version]
- Giger, R.J.; Cloutier, J.-F.; Sahay, A.; Prinjha, R.K.; Levengood, D.V.; Moore, S.E.; Pickering, S.; Simmons, D.; Rastan, S.; Walsh, F.S.; et al. Neuropilin-2 Is Required In Vivo for Selective Axon Guidance Responses to Secreted Semaphorins. Neuron 2000, 25, 29–41. [Google Scholar] [CrossRef] [Green Version]
- Lantuejoul, S.; Constantin, B.; Drabkin, H.; Brambilla, C.; Roche, J.; Brambilla, E. Expression of VEGF, semaphorin SEMA3F, and their common receptors neuropilins NP1 and NP2 in preinvasive bronchial lesions, lung tumours, and cell lines. J. Pathol. 2003, 200, 336–347. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, W.; Cheever, T.; Schwarz, V.; Opperman, K.; Hutter, H.; Koepp, D.; Chen, L. The C. elegans L1CAM homologue LAD-2 functions as a coreceptor in MAB-20/Sema2 mediated axon guidance. J. Cell Biol. 2008, 180, 233–246. [Google Scholar] [CrossRef] [Green Version]
- Matkar, P.N.; Jong, E.D.; Ariyagunarajah, R.; Prud’homme, G.J.; Singh, K.K.; Leong-Poi, H. Jack of many trades: Multifaceted role of neuropilins in pancreatic cancer. Cancer Med. 2018, 7, 5036–5046. [Google Scholar] [CrossRef]
- Strubl, S.; Schubert, U.; Kühnle, A.; Rebl, A.; Ahmadvand, N.; Fischer, S.; Preissner, K.T.; Galuska, S.P. Polysialic acid is released by human umbilical vein endothelial cells (HUVEC) in vitro. Cell Biosci. 2018, 8, 64. [Google Scholar] [CrossRef]
- Grandclement, C.; Pallandre, J.R.; Valmary Degano, S.; Viel, E.; Bouard, A.; Balland, J.; Remy-Martin, J.P.; Simon, B.; Rouleau, A.; Boireau, W.; et al. Neuropilin-2 expression promotes TGF-beta1-mediated epithelial to mesenchymal transition in colorectal cancer cells. PLoS ONE 2011, 6, e20444. [Google Scholar] [CrossRef] [Green Version]
- Glinka, Y.; Stoilova, S.; Mohammed, N.; Prud’homme, G.J. Neuropilin-1 exerts co-receptor function for TGF-beta-1 on the membrane of cancer cells and enhances responses to both latent and active TGF-beta. Carcinogenesis 2011, 32, 613–621. [Google Scholar] [CrossRef] [Green Version]
- Matsushita, A.; Gotze, T.; Korc, M. Hepatocyte growth factor-mediated cell invasion in pancreatic cancer cells is dependent on neuropilin-1. Cancer Res. 2007, 67, 10309–10316. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Theis, T.; Kabat, M.; Loers, G.; Agre, L.A.; Schachner, M. Functions of Small Organic Compounds that Mimic the HNK-1 Glycan. Int. J. Mol. Sci. 2020, 21, 7018. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Vutskits, L.; Calaora, V.; Durbec, P.; Kiss, J.Z. A role for the polysialic acid-neural cell adhesion molecule in PDGF-induced chemotaxis of oligodendrocyte precursor cells. J. Cell Sci. 2004, 117, 93–103. [Google Scholar] [CrossRef] [Green Version]
- Hinek, A.; Bodnaruk, T.D.; Bunda, S.; Wang, Y.; Liu, K. Neuraminidase-1, a subunit of the cell surface elastin receptor, desialylates and functionally inactivates adjacent receptors interacting with the mitogenic growth factors PDGF-BB and IGF-2. Am. J. Pathol. 2008, 173, 1042–1056. [Google Scholar] [CrossRef] [Green Version]
- Muhl, L.; Folestad, E.B.; Gladh, H.; Wang, Y.; Moessinger, C.; Jakobsson, L.; Eriksson, U. Neuropilin 1 binds PDGF-D and is a co-receptor in PDGF-D–PDGFRβ signaling. J. Cell Sci. 2017, 130, 1365–1378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Francavilla, C.; Cattaneo, P.; Berezin, V.; Bock, E.; Ami, D.; de Marco, A.; Christofori, G.; Cavallaro, U. The binding of NCAM to FGFR1 induces a specific cellular response mediated by receptor trafficking. J. Cell Biol. 2009, 187, 1101–1116. [Google Scholar] [CrossRef]
- Christensen, C.; Berezin, V.; Bock, E. Neural cell adhesion molecule differentially interacts with isoforms of the fibroblast growth factor receptor. Neuroreport 2011, 22, 727–732. [Google Scholar] [CrossRef] [PubMed]
- Ono, S.; Hane, M.; Kitajima, K.; Sato, C. Novel regulation of fibroblast growth factor 2 (FGF2)-mediated cell growth by polysialic acid. J. Biol. Chem. 2012, 287, 3710–3722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cirovic, S.; Vjestica, J.; Mueller, C.A.; Tatic, S.; Vasiljevic, J.; Milenkovic, S.; Mueller, G.A.; Markovic-Lipkovski, J. NCAM and FGFR1 coexpression and colocalization in renal tumors. Int. J. Clin. Exp. Pathol. 2014, 7, 1402–1414. [Google Scholar]
- Kanato, Y.; Kitajima, K.; Sato, C. Direct binding of polysialic acid to a brain-derived neurotrophic factor depends on the degree of polymerization. Glycobiology 2008, 18, 1044–1053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monzo, H.J.; Park, T.I.; Dieriks, B.V.; Jansson, D.; Faull, R.L.; Dragunow, M.; Curtis, M.A. Insulin and IGF1 modulate turnover of polysialylated neural cell adhesion molecule (PSA-NCAM) in a process involving specific extracellular matrix components. J. Neurochem. 2013, 126, 758–770. [Google Scholar] [CrossRef]
- Lynch, C.C.; Vargo-Gogola, T.; Martin, M.D.; Fingleton, B.; Crawford, H.C.; Matrisian, L.M. Matrix metalloproteinase 7 mediates mammary epithelial cell tumorigenesis through the ErbB4 receptor. Cancer Res. 2007, 67, 6760–6767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, D.; Wu, Z.; Luo, L.J.; Huang, X.; Qian, W.Q.; Wang, H.; Li, S.H.; Liu, J. E-cadherin maintains the activity of neural stem cells and inhibits the migration. Int. J. Clin. Exp. Pathol. 2015, 8, 14247–14251. [Google Scholar]
- Qian, X.; Karpova, T.; Sheppard, A.M.; McNally, J.; Lowy, D.R. E-cadherin-mediated adhesion inhibits ligand-dependent activation of diverse receptor tyrosine kinases. EMBO J. 2004, 23, 1739–1748. [Google Scholar] [CrossRef] [Green Version]
- Alajati, A.; Guccini, I.; Pinton, S.; Garcia-Escudero, R.; Bernasocchi, T.; Sarti, M.; Montani, E.; Rinaldi, A.; Montemurro, F.; Catapano, C.; et al. Interaction of CDCP1 with HER2 enhances HER2-driven tumorigenesis and promotes trastuzumab resistance in breast cancer. Cell Rep. 2015, 11, 564–576. [Google Scholar] [CrossRef]
- Morath, I.; Jung, C.; Leveque, R.; Linfeng, C.; Toillon, R.A.; Warth, A.; Orian-Rousseau, V. Differential recruitment of CD44 isoforms by ErbB ligands reveals an involvement of CD44 in breast cancer. Oncogene 2018, 37, 1472–1484. [Google Scholar] [CrossRef]
- Donier, E.; Gomez-Sanchez, J.A.; Grijota-Martinez, C.; Lakoma, J.; Baars, S.; Garcia-Alonso, L.; Cabedo, H. L1CAM binds ErbB receptors through Ig-like domains coupling cell adhesion and neuregulin signalling. PLoS ONE 2012, 7, e40674. [Google Scholar] [CrossRef] [Green Version]
- Kawano, S.; Ikeda, W.; Kishimoto, M.; Ogita, H.; Takai, Y. Silencing of ErbB3/ErbB2 signaling by immunoglobulin-like Necl-2. J. Biol. Chem. 2009, 284, 23793–23805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Resovi, A.; Pinessi, D.; Chiorino, G.; Taraboletti, G. Current understanding of the thrombospondin-1 interactome. Matrix Biol. 2014, 37, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Latko, M.; Czyrek, A.; Porebska, N.; Kucinska, M.; Otlewski, J.; Zakrzewska, M.; Opalinski, L. Cross-Talk between Fibroblast Growth Factor Receptors and Other Cell Surface Proteins. Cells 2019, 8, 455. [Google Scholar] [CrossRef] [Green Version]
- Cavallaro, U.; Francavilla, C.; Loeffler, S.; Christofori, G. The NCAM/FGFR signaling complex: A novel player in metastatic dissemination. Clin. Exp. Metastas 2007, 24, 241–242. [Google Scholar]
- Eggers, K.; Werneburg, S.; Schertzinger, A.; Abeln, M.; Schiff, M.; Scharenberg, M.A.; Burkhardt, H.; Muhlenhoff, M.; Hildebrandt, H. Polysialic acid controls NCAM signals at cell-cell contacts to regulate focal adhesion independent from FGF receptor activity. J. Cell Sci. 2011, 124, 3279–3291. [Google Scholar] [CrossRef] [Green Version]
- Zecchini, S.; Bombardelli, L.; Decio, A.; Bianchi, M.; Mazzarol, G.; Sanguineti, F.; Aletti, G.; Maddaluno, L.; Berezin, V.; Bock, E.; et al. The adhesion molecule NCAM promotes ovarian cancer progression via FGFR signalling. EMBO Mol. Med. 2011, 3, 480–494. [Google Scholar] [CrossRef]
- Vales, A.; Kondo, R.; Aichberger, K.J.; Mayerhofer, M.; Kainz, B.; Sperr, W.R.; Sillaber, C.; Jager, U.; Valent, P. Myeloid leukemias express a broad spectrum of VEGF receptors including neuropilin-1 (NRP-1) and NRP-2. Leuk. Lymphoma 2007, 48, 1997–2007. [Google Scholar] [CrossRef]
- Guo, H.F.; Vander Kooi, C.W. Neuropilin Functions as an Essential Cell Surface Receptor. J. Biol. Chem. 2015, 290, 29120–29126. [Google Scholar] [CrossRef] [Green Version]
- Sarabipour, S.; Mac Gabhann, F. VEGF-A121a binding to Neuropilins—A concept revisited. Cell Adh. Migr. 2018, 12, 204–214. [Google Scholar] [CrossRef]
- Favier, B.; Alam, A.; Barron, P.; Bonnin, J.; Laboudie, P.; Fons, P.; Mandron, M.; Herault, J.P.; Neufeld, G.; Savi, P.; et al. Neuropilin-2 interacts with VEGFR-2 and VEGFR-3 and promotes human endothelial cell survival and migration. Blood 2006, 108, 1243–1250. [Google Scholar] [CrossRef]
- Hamerlik, P.; Lathia, J.D.; Rasmussen, R.; Wu, Q.; Bartkova, J.; Lee, M.; Moudry, P.; Bartek, J., Jr.; Fischer, W.; Lukas, J.; et al. Autocrine VEGF-VEGFR2-Neuropilin-1 signaling promotes glioma stem-like cell viability and tumor growth. J. Exp. Med. 2012, 209, 507–520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crnic, I.; Strittmatter, K.; Cavallaro, U.; Kopfstein, L.; Jussila, L.; Alitalo, K.; Christofori, G. Loss of neural cell adhesion molecule induces tumor metastasis by up-regulating lymphangiogenesis. Cancer Res. 2004, 64, 8630–8638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hrgovic, I.; Doll, M.; Pinter, A.; Kaufmann, R.; Kippenberger, S.; Meissner, M. Histone deacetylase inhibitors interfere with angiogenesis by decreasing endothelial VEGFR-2 protein half-life in part via a VE-cadherin-dependent mechanism. Exp. Dermatol. 2017, 26, 194–201. [Google Scholar] [CrossRef]
- Lampugnani, M.G.; Orsenigo, F.; Gagliani, M.C.; Tacchetti, C.; Dejana, E. Vascular endothelial cadherin controls VEGFR-2 internalization and signaling from intracellular compartments. J. Cell Biol. 2006, 174, 593–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shintani, Y.; Takashima, S.; Asano, Y.; Kato, H.; Liao, Y.; Yamazaki, S.; Tsukamoto, O.; Seguchi, O.; Yamamoto, H.; Fukushima, T.; et al. Glycosaminoglycan modification of neuropilin-1 modulates VEGFR2 signaling. EMBO J. 2006, 25, 3045–3055. [Google Scholar] [CrossRef] [Green Version]
- Stanton, M.J.; Dutta, S.; Zhang, H.; Polavaram, N.S.; Leontovich, A.A.; Honscheid, P.; Sinicrope, F.A.; Tindall, D.J.; Muders, M.H.; Datta, K. Autophagy control by the VEGF-C/NRP-2 axis in cancer and its implication for treatment resistance. Cancer Res. 2013, 73, 160–171. [Google Scholar] [CrossRef] [Green Version]
- Koch, S. Neuropilin signalling in angiogenesis. Biochem. Soc. Trans. 2012, 40, 20–25. [Google Scholar] [CrossRef] [Green Version]
- Raimondi, C. Neuropilin-1 enforces extracellular matrix signalling via ABL1 to promote angiogenesis. Biochem. Soc. Trans. 2014, 42, 1429–1434. [Google Scholar] [CrossRef]
- Jastrzebski, K.; Zdzalik-Bielecka, D.; Maminska, A.; Kalaidzidis, Y.; Hellberg, C.; Miaczynska, M. Multiple routes of endocytic internalization of PDGFRbeta contribute to PDGF-induced STAT3 signaling. J. Cell Sci. 2017, 130, 577–589. [Google Scholar] [CrossRef] [Green Version]
- Ueda, Y.; Kedashiro, S.; Maruoka, M.; Mizutani, K.; Takai, Y. Roles of the third Ig-like domain of Necl-5/PVR and the fifth Ig-like domain of the PDGF receptor in its signaling. Genes Cells 2018, 23, 214–224. [Google Scholar] [CrossRef]
- Gopal, S. Syndecans in inflammation at a glance. Front. Immunol. 2020, 11, 227. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Klass, C.; Woods, A. Syndecan-2 regulates transforming growth factor-beta signaling. J. Biol. Chem. 2004, 279, 15715–15718. [Google Scholar] [CrossRef] [Green Version]
- Gomes, C.; Osorio, H.; Pinto, M.T.; Campos, D.; Oliveira, M.J.; Reis, C.A. Expression of ST3GAL4 leads to SLe(x) expression and induces c-Met activation and an invasive phenotype in gastric carcinoma cells. PLoS ONE 2013, 8, e66737. [Google Scholar] [CrossRef]
- Horiguchi, H.; Tsujimoto, H.; Shinomiya, N.; Matsumoto, Y.; Sugasawa, H.; Yamori, T.; Miyazaki, H.; Saitoh, D.; Kishi, Y.; Ueno, H. A Potential Role of Adhesion Molecules on Lung Metastasis Enhanced by Local Inflammation. Anticancer Res. 2020, 40, 6171–6178. [Google Scholar] [CrossRef]
- Zhang, X.; Shao, S.; Li, L. Characterization of Class-3 Semaphorin Receptors, Neuropilins and Plexins, as Therapeutic Targets in a Pan-Cancer Study. Cancers 2020, 12, 1816. [Google Scholar] [CrossRef]
- Janssen, B.J.; Malinauskas, T.; Weir, G.A.; Cader, M.Z.; Siebold, C.; Jones, E.Y. Neuropilins lock secreted semaphorins onto plexins in a ternary signaling complex. Nat. Struct. Mol. Biol 2012, 19, 1293–1299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef] [PubMed]
- Kleene, R.; Schachner, M. Glycans and neural cell interactions. Nat. Rev. Neurosci. 2004, 5, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Chen, S.; Li, Q.; Sheng, Y.; Alvarez, M.R.; Reyes, J.; Xu, G.; Solakyildirim, K.; Lebrilla, C.B. Glycan–protein cross-linking mass spectrometry reveals sialic acid-mediated protein networks on cell surfaces. Chem. Sci. 2021, 12, 8767–8777. [Google Scholar] [CrossRef]
- Buhe, V.; Loisel, S.; Pers, J.O.; Le Ster, K.; Berthou, C.; Youinou, P. Updating the physiology, exploration and disease relevance of complement factor H. Int. J. Immunopath. Pharmacol. 2010, 23, 397–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karlstetter, M.; Kopatz, J.; Aslanidis, A.; Shahraz, A.; Caramoy, A.; Linnartz-Gerlach, B.; Lin, Y.; Luckoff, A.; Fauser, S.; Duker, K.; et al. Polysialic acid blocks mononuclear phagocyte reactivity, inhibits complement activation, and protects from vascular damage in the retina. EMBO Mol. Med. 2017, 9, 154–166. [Google Scholar] [CrossRef]
- Meesmann, H.M.; Fehr, E.M.; Kierschke, S.; Herrmann, M.; Bilyy, R.; Heyder, P.; Blank, N.; Krienke, S.; Lorenz, H.M.; Schiller, M. Decrease of sialic acid residues as an eat-me signal on the surface of apoptotic lymphocytes. J. Cell Sci. 2010, 123, 3347–3356. [Google Scholar] [CrossRef] [Green Version]
- Gohari, B.; Abu-Zahra, N. Polyethersulfone Membranes Prepared with 3-Aminopropyltriethoxysilane Modified Alumina Nanoparticles for Cu(II) Removal from Water. ACS Omega 2018, 3, 10154–10162. [Google Scholar] [CrossRef] [Green Version]
- Jarahian, M.; Watzl, C.; Fournier, P.; Arnold, A.; Djandji, D.; Zahedi, S.; Cerwenka, A.; Paschen, A.; Schirrmacher, V.; Momburg, F. Activation of natural killer cells by newcastle disease virus hemagglutinin-neuraminidase. J. Virol. 2009, 83, 8108–8121. [Google Scholar] [CrossRef] [Green Version]
- Jarahian, M.; Fiedler, M.; Cohnen, A.; Djandji, D.; Hammerling, G.J.; Gati, C.; Cerwenka, A.; Turner, P.C.; Moyer, R.W.; Watzl, C.; et al. Modulation of NKp30- and NKp46-mediated natural killer cell responses by poxviral hemagglutinin. PLoS Pathog. 2011, 7, e1002195. [Google Scholar] [CrossRef] [PubMed]
- Crocker, P.R.; Paulson, J.C.; Varki, A. Siglecs and their roles in the immune system. Nat. Rev. Immunol. 2007, 7, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Yau, T.; Dan, X.; Ng, C.C.; Ng, T.B. Lectins with potential for anti-cancer therapy. Molecules 2015, 20, 3791–3810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, A.; Carvalho, F.C.; Roque-Barreira, M.C.; Bueno, P.R. Impedance-derived electrochemical capacitance spectroscopy for the evaluation of lectin-glycoprotein binding affinity. Biosens. Bioelectron. 2014, 62, 102–105. [Google Scholar] [CrossRef]
- Barrow, A.D.; Colonna, M. Exploiting NK Cell Surveillance Pathways for Cancer Therapy. Cancers 2019, 11, 55. [Google Scholar] [CrossRef] [Green Version]
- Mazalovska, M.; Kouokam, J.C. Lectins as Promising Therapeutics for the Prevention and Treatment of HIV and Other Potential Coinfections. Biomed. Res. Int. 2018, 2018, 3750646. [Google Scholar] [CrossRef]
- Brown, G.D.; Willment, J.A.; Whitehead, L. C-type lectins in immunity and homeostasis. Nat. Rev. Immunol. 2018, 18, 374–389. [Google Scholar] [CrossRef] [PubMed]
- Barthel, S.R.; Gavino, J.D.; Descheny, L.; Dimitroff, C.J. Targeting selectins and selectin ligands in inflammation and cancer. Expert Opin. Ther. Tar. 2007, 11, 1473–1491. [Google Scholar] [CrossRef]
- Zeisig, R.; Stahn, R.; Wenzel, K.; Behrens, D.; Fichtner, I. Effect of sialyl Lewis X-glycoliposomes on the inhibition of E-selectin-mediated tumour cell adhesion in vitro. BBA-Gen. Subj. 2004, 1660, 31–40. [Google Scholar] [CrossRef] [Green Version]
- Barthel, S.R.; Gavino, J.D.; Wiese, G.K.; Jaynes, J.M.; Siddiqui, J.; Dimitroff, C.J. Analysis of glycosyltransferase expression in metastatic prostate cancer cells capable of rolling activity on microvascular endothelial (E)-selectin. Glycobiology 2008, 18, 806–817. [Google Scholar] [CrossRef] [PubMed]
- Silva-Filho, A.F.; Sena, W.L.B.; Lima, L.R.A.; Carvalho, L.V.N.; Pereira, M.C.; Santos, L.G.S.; Santos, R.V.C.; Tavares, L.B.; Pitta, M.G.R.; Rego, M. Glycobiology Modifications in Intratumoral Hypoxia: The Breathless Side of Glycans Interaction. Cell Physiol. Biochem. 2017, 41, 1801–1829. [Google Scholar] [CrossRef]
- Silva, M.; Videira, P.A.; Sackstein, R. E-Selectin Ligands in the Human Mononuclear Phagocyte System: Implications for Infection, Inflammation, and Immunotherapy. Front. Immunol. 2017, 8, 1878. [Google Scholar] [CrossRef] [Green Version]
- Bornhofft, K.F.; Goldammer, T.; Rebl, A.; Galuska, S.P. Siglecs: A journey through the evolution of sialic acid-binding immunoglobulin-type lectins. Dev. Comp. Immunol. 2018, 86, 219–231. [Google Scholar] [CrossRef]
- Macauley, M.S.; Crocker, P.R.; Paulson, J.C. Siglec-mediated regulation of immune cell function in disease. Nat. Rev. Immunol. 2014, 14, 653–666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsubata, T. Ligand Recognition Determines the Role of Inhibitory B Cell Co-receptors in the Regulation of B Cell Homeostasis and Autoimmunity. Front. Immunol. 2018, 9, 2276. [Google Scholar] [CrossRef]
- Baba, Y.; Matsumoto, M.; Kurosaki, T. Calcium signaling in B cells: Regulation of cytosolic Ca2+ increase and its sensor molecules, STIM1 and STIM2. Mol. Immunol. 2014, 62, 339–343. [Google Scholar] [CrossRef] [Green Version]
- Gerlach, J.; Ghosh, S.; Jumaa, H.; Reth, M.; Wienands, J.; Chan, A.C.; Nitschke, L. B cell defects in SLP65/BLNK-deficient mice can be partially corrected by the absence of CD22, an inhibitory coreceptor for BCR signaling. Eur. J. Immunol. 2003, 33, 3418–3426. [Google Scholar] [CrossRef]
- Crocker, P.R.; Redelinghuys, P. Siglecs as positive and negative regulators of the immune system. Biochem. Soc. Trans. 2008, 36, 1467–1471. [Google Scholar] [CrossRef] [PubMed]
- Ram, S.; Gulati, S.; Lewis, L.A.; Chakraborti, S.; Zheng, B.; DeOliveira, R.B.; Reed, G.W.; Cox, A.D.; Li, J.; St Michael, F.; et al. A Novel Sialylation Site on Neisseria gonorrhoeae Lipooligosaccharide Links Heptose II Lactose Expression with Pathogenicity. Infect. Immun. 2018, 86, e00285-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, J.; Li, J.; Liu, S.M.; Feng, X.Y.; Chen, S.; Chen, Y.B.; Zhang, X.S. CD33(+)/p-STAT1(+) double-positive cell as a prognostic factor for stage IIIa gastric cancer. Med. Oncol. 2013, 30, 442. [Google Scholar] [CrossRef] [Green Version]
- Ishikawa, J.; Kano, F.; Ando, Y.; Hibi, H.; Yamamoto, A. Monocyte chemoattractant protein-1 and secreted ectodomain of sialic acid-binding Ig-like lectin-9 enhance bone regeneration by inducing M2 macrophages. J. Oral Maxillofac. Surg. Med. Pathol. 2019, 31, 169–174. [Google Scholar] [CrossRef]
- McMillan, S.J.; Crocker, P.R. CD33-related sialic-acid-binding immunoglobulin-like lectins in health and disease. Carbohyd. Res. 2008, 343, 2050–2056. [Google Scholar] [CrossRef]
- Pillai, S.; Netravali, I.A.; Cariappa, A.; Mattoo, H. Siglecs and immune regulation. Annu. Rev. Immunol. 2012, 30, 357–392. [Google Scholar] [CrossRef] [Green Version]
- Fraschilla, I.; Pillai, S. Viewing Siglecs through the lens of tumor immunology. Immunol. Rev. 2017, 276, 178–191. [Google Scholar] [CrossRef] [Green Version]
- Kakio, A.; Yano, Y.; Takai, D.; Kuroda, Y.; Matsumoto, O.; Kozutsumi, Y.; Matsuzaki, K. Interaction between amyloid beta-protein aggregates and membranes. J. Pept. Sci. 2004, 10, 612–621. [Google Scholar] [CrossRef]
- Cowan, C.B.; Patel, D.A.; Good, T.A. Exploring the mechanism of beta-amyloid toxicity attenuation by multivalent sialic acid polymers through the use of mathematical models. J. Theor. Biol. 2009, 258, 189–197. [Google Scholar] [CrossRef] [Green Version]
- Hollingworth, P.; Harold, D.; Sims, R.; Gerrish, A.; Lambert, J.C.; Carrasquillo, M.M.; Abraham, R.; Hamshere, M.L.; Pahwa, J.S.; Moskvina, V.; et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat. Genet. 2011, 43, 429–435. [Google Scholar] [CrossRef] [Green Version]
- Malik, M.; Simpson, J.F.; Parikh, I.; Wilfred, B.R.; Fardo, D.W.; Nelson, P.T.; Estus, S. CD33 Alzheimer’s risk-altering polymorphism, CD33 expression, and exon 2 splicing. J. Neurosci. 2013, 33, 13320–13325. [Google Scholar] [CrossRef] [Green Version]
- Malik, M.; Chiles, J., 3rd; Xi, H.S.; Medway, C.; Simpson, J.; Potluri, S.; Howard, D.; Liang, Y.; Paumi, C.M.; Mukherjee, S.; et al. Genetics of CD33 in Alzheimer’s disease and acute myeloid leukemia. Hum. Mol. Genet. 2015, 24, 3557–3570. [Google Scholar] [CrossRef] [Green Version]
- Griciuc, A.; Serrano-Pozo, A.; Parrado, A.R.; Lesinski, A.N.; Asselin, C.N.; Mullin, K.; Hooli, B.; Choi, S.H.; Hyman, B.T.; Tanzi, R.E. Alzheimer’s disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron 2013, 78, 631–643. [Google Scholar] [CrossRef] [Green Version]
- Heneka, M.T.; Golenbock, D.T.; Latz, E. Innate immunity in Alzheimer’s disease. Nat. Immunol. 2015, 16, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Jandus, C.; Boligan, K.F.; Chijioke, O.; Liu, H.; Dahlhaus, M.; Demoulins, T.; Schneider, C.; Wehrli, M.; Hunger, R.E.; Baerlocher, G.M.; et al. Interactions between Siglec-7/9 receptors and ligands influence NK cell-dependent tumor immunosurveillance. J. Clin. Investig. 2014, 124, 1810–1820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicoll, G.; Avril, T.; Lock, K.; Furukawa, K.; Bovin, N.; Crocker, P.R. Ganglioside GD3 expression on target cells can modulate NK cell cytotoxicity via siglec-7-dependent and -independent mechanisms. Eur. J. Immunol. 2003, 33, 1642–1648. [Google Scholar] [CrossRef] [PubMed]
- Kitajima, K. Structural Diversity and Evolution of Sialic Acids. Trends Glycosci. Glycotechnol. 2019, 31, SE18–SE20. [Google Scholar] [CrossRef]
- Ali, S.R.; Fong, J.J.; Carlin, A.F.; Busch, T.D.; Linden, R.; Angata, T.; Areschoug, T.; Parast, M.; Varki, N.; Murray, J.; et al. Siglec-5 and Siglec-14 are polymorphic paired receptors that modulate neutrophil and amnion signaling responses to group B Streptococcus. J. Exp. Med. 2014, 211, 1231–1242. [Google Scholar] [CrossRef]
- Avril, T.; Attrill, H.; Zhang, J.; Raper, A.; Crocker, P.R. Negative regulation of leucocyte functions by CD33-related siglecs. Biochem. Soc. Trans. 2006, 34, 1024–1027. [Google Scholar] [CrossRef] [Green Version]
- Lubbers, J.; Rodriguez, E.; van Kooyk, Y. Modulation of Immune Tolerance via Siglec-Sialic Acid Interactions. Front. Immunol. 2018, 9, 2807. [Google Scholar] [CrossRef] [Green Version]
- Stanczak, M.A.; Siddiqui, S.S.; Trefny, M.P.; Thommen, D.S.; Boligan, K.F.; von Gunten, S.; Tzankov, A.; Tietze, L.; Lardinois, D.; Heinzelmann-Schwarz, V.; et al. Self-associated molecular patterns mediate cancer immune evasion by engaging Siglecs on T cells. J. Clin. Investig. 2018, 128, 4912–4923. [Google Scholar] [CrossRef] [Green Version]
- Duan, S.; Paulson, J.C. Siglecs as immune cell checkpoints in disease. Annu. Rev. Immunol. 2020, 38, 365–395. [Google Scholar] [CrossRef] [Green Version]
- Kolar, P.; Knieke, K.; Hegel, J.K.; Quandt, D.; Burmester, G.R.; Hoff, H.; Brunner-Weinzierl, M.C. CTLA-4 (CD152) controls homeostasis and suppressive capacity of regulatory T cells in mice. Arthritis Rheum. 2009, 60, 123–132. [Google Scholar] [CrossRef]
- Syn, N.L.; Teng, M.W.L.; Mok, T.S.K.; Soo, R.A. De-novo and acquired resistance to immune checkpoint targeting. Lancet. Oncol. 2017, 18, e731–e741. [Google Scholar] [CrossRef]
- Philips, G.K.; Atkins, M. Therapeutic uses of anti-PD-1 and anti-PD-L1 antibodies. Int. Immunol. 2015, 27, 39–46. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.F.; Wu, L.; Yang, L.L.; Deng, W.W.; Mao, L.; Wu, H.; Zhang, W.F.; Sun, Z.J. Blockade of TIM3 relieves immunosuppression through reducing regulatory T cells in head and neck cancer. J. Exp. Clin. Canc. Res. 2018, 37, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chien, M.W.; Lin, M.H.; Huang, S.H.; Fu, S.H.; Hsu, C.Y.; Yen, B.L.; Chen, J.T.; Chang, D.M.; Sytwu, H.K. Glucosamine Modulates T Cell Differentiation through Down-regulating N-Linked Glycosylation of CD25. J. Biol. Chem. 2015, 290, 29329–29344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.T.; Chen, C.H.; Ku, K.L.; Hsiao, M.; Chiang, C.P.; Hsu, T.L.; Chen, M.H.; Wong, C.H. Glycoprotein B7-H3 overexpression and aberrant glycosylation in oral cancer and immune response. Proc. Natl. Acad. Sci. USA 2015, 112, 13057–13062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, M.S.; Alves, I.; Vicente, M.; Campar, A.; Silva, M.C.; Padrao, N.A.; Pinto, V.; Fernandes, A.; Dias, A.M.; Pinho, S.S. Glycans as Key Checkpoints of T Cell Activity and Function. Front. Immunol. 2018, 9, 2754. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.S.; Foell, J.; McCausland, M.; Strahotin, S.; Niu, L.; Bapat, A.; Hewes, L.B. Anti-CD137 antibodies in the treatment of autoimmune disease and cancer. Immunol. Res. 2004, 29, 197–208. [Google Scholar] [CrossRef]
- Eastwood, D.; Findlay, L.; Poole, S.; Bird, C.; Wadhwa, M.; Moore, M.; Burns, C.; Thorpe, R.; Stebbings, R. Monoclonal antibody TGN1412 trial failure explained by species differences in CD28 expression on CD4+ effector memory T-cells. Br. J. Pharmacol. 2010, 161, 512–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, R.; Kim, A.M.J.; Lim, S.-O. Glycosylation of Immune Receptors in Cancer. Cells 2021, 10, 1100. [Google Scholar] [CrossRef]
- Liu, Z.; Swindall, A.F.; Kesterson, R.A.; Schoeb, T.R.; Bullard, D.C.; Bellis, S.L. ST6Gal-I regulates macrophage apoptosis via alpha2-6 sialylation of the TNFR1 death receptor. J. Biol. Chem. 2011, 286, 39654–39662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef] [Green Version]
- Aicher, A.; Hayden-Ledbetter, M.; Brady, W.A.; Pezzutto, A.; Richter, G.; Magaletti, D.; Buckwalter, S.; Ledbetter, J.A.; Clark, E.A. Characterization of human inducible costimulator ligand expression and function. J. Immunol. 2000, 164, 4689–4696. [Google Scholar] [CrossRef] [Green Version]
- Rujas, E.; Cui, H.; Sicard, T.; Semesi, A.; Julien, J.-P. Structural characterization of the ICOS/ICOS-L immune complex reveals high molecular mimicry by therapeutic antibodies. Nat. Commun. 2020, 11, 5066. [Google Scholar] [CrossRef]
- Sun, L.; Li, C.-W.; Chung, E.M.; Yang, R.; Kim, Y.-S.; Park, A.H.; Lai, Y.-J.; Yang, Y.; Wang, Y.-H.; Liu, J. Targeting glycosylated PD-1 induces potent antitumor immunity. Cancer Res. 2020, 80, 2298–2310. [Google Scholar]
- Lin, D.Y.; Tanaka, Y.; Iwasaki, M.; Gittis, A.G.; Su, H.P.; Mikami, B.; Okazaki, T.; Honjo, T.; Minato, N.; Garboczi, D.N. The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors. Proc. Natl. Acad. Sci. USA 2008, 105, 3011–3016. [Google Scholar] [CrossRef] [Green Version]
- Zak, K.M.; Grudnik, P.; Magiera, K.; Domling, A.; Dubin, G.; Holak, T.A. Structural Biology of the Immune Checkpoint Receptor PD-1 and Its Ligands PD-L1/PD-L2. Structure 2017, 25, 1163–1174. [Google Scholar] [CrossRef]
- Dong, Y.; Sun, Q.; Zhang, X. PD-1 and its ligands are important immune checkpoints in cancer. Oncotarget 2017, 8, 2171–2186. [Google Scholar] [CrossRef] [Green Version]
- Redelinghuys, P.; Antonopoulos, A.; Liu, Y.; Campanero-Rhodes, M.A.; McKenzie, E.; Haslam, S.M.; Dell, A.; Feizi, T.; Crocker, P.R. Early murine T-lymphocyte activation is accompanied by a switch from N-Glycolyl- to N-acetyl-neuraminic acid and generation of ligands for siglec-E. J. Biol. Chem. 2011, 286, 34522–34532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kavvoura, F.K.; Ioannidis, J.P. CTLA-4 gene polymorphisms and susceptibility to type 1 diabetes mellitus: A HuGE Review and meta-analysis. Am. J. Epidemiol. 2005, 162, 3–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez Allo, V.C.; Hauk, V.; Sarbia, N.; Pinto, N.A.; Croci, D.O.; Dalotto-Moreno, T.; Morales, R.M.; Gatto, S.G.; Manselle Cocco, M.N.; Stupirski, J.C.; et al. Suppression of age-related salivary gland autoimmunity by glycosylation-dependent galectin-1-driven immune inhibitory circuits. Proc. Natl. Acad. Sci. USA 2020, 117, 6630–6639. [Google Scholar] [CrossRef] [PubMed]
- Elola, M.T.; Wolfenstein-Todel, C.; Troncoso, M.F.; Vasta, G.R.; Rabinovich, G.A. Galectins: Matricellular glycan-binding proteins linking cell adhesion, migration, and survival. Cell Mol. Life Sci. 2007, 64, 1679–1700. [Google Scholar] [CrossRef] [PubMed]
- Chou, F.C.; Chen, H.Y.; Kuo, C.C.; Sytwu, H.K. Role of Galectins in Tumors and in Clinical Immunotherapy. Int. J. Mol. Sci. 2018, 19, 430. [Google Scholar] [CrossRef] [Green Version]
- Vasta, G.R.; Feng, C.; Gonzalez-Montalban, N.; Mancini, J.; Yang, L.; Abernathy, K.; Frost, G.; Palm, C. Functions of galectins as ‘self/non-self’-recognition and effector factors. Pathog. Dis. 2017, 75, ftx046. [Google Scholar] [CrossRef] [Green Version]
- Wan, L.; Yang, R.Y.; Liu, F.T. Galectin-12 in Cellular Differentiation, Apoptosis and Polarization. Int. J. Mol. Sci. 2018, 19, 176. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Huang, Y.; Zhang, J.; Xing, B.; Xuan, W.; Wang, H.; Huang, H.; Yang, J.; Tang, J. NRP-2 in tumor lymphangiogenesis and lymphatic metastasis. Cancer Lett. 2018, 418, 176–184. [Google Scholar] [CrossRef]
- Galvan, M.; Tsuboi, S.; Fukuda, M.; Baum, L.G. Expression of a specific glycosyltransferase enzyme regulates T cell death mediated by galectin-1. J. Biol. Chem. 2000, 275, 16730–16737. [Google Scholar] [CrossRef] [Green Version]
- Garin, M.I.; Chu, C.C.; Golshayan, D.; Cernuda-Morollon, E.; Wait, R.; Lechler, R.I. Galectin-1: A key effector of regulation mediated by CD4+CD25+ T cells. Blood 2007, 109, 2058–2065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sturm, A.; Lensch, M.; Andre, S.; Kaltner, H.; Wiedenmann, B.; Rosewicz, S.; Dignass, A.U.; Gabius, H.J. Human galectin-2: Novel inducer of T cell apoptosis with distinct profile of caspase activation. J. Immunol. 2004, 173, 3825–3837. [Google Scholar] [CrossRef] [Green Version]
- Seki, M.; Oomizu, S.; Sakata, K.M.; Sakata, A.; Arikawa, T.; Watanabe, K.; Ito, K.; Takeshita, K.; Niki, T.; Saita, N.; et al. Galectin-9 suppresses the generation of Th17, promotes the induction of regulatory T cells, and regulates experimental autoimmune arthritis. Clin. Immunol. 2008, 127, 78–88. [Google Scholar] [CrossRef]
- Clark, M.C.; Baum, L.G. T cells modulate glycans on CD43 and CD45 during development and activation, signal regulation, and survival. Ann. N. Y. Acad. Sci. 2012, 1253, 58–67. [Google Scholar] [CrossRef] [Green Version]
- Kadaja-Saarepuu, L.; Laos, S.; Jaager, K.; Viil, J.; Balikova, A.; Looke, M.; Hansson, G.C.; Maimets, T. CD43 promotes cell growth and helps to evade FAS-mediated apoptosis in non-hematopoietic cancer cells lacking the tumor suppressors p53 or ARF. Oncogene 2008, 27, 1705–1715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Earl, L.A.; Bi, S.; Baum, L.G. N- and O-glycans modulate galectin-1 binding, CD45 signaling, and T cell death. J. Biol. Chem. 2010, 285, 2232–2244. [Google Scholar] [CrossRef] [Green Version]
- Woodard-Grice, A.V.; McBrayer, A.C.; Wakefield, J.K.; Zhuo, Y.; Bellis, S.L. Proteolytic shedding of ST6Gal-I by BACE1 regulates the glycosylation and function of alpha4beta1 integrins. J. Biol. Chem. 2008, 283, 26364–26373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pretzlaff, R.K.; Xue, V.W.; Rowin, M.E. Sialidase treatment exposes the beta1-integrin active ligand binding site on HL60 cells and increases binding to fibronectin. Cell Adhes. Commun. 2000, 7, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Chang, V.T.; Fernandes, R.A.; Ganzinger, K.A.; Lee, S.F.; Siebold, C.; McColl, J.; Jönsson, P.; Palayret, M.; Harlos, K.; Coles, C.H. Initiation of T cell signaling by CD45 segregation at‘close contacts’. Nat. Immunol. 2016, 17, 574–582. [Google Scholar] [CrossRef] [Green Version]
- Stroop, C.J.; Weber, W.; Gerwig, G.J.; Nimtz, M.; Kamerling, J.P.; Vliegenthart, J.F. Characterization of the carbohydrate chains of the secreted form of the human epidermal growth factor receptor. Glycobiology 2000, 10, 901–917. [Google Scholar] [CrossRef] [Green Version]
- Kitazume, S.; Imamaki, R.; Kurimoto, A.; Ogawa, K.; Kato, M.; Yamaguchi, Y.; Tanaka, K.; Ishida, H.; Ando, H.; Kiso, M.; et al. Interaction of platelet endothelial cell adhesion molecule (PECAM) with alpha2,6-sialylated glycan regulates its cell surface residency and anti-apoptotic role. J. Biol. Chem. 2014, 289, 27604–27613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dall’Olio, F.; Malagolini, N.; Trinchera, M.; Chiricolo, M. Sialosignaling: Sialyltransferases as engines of self-fueling loops in cancer progression. BBA-Gen. Subj. 2014, 1840, 2752–2764. [Google Scholar] [CrossRef] [Green Version]
- Cheung, I.Y.; Vickers, A.; Cheung, N.K. Sialyltransferase STX (ST8SiaII): A novel molecular marker of metastatic neuroblastoma. Int. J. Cancer 2006, 119, 152–156. [Google Scholar] [CrossRef]
- Cazet, A.; Julien, S.; Bobowski, M.; Burchell, J.; Delannoy, P. Tumour-associated carbohydrate antigens in breast cancer. Breast Cancer Res. 2010, 12, 204. [Google Scholar] [CrossRef] [Green Version]
- Schultz, M.J.; Swindall, A.F.; Bellis, S.L. Regulation of the metastatic cell phenotype by sialylated glycans. Cancer Metast Rev. 2012, 31, 501–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, G.G.; Berger, S.B.; Sadighi Akha, A.A.; Miller, R.A. Age-associated changes in glycosylation of CD43 and CD45 on mouse CD4 T cells. Eur. J. Immunol. 2005, 35, 622–631. [Google Scholar] [CrossRef] [PubMed]
- Stillman, B.N.; Hsu, D.K.; Pang, M.; Brewer, C.F.; Johnson, P.; Liu, F.T.; Baum, L.G. Galectin-3 and galectin-1 bind distinct cell surface glycoprotein receptors to induce T cell death. J. Immunol. 2006, 176, 778–789. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Dong, L.; Chang, P. CD44v6 engages in colorectal cancer progression. Cell Death Dis. 2019, 10, 30. [Google Scholar] [CrossRef]
- Zhao, Y.; Sato, Y.; Isaji, T.; Fukuda, T.; Matsumoto, A.; Miyoshi, E.; Gu, J.; Taniguchi, N. Branched N-glycans regulate the biological functions of integrins and cadherins. FEBS J. 2008, 275, 1939–1948. [Google Scholar] [CrossRef]
- Fukumori, T.; Takenaka, Y.; Yoshii, T.; Kim, H.R.; Hogan, V.; Inohara, H.; Kagawa, S.; Raz, A. CD29 and CD7 mediate galectin-3-induced type II T-cell apoptosis. Cancer Res. 2003, 63, 8302–8311. [Google Scholar] [PubMed]
- Baycin-Hizal, D.; Gottschalk, A.; Jacobson, E.; Mai, S.; Wolozny, D.; Zhang, H.; Krag, S.S.; Betenbaugh, M.J. Physiologic and pathophysiologic consequences of altered sialylation and glycosylation on ion channel function. Biochem. Biophys. Res. Commun. 2014, 453, 243–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, C.; Kitajima, K. Polysialylation and disease. Mol. Asp. Med. 2020, 79, 100892. [Google Scholar] [CrossRef] [PubMed]
- Bull, C.; Collado-Camps, E.; Kers-Rebel, E.D.; Heise, T.; Sondergaard, J.N.; den Brok, M.H.; Schulte, B.M.; Boltje, T.J.; Adema, G.J. Metabolic sialic acid blockade lowers the activation threshold of moDCs for TLR stimulation. Immunol. Cell Biol. 2017, 95, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.; Silva, Z.; Marques, G.; Ferro, T.; Goncalves, M.; Monteiro, M.; van Vliet, S.J.; Mohr, E.; Lino, A.C.; Fernandes, A.R.; et al. Sialic acid removal from dendritic cells improves antigen cross-presentation and boosts anti-tumor immune responses. Oncotarget 2016, 7, 41053–41066. [Google Scholar] [CrossRef]
- Bull, C.; Heise, T.; Adema, G.J.; Boltje, T.J. Sialic Acid Mimetics to Target the Sialic Acid-Siglec Axis. Trends Biochem. Sci. 2016, 41, 519–531. [Google Scholar] [CrossRef] [PubMed]
- Cerliani, J.P.; Blidner, A.G.; Toscano, M.A.; Croci, D.O.; Rabinovich, G.A. Translating the ‘Sugar Code’ into Immune and Vascular Signaling Programs. Trends Biochem. Sci. 2017, 42, 255–273. [Google Scholar] [CrossRef]
- Adams, R.; Brown, E.; Brown, L.; Butler, R.; Falk, S.; Fisher, D.; Kaplan, R.; Quirke, P.; Richman, S.; Samuel, L.; et al. Inhibition of EGFR, HER2, and HER3 signalling in patients with colorectal cancer wild-type for BRAF, PIK3CA, KRAS, and NRAS (FOCUS4-D): A phase 2–3 randomised trial. Lancet Gastroenterol. Hepatol. 2018, 3, 162–171. [Google Scholar] [CrossRef] [Green Version]
- Wittmann, P.; Grubinger, M.; Groger, C.; Huber, H.; Sieghart, W.; Peck-Radosavljevic, M.; Mikulits, W. Neuropilin-2 induced by transforming growth factor-beta augments migration of hepatocellular carcinoma cells. BMC Cancer 2015, 15, 909. [Google Scholar] [CrossRef] [Green Version]
- Patel, D.A.; Henry, J.E.; Good, T.A. Attenuation of beta-amyloid-induced toxicity by sialic-acid-conjugated dendrimers: Role of sialic acid attachment. Brain Res. 2007, 1161, 95–105. [Google Scholar] [CrossRef] [Green Version]
- Malicdan, M.C.; Noguchi, S.; Nonaka, I.; Hayashi, Y.K.; Nishino, I. A Gne knockout mouse expressing human GNE D176V mutation develops features similar to distal myopathy with rimmed vacuoles or hereditary inclusion body myopathy. Hum. Mol. Genet. 2007, 16, 2669–2682. [Google Scholar] [CrossRef] [Green Version]
- Murray, H.C.; Low, V.F.; Swanson, M.E.; Dieriks, B.V.; Turner, C.; Faull, R.L.; Curtis, M.A. Distribution of PSA-NCAM in normal, Alzheimer’s and Parkinson’s disease human brain. Neuroscience 2016, 330, 359–375. [Google Scholar] [CrossRef] [PubMed]
- Cvetko, A.; Kifer, D.; Gornik, O.; Klarić, L.; Visser, E.; Lauc, G.; Wilson, J.F.; Štambuk, T. Glycosylation alterations in multiple sclerosis show increased proinflammatory potential. Biomedicines 2020, 8, 410. [Google Scholar] [CrossRef]
- Nishino, I.; Wu, S.W.; Ibarra, C.A.; Malicdan, M.C.V.; Murayama, K.; Noguchi, S.; Hayashi, Y.K.; Nonaka, I. Central core disease is due to RYR1 mutations in more than 90% of patients. Brain 2006, 129, 1470–1480. [Google Scholar]
- Oizumi, H.; Hayashita-Kinoh, H.; Hayakawa, H.; Arai, H.; Furuya, T.; Ren, Y.R.; Yasuda, T.; Seki, T.; Mizuno, Y.; Mochizuki, H. Alteration in the differentiation-related molecular expression in the subventricular zone in a mouse model of Parkinson’s disease. Neurosci. Res. 2008, 60, 15–21. [Google Scholar] [CrossRef]
- Albrecht, A.; Stork, O. Are NCAM deficient mice an animal model for schizophrenia? Front. Behav. Neurosci. 2012, 6, 43. [Google Scholar] [CrossRef] [Green Version]
- McHugh, J. Autoimmunity: Glycoengineering has therapeutic potential. Nat. Rev. Rheumatol. 2018, 14, 121. [Google Scholar] [CrossRef] [PubMed]
- Opferman, J.T. Apoptosis in the development of the immune system. Cell Death Differ. 2008, 15, 234–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovacs-Solyom, F.; Blasko, A.; Fajka-Boja, R.; Katona, R.L.; Vegh, L.; Novak, J.; Szebeni, G.J.; Krenacs, L.; Uher, F.; Tubak, V.; et al. Mechanism of tumor cell-induced T-cell apoptosis mediated by galectin-1. Immunol. Lett. 2010, 127, 108–118. [Google Scholar] [CrossRef]
- Toscano, M.A.; Bianco, G.A.; Ilarregui, J.M.; Croci, D.O.; Correale, J.; Hernandez, J.D.; Zwirner, N.W.; Poirier, F.; Riley, E.M.; Baum, L.G.; et al. Differential glycosylation of TH1, TH2 and TH-17 effector cells selectively regulates susceptibility to cell death. Nat. Immunol. 2007, 8, 825–834. [Google Scholar] [CrossRef]
- Bi, S.; Baum, L.G. Sialic acids in T cell development and function. BBA-Gen. Subj. 2009, 1790, 1599–1610. [Google Scholar] [CrossRef]
- Liao, W.; Lin, J.X.; Wang, L.; Li, P.; Leonard, W.J. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nat. Immunol. 2011, 12, 551–559. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Kim, Y.C.; Laurence, A.; Punkosdy, G.A.; Shevach, E.M. IL-2 controls the stability of Foxp3 expression in TGF-beta-induced Foxp3+ T cells in vivo. J. Immunol. 2011, 186, 6329–6337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Earl, L.A.; Baum, L.G. CD45 glycosylation controls T-cell life and death. Immunol. Cell Biol. 2008, 86, 608–615. [Google Scholar] [CrossRef] [PubMed]
- Marth, J.D.; Grewal, P.K. Mammalian glycosylation in immunity. Nat. Rev. Immunol. 2008, 8, 874–887. [Google Scholar] [CrossRef] [Green Version]
- Gascoigne, N.R. T-cell differentiation: MHC class I’s sweet tooth lost on maturity. Curr. Biol. 2002, 12, R99–R101. [Google Scholar] [CrossRef] [Green Version]
- Dings, R.P.M.; Miller, M.C.; Griffin, R.J.; Mayo, K.H. Galectins as Molecular Targets for Therapeutic Intervention. Int. J. Mol. Sci. 2018, 19, 905. [Google Scholar] [CrossRef] [Green Version]
- Grigorian, A.; Torossian, S.; Demetriou, M. T-cell growth, cell surface organization, and the galectin-glycoprotein lattice. Immunol. Rev. 2009, 230, 232–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kizuka, Y.; Oka, S. Regulated expression and neural functions of human natural killer-1 (HNK-1) carbohydrate. Cell Mol. Life Sci. 2012, 69, 4135–4147. [Google Scholar] [CrossRef] [Green Version]
- Morise, J.; Kizuka, Y.; Yabuno, K.; Tonoyama, Y.; Hashii, N.; Kawasaki, N.; Manya, H.; Miyagoe-Suzuki, Y.; Takeda, S.i.; Endo, T. Structural and biochemical characterization of O-mannose-linked human natural killer-1 glycan expressed on phosphacan in developing mouse brains. Glycobiology 2014, 24, 314–324. [Google Scholar] [CrossRef] [Green Version]
- Yu, R.K.; Yanagisawa, M. Glycobiology of neural stem cells: Functional aspects. J. Neurochem. 2006, 5, 415–423. [Google Scholar]
- Ribeiro, M.; Levay, K.; Yon, B.; Ayupe, A.C.; Salgueiro, Y.; Park, K.K. Neural cadherin plays distinct roles for neuronal survival and axon growth under different regenerative conditions. Eneuro 2020, 7. [Google Scholar] [CrossRef]
- Hamlin, J.A.; Fang, H.; Schwob, J.E. Differential expression of the mammalian homologue of fasciclin II during olfactory development in vivo and in vitro. J. Comp. Neurol. 2004, 474, 438–452. [Google Scholar] [CrossRef] [PubMed]
- Winther, M.; Walmod, P.S. Neural cell adhesion molecules belonging to the family of leucine-rich repeat proteins. Cell Adhes. Mol. 2014, 315–395. [Google Scholar]
- Takahashi, S.; Kato, K.; Nakamura, K.; Nakano, R.; Kubota, K.; Hamada, H. Neural cell adhesion molecule 2 as a target molecule for prostate and breast cancer gene therapy. Cancer Sci. 2011, 102, 808–814. [Google Scholar] [CrossRef] [PubMed]
- Edwards, S.; Campbell, C.; Flohr, P.; Shipley, J.; Giddings, I.; Te-Poele, R.; Dodson, A.; Foster, C.; Clark, J.; Jhavar, S.; et al. Expression analysis onto microarrays of randomly selected cDNA clones highlights HOXB13 as a marker of human prostate cancer. Brit. J. Cancer 2005, 92, 376–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, E.A.; Walker, S.R.; Li, W.; Liu, X.S.; Frank, D.A. Identification of human STAT5-dependent gene regulatory elements based on interspecies homology. J. Biol. Chem. 2006, 281, 26216–26224. [Google Scholar] [CrossRef] [Green Version]
- Cirielli, V.; Cima, L.; Chindemi, C.; Danzi, O.; Ghimenton, C.; Eccher, A.; Mauriello, S.; Bortolotti, F.; De Leo, D.; Brunelli, M. Cortical Expression of the Polysialylated Isoform of the Neural Cell Adhesion Molecule on Brain Tissue to Recognize Drug-Related Death: An Exploratory Analysis. Am. J. Forensic Med. Pathol. 2018, 39, 8–13. [Google Scholar] [CrossRef]
- Wuhrer, M.; Geyer, H.; von der Ohe, M.; Gerardy-Schahn, R.; Schachner, M.; Geyer, R. Localization of defined carbohydrate epitopes in bovine polysialylated NCAM. Biochimie 2003, 85, 207–218. [Google Scholar] [CrossRef]
- Ong, E.; Suzuki, M.; Belot, F.; Yeh, J.C.; Franceschini, I.; Angata, K.; Hindsgaul, O.; Fukuda, M. Biosynthesis of HNK-1 glycans on O-linked oligosaccharides attached to the neural cell adhesion molecule (NCAM): The requirement for core 2 beta 1,6-N-acetylglucosaminyltransferase and the muscle-specific domain in NCAM. J. Biol. Chem. 2002, 277, 18182–18190. [Google Scholar] [CrossRef] [Green Version]
- Liedtke, S.; Geyer, H.; Wuhrer, M.; Geyer, R.; Frank, G.; Gerardy-Schahn, R.; Zahringer, U.; Schachner, M. Characterization of N-glycans from mouse brain neural cell adhesion molecule. Glycobiology 2001, 11, 373–384. [Google Scholar] [CrossRef]
- Eberhardt, K.A.; Irintchev, A.; Al-Majed, A.A.; Simova, O.; Brushart, T.M.; Gordon, T.; Schachner, M. BDNF/TrkB signaling regulates HNK-1 carbohydrate expression in regenerating motor nerves and promotes functional recovery after peripheral nerve repair. Exp. Neurol. 2006, 198, 500–510. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Ferrer, M.; Dityatev, A. Shaping Synapses by the Neural Extracellular Matrix. Front. Neuroanat. 2018, 12, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bukalo, O.; Schachner, M.; Dityatev, A. Hippocampal metaplasticity induced by deficiency in the extracellular matrix glycoprotein tenascin-R. J. Neurosci. 2007, 27, 6019–6028. [Google Scholar] [CrossRef]
- Saito, H.; Nakao, Y.; Takayama, S.; Toyama, Y.; Asou, H. Specific expression of an HNK-1 carbohydrate epitope and NCAM on femoral nerve Schwann cells in mice. Neurosci. Res. 2005, 53, 314–322. [Google Scholar] [CrossRef]
- Irintchev, A.; Schachner, M. The injured and regenerating nervous system: Immunoglobulin superfamily members as key players. Neuroscientist. 2012, 18, 452–466. [Google Scholar] [CrossRef]
- Makhina, T.; Loers, G.; Schulze, C.; Ueberle, B.; Schachner, M.; Kleene, R. Extracellular GAPDH binds to L1 and enhances neurite outgrowth. Mol. Cell. Neurosci. 2009, 41, 206–218. [Google Scholar] [CrossRef]
- Hillenbrand, R.; Molthagen, M.; Montag, D.; Schachner, M. The close homologue of the neural adhesion molecule L1 (CHL1): Patterns of expression and promotion of neurite outgrowth by heterophilic interactions. Eur. J. Neurosci. 1999, 11, 813–826. [Google Scholar] [CrossRef]
- Franceschini, I.; Vitry, S.; Padilla, F.; Casanova, P.; Tham, T.N.; Fukuda, M.; Rougon, G.; Durbec, P.; Dubois-Dalcq, M. Migrating and myelinating potential of neural precursors engineered to overexpress PSA-NCAM. Mol. Cell. Neurosci. 2004, 27, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Morise, J.; Yabuno-Nakagawa, K.; Hashimoto, Y.; Takematsu, H.; Oka, S. Site-specific HNK-1 epitope on alternatively spliced fibronectin type-III repeats in tenascin-C promotes neurite outgrowth of hippocampal neurons through contactin-1. PLoS ONE 2019, 14, e0210193. [Google Scholar] [CrossRef]
- Valdembri, D.; Caswell, P.T.; Anderson, K.I.; Schwarz, J.P.; Konig, I.; Astanina, E.; Caccavari, F.; Norman, J.C.; Humphries, M.J.; Bussolino, F.; et al. Neuropilin-1/GIPC1 signaling regulates alpha5beta1 integrin traffic and function in endothelial cells. PLoS Biol. 2009, 7, e25. [Google Scholar] [CrossRef]
- Robinson, S.D.; Reynolds, L.E.; Kostourou, V.; Reynolds, A.R.; da Silva, R.G.; Tavora, B.; Baker, M.; Marshall, J.F.; Hodivala-Dilke, K.M. Alphav beta3 integrin limits the contribution of neuropilin-1 to vascular endothelial growth factor-induced angiogenesis. J. Biol. Chem. 2009, 284, 33966–33981. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Zhao, S.; Karnad, A.; Freeman, J.W. The biology and role of CD44 in cancer progression: Therapeutic implications. J. Hematol. Oncol. 2018, 11, 64. [Google Scholar] [CrossRef] [Green Version]
- Uniewicz, K.A.; Ori, A.; Ahmed, Y.A.; Yates, E.A.; Fernig, D.G. Characterisation of the interaction of neuropilin-1 with heparin and a heparan sulfate mimetic library of heparin-derived sugars. Peerj 2014, 2, e461. [Google Scholar] [CrossRef] [Green Version]
- Soares, M.A.; Teixeira, F.C.; Fontes, M.; Areas, A.L.; Leal, M.G.; Pavao, M.S.; Stelling, M.P. Heparan Sulfate Proteoglycans May Promote or Inhibit Cancer Progression by Interacting with Integrins and Affecting Cell Migration. Biomed. Res. Int. 2015, 2015, 453801. [Google Scholar] [CrossRef] [Green Version]
- Cai, Y.; Wang, R.; Zhao, Y.F.; Jia, J.; Sun, Z.J.; Chen, X.M. Expression of Neuropilin-2 in salivary adenoid cystic carcinoma: Its implication in tumor progression and angiogenesis. Pathol. Res. Pract. 2010, 206, 793–799. [Google Scholar] [CrossRef]
- Tu, D.G.; Chang, W.W.; Jan, M.S.; Tu, C.W.; Lu, Y.C.; Tai, C.K. Promotion of metastasis of thyroid cancer cells via NRP-2-mediated induction. Oncol. Lett. 2016, 12, 4224–4230. [Google Scholar] [CrossRef] [Green Version]
- Dewan, M.Z.; Takamatsu, N.; Hidaka, T.; Hatakeyama, K.; Nakahata, S.; Fujisawa, J.; Katano, H.; Yamamoto, N.; Morishita, K. Critical role for TSLC1 expression in the growth and organ infiltration of adult T-cell leukemia cells in vivo. J. Virol. 2008, 82, 11958–11963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galuska, S.P.; Rollenhagen, M.; Kaup, M.; Eggers, K.; Oltmann-Norden, I.; Schiff, M.; Hartmann, M.; Weinhold, B.; Hildebrandt, H.; Geyer, R.; et al. Synaptic cell adhesion molecule SynCAM 1 is a target for polysialylation in postnatal mouse brain. Proc. Natl. Acad. Sci. USA 2010, 107, 10250–10255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caunt, M.; Mak, J.; Liang, W.C.; Stawicki, S.; Pan, Q.; Tong, R.K.; Kowalski, J.; Ho, C.; Reslan, H.B.; Ross, J.; et al. Blocking neuropilin-2 function inhibits tumor cell metastasis. Cancer Cell 2008, 13, 331–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prud’homme, G.J. Cancer stem cells and novel targets for antitumor strategies. Curr. Pharm. Des. 2012, 18, 2838–2849. [Google Scholar] [CrossRef]
- Schellenburg, S.; Schulz, A.; Poitz, D.M.; Muders, M.H. Role of neuropilin-2 in the immune system. Mol. Immunol. 2017, 90, 239–244. [Google Scholar] [CrossRef]
- Rossignol, M.; Gagnon, M.L.; Klagsbrun, M. Genomic organization of human neuropilin-1 and neuropilin-2 genes: Identification and distribution of splice variants and soluble isoforms. Genomics 2000, 70, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Handa, A.; Tokunaga, T.; Tsuchida, T.; Lee, Y.H.; Kijima, H.; Yamazaki, H.; Ueyama, Y.; Fukuda, H.; Nakamura, M. Neuropilin-2 expression affects the increased vascularization and is a prognostic factor in osteosarcoma. Int. J. Oncol. 2000, 17, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yang, R.; Yang, H.; Chen, S.; Wang, L.; Li, M.; Yang, S.; Feng, Z.; Bi, J. NCAM regulates the proliferation, apoptosis, autophagy, EMT, and migration of human melanoma cells via the Src/Akt/mTOR/cofilin signaling pathway. J. Cell Biochem. 2020, 121, 1192–1204. [Google Scholar] [CrossRef]
- Yasuoka, H.; Kodama, R.; Tsujimoto, M.; Yoshidome, K.; Akamatsu, H.; Nakahara, M.; Inagaki, M.; Sanke, T.; Nakamura, Y. Neuropilin-2 expression in breast cancer: Correlation with lymph node metastasis, poor prognosis, and regulation of CXCR4 expression. Bmc. Cancer 2009, 9, 220. [Google Scholar] [CrossRef] [Green Version]
- Nasarre, P.; Gemmill, R.M.; Potiron, V.A.; Roche, J.; Lu, X.; Baron, A.E.; Korch, C.; Garrett-Mayer, E.; Lagana, A.; Howe, P.H.; et al. Neuropilin-2 Is upregulated in lung cancer cells during TGF-beta1-induced epithelial-mesenchymal transition. Cancer Res. 2013, 73, 7111–7121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rieger, J.; Wick, W.; Weller, M. Human malignant glioma cells express semaphorins and their receptors, neuropilins and plexins. Glia 2003, 42, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Fakhari, M.; Pullirsch, D.; Abraham, D.; Paya, K.; Hofbauer, R.; Holzfeind, P.; Hofmann, M.; Aharinejad, S. Selective upregulation of vascular endothelial growth factor receptors neuropilin-1 and -2 in human neuroblastoma. Cancer-Am. Cancer Soc. 2002, 94, 258–263. [Google Scholar] [CrossRef]
- Hansel, D.E.; Wilentz, R.E.; Yeo, C.J.; Schulick, R.D.; Montgomery, E.; Maitra, A. Expression of neuropilin-1 in high-grade dysplasia, invasive cancer, and metastases of the human gastrointestinal tract. Am. J. Surg. Pathol. 2004, 28, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Dallas, N.A.; Gray, M.J.; Xia, L.; Fan, F.; van Buren, G., 2nd; Gaur, P.; Samuel, S.; Lim, S.J.; Arumugam, T.; Ramachandran, V.; et al. Neuropilin-2-mediated tumor growth and angiogenesis in pancreatic adenocarcinoma. Clin. Cancer Res. 2008, 14, 8052–8060. [Google Scholar] [CrossRef] [Green Version]
- Cohen, T.; Herzog, Y.; Brodzky, A.; Greenson, J.K.; Eldar, S.; Gluzman-Poltorak, Z.; Neufeld, G.; Resnick, M.B. Neuropilin-2 is a novel marker expressed in pancreatic islet cells and endocrine pancreatic tumours. J. Pathol. 2002, 198, 77–82. [Google Scholar] [CrossRef]
- Parikh, A.A.; Liu, W.B.; Fan, F.; Stoeltzing, O.; Reinmuth, N.; Bruns, C.J.; Bucana, C.D.; Evans, D.B.; Ellis, L.M. Expression and regulation of the novel vascular endothelial growth factor receptor neuropilin-1 by epidermal growth factor in human pancreatic carcinoma. Cancer-Am. Cancer Soc. 2003, 98, 720–729. [Google Scholar] [CrossRef] [PubMed]
- Calicchio, M.L.; Collins, T.; Kozakewich, H.P. Identification of signaling systems in proliferating and involuting phase infantile hemangiomas by genome-wide transcriptional profiling. Am. J. Pathol. 2009, 174, 1638–1649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulz, A.; Gorodetska, I.; Behrendt, R.; Fuessel, S.; Erdmann, K.; Foerster, S.; Datta, K.; Mayr, T.; Dubrovska, A.; Muders, M.H. Linking NRP2 with EMT and chemoradioresistance in bladder cancer. Front. Oncol. 2020, 9, 1461. [Google Scholar] [CrossRef] [PubMed]
- Polavaram, N.S.; Dutta, S.; Islam, R.; Bag, A.K.; Roy, S.; Poitz, D.; Karnes, J.; Hofbauer, L.C.; Kohli, M.; Costello, B.A. Tumor-and osteoclast-derived NRP2 in prostate cancer bone metastases. Bone Res. 2021, 9, 1–16. [Google Scholar] [CrossRef]
- Butti, R.; Kumar, T.V.; Nimma, R.; Kundu, G.C. Impact of semaphorin expression on prognostic characteristics in breast cancer. Breast Cancer 2018, 10, 79–88. [Google Scholar] [CrossRef] [Green Version]
- Hildebrandt, H.; Dityatev, A. Polysialic acid in brain development and synaptic plasticity. SialoGlyco Chem. Biol. I 2013, 366, 55–96. [Google Scholar]
- Windisch, R.; Pirschtat, N.; Kellner, C.; Chen-Wichmann, L.; Lausen, J.; Humpe, A.; Krause, D.S.; Wichmann, C. Oncogenic deregulation of cell adhesion molecules in leukemia. Cancers 2019, 11, 311. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, H.; Nishikata, I.; Shiraga, T.; Akamatsu, E.; Fukami, T.; Hidaka, T.; Kubuki, Y.; Okayama, A.; Hamada, K.; Okabe, H.; et al. Overexpression of a cell adhesion molecule, TSLC1, as a possible molecular marker for acute-type adult T-cell leukemia. Blood 2005, 105, 1204–1213. [Google Scholar] [CrossRef]
- Qin, L.; Zhu, W.; Xu, T.; Hao, Y.; Zhang, Z.; Tian, Y.; Yang, D. Effect of TSLC1 gene on proliferation, invasion and apoptosis of human hepatocellular carcinoma cell line HepG2. J. Huazhong Univ. Sci. Technol. 2007, 27, 535–537. [Google Scholar] [CrossRef] [PubMed]
- Usami, Y.; Ito, A.; Ohnuma, K.; Fuku, T.; Komori, T.; Yokozaki, H. Tumor suppressor in lung cancer-1 as a novel ameloblast adhesion molecule and its downregulation in ameloblastoma. Pathol. Int. 2007, 57, 68–75. [Google Scholar] [CrossRef]
- Werneburg, S.; Buettner, F.F.; Muhlenhoff, M.; Hildebrandt, H. Polysialic acid modification of the synaptic cell adhesion molecule SynCAM 1 in human embryonic stem cell-derived oligodendrocyte precursor cells. Stem. Cell Res. 2015, 14, 339–346. [Google Scholar] [CrossRef] [Green Version]
- Fogel, A.I.; Akins, M.R.; Krupp, A.J.; Stagi, M.; Stein, V.; Biederer, T. SynCAMs organize synapses through heterophilic adhesion. J. Neurosci. 2007, 27, 12516–12530. [Google Scholar] [CrossRef] [Green Version]
- Takase, N.; Koma, Y.; Urakawa, N.; Nishio, M.; Arai, N.; Akiyama, H.; Shigeoka, M.; Kakeji, Y.; Yokozaki, H. NCAM- and FGF-2-mediated FGFR1 signaling in the tumor microenvironment of esophageal cancer regulates the survival and migration of tumor-associated macrophages and cancer cells. Cancer Lett. 2016, 380, 47–58. [Google Scholar] [CrossRef] [Green Version]
- Britain, C.M.; Holdbrooks, A.T.; Anderson, J.C.; Willey, C.D.; Bellis, S.L. Sialylation of EGFR by the ST6Gal-I sialyltransferase promotes EGFR activation and resistance to gefitinib-mediated cell death. J. Ovarian Res. 2018, 11, 12. [Google Scholar] [CrossRef] [Green Version]
- Karpanen, T.; Heckman, C.A.; Keskitalo, S.; Jeltsch, M.; Ollila, H.; Neufeld, G.; Tamagnone, L.; Alitalo, K. Functional interaction of VEGF-C and VEGF-D with neuropilin receptors. FASEB J. 2006, 20, 1462–1472. [Google Scholar] [CrossRef] [Green Version]
- Seidenfaden, R.; Krauter, A.; Schertzinger, F.; Gerardy-Schahn, R.; Hildebrandt, H. Polysialic acid directs tumor cell growth by controlling heterophilic neural cell adhesion molecule interactions. Mol. Cell Biol. 2003, 23, 5908–5918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vempati, P.; Popel, A.S.; Mac Gabhann, F. Extracellular regulation of VEGF: Isoforms, proteolysis, and vascular patterning. Cytokine Growth Factor Rev. 2014, 25, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Grunewald, F.S.; Prota, A.E.; Giese, A.; Ballmer-Hofer, K. Structure-function analysis of VEGF receptor activation and the role of coreceptors in angiogenic signaling. BBA-Gen. Subj. 2010, 1804, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Shraga-Heled, N.; Kessler, O.; Prahst, C.; Kroll, J.; Augustin, H.; Neufeld, G. Neuropilin-1 and neuropilin-2 enhance VEGF121 stimulated signal transduction by the VEGFR-2 receptor. FASEB J. 2007, 21, 915–926. [Google Scholar] [CrossRef] [PubMed]
- Ellis, L.M.; Rosen, L.; Gordon, M.S. Overview of anti-VEGF therapy and angiogenesis. Part 1: Angiogenesis inhibition in solid tumor malignancies. Clin. Adv. Hematol. Oncol. 2006, 4 (Suppl. 1–10). [Google Scholar]
- Elola, M.T.; Blidner, A.G.; Ferragut, F.; Bracalente, C.; Rabinovich, G.A. Assembly, organization and regulation of cell-surface receptors by lectin-glycan complexes. Biochem. J. 2015, 469, 1–16. [Google Scholar] [CrossRef]
- Domigan, C.K.; Ziyad, S.; Iruela-Arispe, M.L. Canonical and noncanonical vascular endothelial growth factor pathways: New developments in biology and signal transduction. Arter. Thromb. Vasc. Biol. 2015, 35, 30–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basagiannis, D.; Zografou, S.; Murphy, C.; Fotsis, T.; Morbidelli, L.; Ziche, M.; Bleck, C.; Mercer, J.; Christoforidis, S. VEGF induces signalling and angiogenesis by directing VEGFR2 internalisation through macropinocytosis. J. Cell Sci. 2016, 129, 4091–4104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Croci, D.O.; Cerliani, J.P.; Pinto, N.A.; Morosi, L.G.; Rabinovich, G.A. Regulatory role of glycans in the control of hypoxia-driven angiogenesis and sensitivity to anti-angiogenic treatment. Glycobiology 2014, 24, 1283–1290. [Google Scholar] [CrossRef] [Green Version]
- Povlsen, G.K.; Berezin, V.; Bock, E. Neural cell adhesion molecule-180-mediated homophilic binding induces epidermal growth factor receptor (EGFR) down-regulation and uncouples the inhibitory function of EGFR in neurite outgrowth. J. Neurochem. 2008, 104, 624–639. [Google Scholar] [CrossRef] [PubMed]
- Islamov, R.R.; Izmailov, A.A.; Sokolov, M.E.; Fadeev, P.O.; Bashirov, F.V.; Eremeev, A.A.; Shaymardanova, G.F.; Shmarov, M.M.; Naroditskiy, B.S.; Chelyshev, Y.A.; et al. Evaluation of direct and cell-mediated triple-gene therapy in spinal cord injury in rats. Brain Res. Bull. 2017, 132, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Islamov, R.R.; Rizvanov, A.A.; Mukhamedyarov, M.A.; Salafutdinov, I.I.; Garanina, E.E.; Fedotova, V.Y.; Solovyeva, V.V.; Mukhamedshina, Y.O.; Safiullov, Z.Z.; Izmailov, A.A.; et al. Symptomatic improvement, increased life-span and sustained cell homing in amyotrophic lateral sclerosis after transplantation of human umbilical cord blood cells genetically modified with adeno-viral vectors expressing a neuro-protective factor and a neural cell adhesion molecule. Curr. Gene Ther. 2015, 15, 266–276. [Google Scholar] [CrossRef]
- Izmailov, A.A.; Povysheva, T.V.; Bashirov, F.V.; Sokolov, M.E.; Fadeev, F.O.; Garifulin, R.R.; Naroditsky, B.S.; Logunov, D.Y.; Salafutdinov, I.I.; Chelyshev, Y.A.; et al. Spinal Cord Molecular and Cellular Changes Induced by Adenoviral Vector- and Cell-Mediated Triple Gene Therapy after Severe Contusion. Front. Pharm. 2017, 8, 813. [Google Scholar] [CrossRef] [PubMed]
- Jarahian, M.; Watzl, C.; Issa, Y.; Altevogt, P.; Momburg, F. Blockade of natural killer cell-mediated lysis by NCAM140 expressed on tumor cells. Int. J. Cancer 2007, 120, 2625–2634. [Google Scholar] [CrossRef]
- Bonfanti, L.; Theodosis, D.T. Polysialic acid and activity-dependent synapse remodeling. Cell Adh. Migr. 2009, 3, 43–50. [Google Scholar] [CrossRef] [Green Version]
- Plein, A.; Fantin, A.; Ruhrberg, C. Neuropilin regulation of angiogenesis, arteriogenesis, and vascular permeability. Microcirculation 2014, 21, 315–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wee, P.; Wang, Z. Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways. Cancers 2017, 9, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, P.; Hu, J.; Wang, X.; Wang, J.; Zhang, Y.; Wang, C. Epidermal growth factor receptor expression affects proliferation and apoptosis in non-small cell lung cancer cells via the extracellular signal-regulated kinase/microRNA 200a signaling pathway. Oncol. Lett. 2018, 15, 5201–5207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berger, R.P.; Sun, Y.H.; Kulik, M.; Lee, J.K.; Nairn, A.V.; Moremen, K.W.; Pierce, M.; Dalton, S. ST8SIA4-Dependent Polysialylation is Part of a Developmental Program Required for Germ Layer Formation from Human Pluripotent Stem Cells. Stem Cells 2016, 34, 1742–1752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Young, N.M.; Tropp, S.; Hu, D.; Xu, Y.; Hallgrimsson, B.; Marcucio, R.S. Quantification of shape and cell polarity reveals a novel mechanism underlying malformations resulting from related FGF mutations during facial morphogenesis. Hum. Mol. Genet. 2013, 22, 5160–5172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colombo, F.; Meldolesi, J. L1-CAM and N-CAM: From Adhesion Proteins to Pharmacological Targets. Trends Pharm. Sci. 2015, 36, 769–781. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Dai, G.; Cheng, Y.B.; Qi, X.; Geng, M.Y. Polysialylation promotes neural cell adhesion molecule-mediated cell migration in a fibroblast growth factor receptor-dependent manner, but independent of adhesion capability. Glycobiology 2011, 21, 1010–1018. [Google Scholar] [CrossRef] [Green Version]
- Steenbergen, S.M.; Vimr, E.R. Chromatographic analysis of the Escherichia coli polysialic acid capsule. Methods Mol. Biol. 2013, 966, 109–120. [Google Scholar] [CrossRef] [Green Version]
- Bull, C.; den Brok, M.H.; Adema, G.J. Sweet escape: Sialic acids in tumor immune evasion. BBA-Gen. Subj. 2014, 1846, 238–246. [Google Scholar] [CrossRef]
- Roy, S.; Bag, A.K.; Singh, R.K.; Talmadge, J.E.; Batra, S.K.; Datta, K. Multifaceted Role of Neuropilins in the Immune System: Potential Targets for Immunotherapy. Front. Immunol. 2017, 8, 1228. [Google Scholar] [CrossRef] [Green Version]
- Lemmon, M.A.; Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 2010, 141, 1117–1134. [Google Scholar] [CrossRef] [Green Version]
- Sjostrand, D.; Ibanez, C.F. Insights into GFRalpha1 regulation of neural cell adhesion molecule (NCAM) function from structure-function analysis of the NCAM/GFRalpha1 receptor complex. J. Biol. Chem. 2008, 283, 13792–13798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandhya, V.K.; Raju, R.; Verma, R.; Advani, J.; Sharma, R.; Radhakrishnan, A.; Nanjappa, V.; Narayana, J.; Somani, B.L.; Mukherjee, K.K.; et al. A network map of BDNF/TRKB and BDNF/p75NTR signaling system. J. Cell Commun. Signal. 2013, 7, 301–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Zhao, H.; Zhang, X.; Chen, L.; Zhao, X.; Bai, X.; Zhang, J. Nobiletin protects against cerebral ischemia via activating the p-Akt, p-CREB, BDNF and Bcl-2 pathway and ameliorating BBB permeability in rat. Brain Res. Bull. 2013, 96, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Ren, Z.; Zhang, J.; Chuang, C.C.; Kandaswamy, E.; Zhou, T.; Zuo, L. Role of ROS and Nutritional Antioxidants in Human Diseases. Front. Physiol. 2018, 9, 477. [Google Scholar] [CrossRef] [Green Version]
- Muramatsu, T. Midkine and pleiotrophin: Two related proteins involved in development, survival, inflammation and tumorigenesis. J. Biochem. 2002, 132, 359–371. [Google Scholar] [CrossRef]
- Palmer, R.H.; Vernersson, E.; Grabbe, C.; Hallberg, B. Anaplastic lymphoma kinase: Signalling in development and disease. Biochem. J. 2009, 420, 345–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berge, E.M.; Doebele, R.C. Targeted therapies in non-small cell lung cancer: Emerging oncogene targets following the success of epidermal growth factor receptor. Semin. Oncol. 2014, 41, 110–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ou, S.H.; Tan, J.; Yen, Y.; Soo, R.A. ROS1 as a ‘druggable’ receptor tyrosine kinase: Lessons learned from inhibiting the ALK pathway. Expert Rev. Anticancer. Ther. 2012, 12, 447–456. [Google Scholar] [CrossRef]
- Davies, K.D.; Le, A.T.; Theodoro, M.F.; Skokan, M.C.; Aisner, D.L.; Berge, E.M.; Terracciano, L.M.; Cappuzzo, F.; Incarbone, M.; Roncalli, M.; et al. Identifying and targeting ROS1 gene fusions in non-small cell lung cancer. Clin. Cancer Res. 2012, 18, 4570–4579. [Google Scholar] [CrossRef] [Green Version]
- Acquaviva, J.; Wong, R.; Charest, A. The multifaceted roles of the receptor tyrosine kinase ROS in development and cancer. BBA-Gen. Subj. 2009, 1795, 37–52. [Google Scholar] [CrossRef]
- Charest, A.; Wilker, E.W.; McLaughlin, M.E.; Lane, K.; Gowda, R.; Coven, S.; McMahon, K.; Kovach, S.; Feng, Y.; Yaffe, M.B.; et al. ROS fusion tyrosine kinase activates a SH2 domain-containing phosphatase-2/phosphatidylinositol 3-kinase/mammalian target of rapamycin signaling axis to form glioblastoma in mice. Cancer Res. 2006, 66, 7473–7481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiarle, R.; Voena, C.; Ambrogio, C.; Piva, R.; Inghirami, G. The anaplastic lymphoma kinase in the pathogenesis of cancer. Nat. Rev. Cancer 2008, 8, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Chiarle, R.; Simmons, W.J.; Cai, H.; Dhall, G.; Zamo, A.; Raz, R.; Karras, J.G.; Levy, D.E.; Inghirami, G. Stat3 is required for ALK-mediated lymphomagenesis and provides a possible therapeutic target. Nat. Med. 2005, 11, 623–629. [Google Scholar] [CrossRef]
- Soda, M.; Takada, S.; Takeuchi, K.; Ishikawa, Y.; Sugiyama, Y.; Mano, H. Analysis of a Mouse Model for EML4-ALK-Positive Lung Cancer. Am. J. Respir. Crit. Care Med. 2009, 179, A2693. [Google Scholar]
- Nguyen, K.T.; Zong, C.S.; Uttamsingh, S.; Sachdev, P.; Bhanot, M.; Le, M.T.; Chan, J.L.; Wang, L.H. The role of phosphatidylinositol 3-kinase, rho family GTPases, and STAT3 in Ros-induced cell transformation. J. Biol. Chem. 2002, 277, 11107–11115. [Google Scholar] [CrossRef] [Green Version]
- Charest, A.; Lane, K.; McMahon, K.; Park, J.; Preisinger, E.; Conroy, H.; Housman, D. Fusion of FIG to the receptor tyrosine kinase ROS in a glioblastoma with an interstitial del(6)(q21q21). Genes Chromosomes Cancer 2003, 37, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Stransky, N.; Cerami, E.; Schalm, S.; Kim, J.L.; Lengauer, C. The landscape of kinase fusions in cancer. Nat. Commun. 2014, 5, 4846. [Google Scholar] [CrossRef] [Green Version]
- Lovly, C.M.; Gupta, A.; Lipson, D.; Otto, G.; Brennan, T.; Chung, C.T.; Borinstein, S.C.; Ross, J.S.; Stephens, P.J.; Miller, V.A.; et al. Inflammatory myofibroblastic tumors harbor multiple potentially actionable kinase fusions. Cancer Discov. 2014, 4, 889–895. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, H.; Yoshida, A.; Taguchi, K.; Kohashi, K.; Hatanaka, Y.; Yamashita, A.; Mori, D.; Oda, Y. ALK, ROS1 and NTRK3 gene rearrangements in inflammatory myofibroblastic tumours. Histopathology 2016, 69, 72–83. [Google Scholar] [CrossRef]
- Gu, T.L.; Deng, X.; Huang, F.; Tucker, M.; Crosby, K.; Rimkunas, V.; Wang, Y.; Deng, G.; Zhu, L.; Tan, Z.; et al. Survey of tyrosine kinase signaling reveals ROS kinase fusions in human cholangiocarcinoma. PLoS ONE 2011, 6, e15640. [Google Scholar] [CrossRef] [Green Version]
- Birch, A.H.; Arcand, S.L.; Oros, K.K.; Rahimi, K.; Watters, A.K.; Provencher, D.; Greenwood, C.M.; Mes-Masson, A.M.; Tonin, P.N. Chromosome 3 anomalies investigated by genome wide SNP analysis of benign, low malignant potential and low grade ovarian serous tumours. PLoS ONE 2011, 6, e28250. [Google Scholar] [CrossRef]
- Lee, J.; Lee, S.E.; Kang, S.Y.; Do, I.G.; Lee, S.; Ha, S.Y.; Cho, J.; Kang, W.K.; Jang, J.; Ou, S.H.; et al. Identification of ROS1 rearrangement in gastric adenocarcinoma. Cancer-Am. Cancer Soc. 2013, 119, 1627–1635. [Google Scholar] [CrossRef]
- Aisner, D.L.; Nguyen, T.T.; Paskulin, D.D.; Le, A.T.; Haney, J.; Schulte, N.; Chionh, F.; Hardingham, J.; Mariadason, J.; Tebbutt, N.; et al. ROS1 and ALK fusions in colorectal cancer, with evidence of intratumoral heterogeneity for molecular drivers. Mol. Cancer Res. 2014, 12, 111–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giacomini, C.P.; Sun, S.; Varma, S.; Shain, A.H.; Giacomini, M.M.; Balagtas, J.; Sweeney, R.T.; Lai, E.; Del Vecchio, C.A.; Forster, A.D.; et al. Breakpoint analysis of transcriptional and genomic profiles uncovers novel gene fusions spanning multiple human cancer types. PLoS Genet. 2013, 9, e1003464. [Google Scholar] [CrossRef] [PubMed]
- Wiesner, T.; He, J.; Yelensky, R.; Esteve-Puig, R.; Botton, T.; Yeh, I.; Lipson, D.; Otto, G.; Brennan, K.; Murali, R.; et al. Kinase fusions are frequent in Spitz tumours and spitzoid melanomas. Nat. Commun. 2014, 5, 3116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaw, A.; Riley, G.J.; Bang, Y.J.; Kim, D.W.; Camidge, D.R.; Varella-Garcia, M.; Lafrate, A.J.; Shapiro, G.; Winter, M.; Usari, T.; et al. Crizotinib in advanced ROS1-rearranged non-small cell lung cancer (NSCLC): Updated results from PROFILE 1001. Ann. Oncol. 2016, 27, vi418. [Google Scholar] [CrossRef]
- Collisson, E.A.; Campbell, J.D.; Brooks, A.N.; Berger, A.H.; Lee, W.; Chmielecki, J.; Beer, D.G.; Cope, L.; Creighton, C.J.; Danilova, L.; et al. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014, 511, 543–550. [Google Scholar] [CrossRef]
- Zhu, V.W.; Upadhyay, D.; Schrock, A.B.; Gowen, K.; Ali, S.M.; Ou, S.H. TPD52L1-ROS1, a new ROS1 fusion variant in lung adenosquamous cell carcinoma identified by comprehensive genomic profiling. Lung Cancer 2016, 97, 48–50. [Google Scholar] [CrossRef] [PubMed]
- Farago, A.F.; Zheng, Z.L.; Muzikansky, A.; Gainor, J.F.; Iafrate, A.J.; Engelman, J.A.; Le, L.P.; Shaw, A.T. Clinical implementation of anchored multiplex PCR with targeted next-generation sequencing for detection of ALK, ROS1, RET and NTRK1 fusions in non-small cell lung carcinoma. J. Clin. Oncol. 2015, 33, 8095. [Google Scholar] [CrossRef]
- Rikova, K.; Guo, A.; Zeng, Q.; Possemato, A.; Yu, J.; Haack, H.; Nardone, J.; Lee, K.; Reeves, C.; Li, Y.; et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007, 131, 1190–1203. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, K.; Soda, M.; Togashi, Y.; Suzuki, R.; Sakata, S.; Hatano, S.; Asaka, R.; Hamanaka, W.; Ninomiya, H.; Uehara, H.; et al. RET, ROS1 and ALK fusions in lung cancer. Nat. Med. 2012, 18, 378–381. [Google Scholar] [CrossRef] [PubMed]
- Rimkunas, V.M.; Crosby, K.E.; Li, D.; Hu, Y.; Kelly, M.E.; Gu, T.L.; Mack, J.S.; Silver, M.R.; Zhou, X.; Haack, H. Analysis of receptor tyrosine kinase ROS1-positive tumors in non-small cell lung cancer: Identification of a FIG-ROS1 fusion. Clin. Cancer Res. 2012, 18, 4449–4457. [Google Scholar] [CrossRef] [Green Version]
- Al-Saraireh, Y.M.; Sutherland, M.; Springett, B.R.; Freiberger, F.; Ribeiro Morais, G.; Loadman, P.M.; Errington, R.J.; Smith, P.J.; Fukuda, M.; Gerardy-Schahn, R.; et al. Pharmacological inhibition of polysialyltransferase ST8SiaII modulates tumour cell migration. PLoS ONE 2013, 8, e73366. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, A.; Kohno, T.; Tsuta, K.; Wakai, S.; Arai, Y.; Shimada, Y.; Asamura, H.; Furuta, K.; Shibata, T.; Tsuda, H. ROS1-rearranged lung cancer: A clinicopathologic and molecular study of 15 surgical cases. Am. J. Surg. Pathol. 2013, 37, 554–562. [Google Scholar] [CrossRef]
- Suehara, Y.; Arcila, M.; Wang, L.; Hasanovic, A.; Ang, D.; Ito, T.; Kimura, Y.; Drilon, A.; Guha, U.; Rusch, V.; et al. Identification of KIF5B-RET and GOPC-ROS1 fusions in lung adenocarcinomas through a comprehensive mRNA-based screen for tyrosine kinase fusions. Clin. Cancer Res. 2012, 18, 6599–6608. [Google Scholar] [CrossRef] [Green Version]
- Govindan, R.; Ding, L.; Griffith, M.; Subramanian, J.; Dees, N.D.; Kanchi, K.L.; Maher, C.A.; Fulton, R.; Fulton, L.; Wallis, J.; et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 2012, 150, 1121–1134. [Google Scholar] [CrossRef] [Green Version]
- Seo, J.S.; Ju, Y.S.; Lee, W.C.; Shin, J.Y.; Lee, J.K.; Bleazard, T.; Lee, J.; Jung, Y.J.; Kim, J.O.; Shin, J.Y.; et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res. 2012, 22, 2109–2119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gainor, J.F.; Shaw, A.T. Novel targets in non-small cell lung cancer: ROS1 and RET fusions. Oncologist 2013, 18, 865–875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, J.J.; Shaw, A.T. Recent Advances in Targeting ROS1 in Lung Cancer. J. Thorac. Oncol. 2017, 12, 1611–1625. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, Y.; Jinnou, H.; Sawamoto, K.; Hitoshi, S. Adult neurogenesis and its role in brain injury and psychiatric diseases. J. Neurochem. 2018, 147, 584–594. [Google Scholar] [CrossRef] [Green Version]
- Quartu, M.; Serra, M.P.; Boi, M.; Ibba, V.; Melis, T.; Del Fiacco, M. Polysialylated-neural cell adhesion molecule (PSA-NCAM) in the human trigeminal ganglion and brainstem at prenatal and adult ages. BMC Neurosci. 2008, 9, 108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coviello, S.; Benedetti, B.; Jakubecova, D.; Belles, M.; Klimczak, P.; Gramuntell, Y.; Couillard-Despres, S.; Nacher, J. PSA Depletion Induces the Differentiation of Immature Neurons in the Piriform Cortex of Adult Mice. Int. J. Mol. Sci. 2021, 22, 5733. [Google Scholar] [CrossRef] [PubMed]
- Varea, E.; Castillo-Gomez, E.; Gomez-Climent, M.A.; Guirado, R.; Blasco-Ibanez, J.M.; Crespo, C.; Martinez-Guijarro, F.J.; Nacher, J. Differential evolution of PSA-NCAM expression during aging of the rat telencephalon. Neurobiol. Aging 2009, 30, 808–818. [Google Scholar] [CrossRef] [PubMed]
- Aonurm-Helm, A.; Jaako, K.; Jürgenson, M.; Zharkovsky, A. Pharmacological approach for targeting dysfunctional brain plasticity: Focus on neural cell adhesion molecule (NCAM). Pharmacol. Res. 2016, 113, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Gattenlohner, S.; Stuhmer, T.; Leich, E.; Reinhard, M.; Etschmann, B.; Volker, H.U.; Rosenwald, A.; Serfling, E.; Bargou, R.C.; Ertl, G.; et al. Specific detection of CD56 (NCAM) isoforms for the identification of aggressive malignant neoplasms with progressive development. Am. J. Pathol. 2009, 174, 1160–1171. [Google Scholar] [CrossRef] [Green Version]
- Galuska, C.E.; Lutteke, T.; Galuska, S.P. Is Polysialylated NCAM Not Only a Regulator during Brain Development But also during the Formation of Other Organs? Biology 2017, 6, 27. [Google Scholar] [CrossRef] [Green Version]
- Ono, T.; Takeshita, A.; Kishimoto, Y.; Kiyoi, H.; Okada, M.; Yamauchi, T.; Emi, N.; Horikawa, K.; Matsuda, M.; Shinagawa, K.; et al. Expression of CD56 is an unfavorable prognostic factor for acute promyelocytic leukemia with higher initial white blood cell counts. Cancer Sci. 2014, 105, 97–104. [Google Scholar] [CrossRef]
- Murray, N.; Salgia, R.; Fossella, F.V. Targeted molecules in small cell lung cancer. Semin. Oncol. 2004, 31, 106–111. [Google Scholar] [CrossRef]
- Rushing, E.C.; Stine, M.J.; Hahn, S.J.; Shea, S.; Eller, M.S.; Naif, A.; Khanna, S.; Westra, W.H.; Jungbluth, A.A.; Busam, K.J.; et al. Neuropilin-2: A novel biomarker for malignant melanoma? Hum. Pathol. 2012, 43, 381–389. [Google Scholar] [CrossRef] [Green Version]
- Vossen, L.I.; Markovsky, E.; Eldar-Boock, A.; Tschiche, H.R.; Wedepohl, S.; Pisarevsky, E.; Satchi-Fainaro, R.; Calderon, M. PEGylated dendritic polyglycerol conjugate targeting NCAM-expressing neuroblastoma: Limitations and challenges. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 1169–1179. [Google Scholar] [CrossRef]
- Wachowiak, R.; Krause, M.; Mayer, S.; Peukert, N.; Suttkus, A.; Muller, W.C.; Lacher, M.; Meixensberger, J.; Nestler, U. Increased L1CAM (CD171) levels are associated with glioblastoma and metastatic brain tumors. Medicine 2018, 97, e12396. [Google Scholar] [CrossRef] [PubMed]
- Ardizzone, A.; Scuderi, S.A.; Giuffrida, D.; Colarossi, C.; Puglisi, C.; Campolo, M.; Cuzzocrea, S.; Esposito, E.; Paterniti, I. Role of Fibroblast Growth Factors Receptors (FGFRs) in Brain Tumors, Focus on Astrocytoma and Glioblastoma. Cancers 2020, 12, 3825. [Google Scholar] [CrossRef] [PubMed]
- Gattenloehner, S.; Chuvpilo, S.; Langebrake, C.; Reinhardt, D.; Muller-Hermelink, H.K.; Serfling, E.; Vincent, A.; Marx, A. Novel RUNX1 isoforms determine the fate of acute myeloid leukemia cells by controlling CD56 expression. Blood 2007, 110, 2027–2033. [Google Scholar] [CrossRef] [Green Version]
- Montesinos, P.; Rayon, C.; Vellenga, E.; Brunet, S.; Gonzalez, J.; Gonzalez, M.; Holowiecka, A.; Esteve, J.; Bergua, J.; Gonzalez, J.D.; et al. Clinical significance of CD56 expression in patients with acute promyelocytic leukemia treated with all-trans retinoic acid and anthracycline-based regimens. Blood 2011, 117, 1799–1805. [Google Scholar] [CrossRef] [Green Version]
- Paietta, E.; Neuberg, D.; Richards, S.; Bennett, J.M.; Han, L.; Racevskis, J.; Dewald, G.; Rowe, J.M.; Wiernik, P.H. Rare adult acute lymphocytic leukemia with CD56 expression in the ECOG experience shows unexpected phenotypic and genotypic heterogeneity. Am. J. Hematol. 2001, 66, 189–196. [Google Scholar] [CrossRef]
- Fischer, L.; Gokbuget, N.; Schwartz, S.; Burmeister, T.; Rieder, H.; Bruggemann, M.; Hoelzer, D.; Thiel, E. CD56 expression in T-cell acute lymphoblastic leukemia is associated with non-thymic phenotype and resistance to induction therapy but no inferior survival after risk-adapted therapy. Haematologica 2009, 94, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Wielgat, P.; Rogowski, K.; Niemirowicz-Laskowska, K.; Car, H. Sialic acid-siglec axis as molecular checkpoints targeting of immune system: Smart players in pathology and conventional therapy. Int. J. Mol. Sci. 2020, 21, 4361. [Google Scholar] [CrossRef]
- Rose, M.G.; Berliner, N. T-cell large granular lymphocyte leukemia and related disorders. Oncologist 2004, 9, 247–258. [Google Scholar] [CrossRef]
- Kawasaki, T.; Suzuki, M.; Sato, A.; Yashima-Abo, A.; Satoh, T.; Kato, R.; Kato, Y.; Obara, W.; Shimoyama, T.; Ishida, Y. Neural cell adhesion molecule (CD56)-positive B cell lymphoma of the urinary bladder. J. Clin. Pathol. 2016, 69, 89–92. [Google Scholar] [CrossRef]
- Guan, F.; Wang, X.; He, F. Promotion of cell migration by neural cell adhesion molecule (NCAM) is enhanced by PSA in a polysialyltransferase-specific manner. PLoS ONE 2015, 10, e0124237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehembre, F.; Yilmaz, M.; Wicki, A.; Schomber, T.; Strittmatter, K.; Ziegler, D.; Kren, A.; Went, P.; Derksen, P.W.; Berns, A.; et al. NCAM-induced focal adhesion assembly: A functional switch upon loss of E-cadherin. EMBO J. 2008, 27, 2603–2615. [Google Scholar] [CrossRef] [Green Version]
- Schreiber, S.C.; Giehl, K.; Kastilan, C.; Hasel, C.; Muhlenhoff, M.; Adler, G.; Wedlich, D.; Menke, A. Polysialylated NCAM represses E-cadherin-mediated cell-cell adhesion in pancreatic tumor cells. Gastroenterology 2008, 134, 1555–1566. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Parrish, A.R.; Hill, M.A.; Meininger, G.A. N-cadherin, A Vascular Smooth Muscle Cell–Cell Adhesion Molecule: Function and Signaling for Vasomotor Control. Microcirculation 2014, 21, 208–218. [Google Scholar] [CrossRef]
- Cagnoni, A.J.; Perez Saez, J.M.; Rabinovich, G.A.; Marino, K.V. Turning-Off Signaling by Siglecs, Selectins, and Galectins: Chemical Inhibition of Glycan-Dependent Interactions in Cancer. Front. Oncol. 2016, 6, 109. [Google Scholar] [CrossRef] [Green Version]
- Ge, H.; Mu, L.; Jin, L.; Yang, C.; Chang, Y.E.; Long, Y.; DeLeon, G.; Deleyrolle, L.; Mitchell, D.A.; Kubilis, P.S.; et al. Tumor associated CD70 expression is involved in promoting tumor migration and macrophage infiltration in GBM. Int. J. Cancer 2017, 141, 1434–1444. [Google Scholar] [CrossRef] [Green Version]
- Micheau, O. Regulation of TNF-Related Apoptosis-Inducing Ligand Signaling by Glycosylation. Int. J. Mol. Sci. 2018, 19, 715. [Google Scholar] [CrossRef] [Green Version]
- Beatty, G.L.; Gladney, W.L. Immune escape mechanisms as a guide for cancer immunotherapy. Clin. Cancer Res. 2015, 21, 687–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeuchi, H.; Haltiwanger, R.S. Significance of glycosylation in Notch signaling. Biochem. Biophys. Res. Commun. 2014, 453, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Wu, A.A.; Drake, V.; Huang, H.S.; Chiu, S.; Zheng, L. Reprogramming the tumor microenvironment: Tumor-induced immunosuppressive factors paralyze T cells. Oncoimmunology 2015, 4, e1016700. [Google Scholar] [CrossRef] [PubMed]
- Klein, D. The Tumor Vascular Endothelium as Decision Maker in Cancer Therapy. Front. Oncol. 2018, 8, 367. [Google Scholar] [CrossRef] [PubMed]
- Weishaupt, C.; Munoz, K.N.; Buzney, E.; Kupper, T.S.; Fuhlbrigge, R.C. T-cell distribution and adhesion receptor expression in metastatic melanoma. Clin. Cancer Res. 2007, 13, 2549–2556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afanasiev, O.K.; Nagase, K.; Simonson, W.; Vandeven, N.; Blom, A.; Koelle, D.M.; Clark, R.; Nghiem, P. Vascular E-selectin expression correlates with CD8 lymphocyte infiltration and improved outcome in Merkel cell carcinoma. J. Investig. Dermatol. 2013, 133, 2065–2073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iozzo, R.V.; Schaefer, L. Proteoglycan form and function: A comprehensive nomenclature of proteoglycans. Matrix Biol. 2015, 42, 11–55. [Google Scholar] [CrossRef]
- Haspel, J.; Grumet, M. The L1CAM extracellular region: A multi-domain protein with modular and cooperative binding modes. Front. Biosci. 2003, 8, s1210–s1225. [Google Scholar] [CrossRef] [Green Version]
- Pollerberg, G.E.; Thelen, K.; Theiss, M.O.; Hochlehnert, B.C. The role of cell adhesion molecules for navigating axons: Density matters. Mech. Dev. 2013, 130, 359–372. [Google Scholar] [CrossRef]
- Fujimoto, I.; Bruses, J.L.; Rutishauser, U. Regulation of cell adhesion by polysialic acid. Effects on cadherin, immunoglobulin cell adhesion molecule, and integrin function and independence from neural cell adhesion molecule binding or signaling activity. J. Biol. Chem. 2001, 276, 31745–31751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nabatov, A.A.; Raginov, I.S. The DC-SIGN-CD56 interaction inhibits the anti-dendritic cell cytotoxicity of CD56 expressing cells. Infect. Agents Cancer 2015, 10, 49. [Google Scholar] [CrossRef]
- Chung, K.Y.; Leung, K.M.; Lin, C.C.; Tam, K.C.; Hao, Y.L.; Taylor, J.S.; Chan, S.O. Regionally specific expression of L1 and sialylated NCAM in the retinofugal pathway of mouse embryos. J. Comp. Neurol. 2004, 471, 482–498. [Google Scholar] [CrossRef]
- Trouillas, J.; Daniel, L.; Guigard, M.P.; Tong, S.; Gouvernet, J.; Jouanneau, E.; Jan, M.; Perrin, G.; Fischer, G.; Tabarin, A.; et al. Polysialylated neural cell adhesion molecules expressed in human pituitary tumors and related to extrasellar invasion. J. Neurosurg. 2003, 98, 1084–1093. [Google Scholar] [CrossRef]
- Fernandez-Briera, A.; Garcia-Parceiro, I.; Cuevas, E.; Gil-Martin, E. Effect of human colorectal carcinogenesis on the neural cell adhesion molecule expression and polysialylation. Oncology-Basel 2010, 78, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Binder, C.; Cvetkovski, F.; Sellberg, F.; Berg, S.; Paternina Visbal, H.; Sachs, D.H.; Berglund, E.; Berglund, D. CD2 Immunobiology. Front. Immunol. 2020, 11, 1090. [Google Scholar] [CrossRef] [PubMed]
- Hamai, A.; Meslin, F.; Benlalam, H.; Jalil, A.; Mehrpour, M.; Faure, F.; Lecluse, Y.; Vielh, P.; Avril, M.F.; Robert, C.; et al. ICAM-1 has a critical role in the regulation of metastatic melanoma tumor susceptibility to CTL lysis by interfering with PI3K/AKT pathway. Cancer Res. 2008, 68, 9854–9864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valgardsdottir, R.; Capitanio, C.; Texido, G.; Pende, D.; Cantoni, C.; Pesenti, E.; Rambaldi, A.; Golay, J.; Introna, M. Direct involvement of CD56 in cytokine-induced killer-mediated lysis of CD56+ hematopoietic target cells. Exp. Hematol. 2014, 42, 1013–1021.e1011. [Google Scholar] [CrossRef]
- McGreal, E.P.; Miller, J.L.; Gordon, S. Ligand recognition by antigen-presenting cell C-type lectin receptors. Curr. Opin. Immunol. 2005, 17, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Khoo, U.S.; Chan, K.Y.; Chan, V.S.; Lin, C.L. DC-SIGN and L-SIGN: The SIGNs for infection. J. Mol. Med. 2008, 86, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Leger, P.; Tetard, M.; Youness, B.; Cordes, N.; Rouxel, R.N.; Flamand, M.; Lozach, P.Y. Differential Use of the C-Type Lectins L-SIGN and DC-SIGN for Phlebovirus Endocytosis. Traffic 2016, 17, 639–656. [Google Scholar] [CrossRef]
- Lozach, P.Y.; Amara, A.; Bartosch, B.; Virelizier, J.L.; Arenzana-Seisdedos, F.; Cosset, F.L.; Altmeyer, R. C-type lectins L-SIGN and DC-SIGN capture and transmit infectious hepatitis C virus pseudotype particles. J. Biol. Chem. 2004, 279, 32035–32045. [Google Scholar] [CrossRef] [Green Version]
- Lozach, P.Y.; Kuhbacher, A.; Meier, R.; Mancini, R.; Bitto, D.; Bouloy, M.; Helenius, A. DC-SIGN as a receptor for phleboviruses. Cell Host Microbe 2011, 10, 75–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goncalves, A.R.; Moraz, M.L.; Pasquato, A.; Helenius, A.; Lozach, P.Y.; Kunz, S. Role of DC-SIGN in Lassa virus entry into human dendritic cells. J. Virol. 2013, 87, 11504–11515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curreli, S.; Arany, Z.; Gerardy-Schahn, R.; Mann, D.; Stamatos, N.M. Polysialylated neuropilin-2 is expressed on the surface of human dendritic cells and modulates dendritic cell-T lymphocyte interactions. J. Biol. Chem. 2007, 282, 30346–30356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abel, A.M.; Yang, C.; Thakar, M.S.; Malarkannan, S. Natural Killer Cells: Development, Maturation, and Clinical Utilization. Front. Immunol. 2018, 9, 1869. [Google Scholar] [CrossRef] [Green Version]
- Gumbiner, B.M. Regulation of cadherin-mediated adhesion in morphogenesis. Nat. Rev. Mol. Cell Biol. 2005, 6, 622–634. [Google Scholar] [CrossRef]
- Shapiro, L.; Fannon, A.M.; Kwong, P.D.; Thompson, A.; Lehmann, M.S.; Grubel, G.; Legrand, J.F.; Als-Nielsen, J.; Colman, D.R.; Hendrickson, W.A. Structural basis of cell-cell adhesion by cadherins. Nature 1995, 374, 327–337. [Google Scholar] [CrossRef]
- Marie, P.J.; Hay, E.; Modrowski, D.; Revollo, L.; Mbalaviele, G.; Civitelli, R. Cadherin-mediated cell-cell adhesion and signaling in the skeleton. Calcif. Tissue Int. 2014, 94, 46–54. [Google Scholar] [CrossRef] [Green Version]
- Takei, R.; Suzuki, D.; Hoshiba, T.; Nagaoka, M.; Seo, S.J.; Cho, C.S.; Akaike, T. Role of E-cadherin molecules in spheroid formation of hepatocytes adhered on galactose-carrying polymer as an artificial asialoglycoprotein model. Biotechnol. Lett. 2005, 27, 1149–1156. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, Y.; Sakane, F.; Hashimoto, K. N-cadherin-based adherens junction regulates the maintenance, proliferation, and differentiation of neural progenitor cells during development. Cell Adh. Migr. 2015, 9, 183–192. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Guo, Z.; Yang, Q.; Sad, S.; Jennings, H.J. Biochemical engineering of surface alpha 2-8 polysialic acid for immunotargeting tumor cells. J. Biol. Chem. 2000, 275, 32832–32836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crossin, K.L.; Krushel, L.A. Cellular signaling by neural cell adhesion molecules of the immunoglobulin superfamily. Dev. Dynam. 2000, 218, 260–279. [Google Scholar] [CrossRef]
- Pruneri, G.; Vingiani, A.; Denkert, C. Tumor infiltrating lymphocytes in early breast cancer. Breast 2018, 37, 207–214. [Google Scholar] [CrossRef] [Green Version]
- Krpina, K.; Babarovic, E.; Spanjol, J.; Dordevic, G.; Maurer, T.; Jonjic, N. Correlation of tumor-associated macrophages and NK cells with bladder cancer size and T stage in patients with solitary low-grade urothelial carcinoma. Wien. Klin. Wochenschr. 2016, 128, 248–252. [Google Scholar] [CrossRef]
- Briercheck, E.L.; Trotta, R.; Chen, L.; Hartlage, A.S.; Cole, J.P.; Cole, T.D.; Mao, C.; Banerjee, P.P.; Hsu, H.T.; Mace, E.M.; et al. PTEN is a negative regulator of NK cell cytolytic function. J. Immunol. 2015, 194, 1832–1840. [Google Scholar] [CrossRef] [Green Version]
- Peng, L.X.; Liu, X.H.; Lu, B.; Liao, S.M.; Zhou, F.; Huang, J.M.; Chen, D.; Troy, F.A., II; Zhou, G.P.; Huang, R.B. The Inhibition of Polysialyltranseferase ST8SiaIV Through Heparin Binding to Polysialyltransferase Domain (PSTD). Med. Chem. 2019, 15, 486–495. [Google Scholar] [CrossRef] [PubMed]
- Dityatev, A.; Bukalo, O.; Schachner, M. Modulation of synaptic transmission and plasticity by cell adhesion and repulsion molecules. Neuron Glia Biol. 2008, 4, 197–209. [Google Scholar] [CrossRef] [PubMed]
- Schmid, R.S.; Maness, P.F. L1 and NCAM adhesion molecules as signaling coreceptors in neuronal migration and process outgrowth. Curr. Opin. Neurobiol. 2008, 18, 245–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmid, R.S.; Pruitt, W.M.; Maness, P.F. A MAP kinase-signaling pathway mediates neurite outgrowth on L1 and requires Src-dependent endocytosis. J. Neurosci. 2000, 20, 4177–4188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angiolini, F.; Cavallaro, U. The Pleiotropic Role of L1CAM in Tumor Vasculature. Int. J. Mol. Sci. 2017, 18, 254. [Google Scholar] [CrossRef] [Green Version]
- Kleene, R.; Yang, H.; Kutsche, M.; Schachner, M. The neural recognition molecule L1 is a sialic acid-binding lectin for CD24, which induces promotion and inhibition of neurite outgrowth. J. Biol. Chem. 2001, 276, 21656–21663. [Google Scholar] [CrossRef] [Green Version]
- Buhusi, M.; Midkiff, B.R.; Gates, A.M.; Richter, M.; Schachner, M.; Maness, P.F. Close homolog of L1 is an enhancer of integrin-mediated cell migration. J. Biol. Chem. 2003, 278, 25024–25031. [Google Scholar] [CrossRef] [Green Version]
- Takei, K.; Chan, T.A.; Wang, F.S.; Deng, H.; Rutishauser, U.; Jay, D.G. The neural cell adhesion molecules L1 and NCAM-180 act in different steps of neurite outgrowth. J. Neurosci. 1999, 19, 9469–9479. [Google Scholar] [CrossRef]
- Mechtersheimer, S.; Gutwein, P.; Agmon-Levin, N.; Stoeck, A.; Oleszewski, M.; Riedle, S.; Postina, R.; Fahrenholz, F.; Fogel, M.; Lemmon, V.; et al. Ectodomain shedding of L1 adhesion molecule promotes cell migration by autocrine binding to integrins. J. Cell Biol. 2001, 155, 661–673. [Google Scholar] [CrossRef] [Green Version]
- Kiefel, H.; Bondong, S.; Pfeifer, M.; Schirmer, U.; Erbe-Hoffmann, N.; Schafer, H.; Sebens, S.; Altevogt, P. EMT-associated up-regulation of L1CAM provides insights into L1CAM-mediated integrin signalling and NF-kappaB activation. Carcinogenesis 2012, 33, 1919–1929. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C.; Künemund, V.; Wintergerst, E.S.; Schmitz, B.; Schachner, M. CD9 of mouse brain is implicated in neurite outgrowth and cell migration in vitro and is associated with the α6/β1 integrin and the neural adhesion molecule L1. J. Neurosci. Res. 1996, 43, 12–31. [Google Scholar] [CrossRef] [PubMed]
- Moulla, A.; Miliaras, D.; Sioga, A.; Kaidoglou, A.; Economou, L. The immunohistochemical expression of CD24 and CD171 adhesion molecules in borderline ovarian tumors. Pol. J. Pathol. 2013, 64, 180–184. [Google Scholar] [CrossRef] [PubMed]
- Stoeck, A.; Schlich, S.; Issa, Y.; Gschwend, V.; Wenger, T.; Herr, I.; Marme, A.; Bourbie, S.; Altevogt, P.; Gutwein, P. L1 on ovarian carcinoma cells is a binding partner for Neuropilin-1 on mesothelial cells. Cancer Lett. 2006, 239, 212–226. [Google Scholar] [CrossRef] [PubMed]
- Castellani, V.; De Angelis, E.; Kenwrick, S.; Rougon, G. Cis and trans interactions of L1 with neuropilin-1 control axonal responses to semaphorin 3A. EMBO J. 2002, 21, 6348–6357. [Google Scholar] [CrossRef] [Green Version]
- Mohanan, V.; Temburni, M.K.; Kappes, J.C.; Galileo, D.S. L1CAM stimulates glioma cell motility and proliferation through the fibroblast growth factor receptor. Clin. Exp. Metastasis 2013, 30, 507–520. [Google Scholar] [CrossRef]
- Shtutman, M.; Levina, E.; Ohouo, P.; Baig, M.; Roninson, I.B. Cell adhesion molecule L1 disrupts E-cadherin-containing adherens junctions and increases scattering and motility of MCF7 breast carcinoma cells. Cancer Res. 2006, 66, 11370–11380. [Google Scholar] [CrossRef] [Green Version]
- Nagasundaram, M.; Horstkorte, R.; Gnanapragassam, V.S. Sialic Acid Metabolic Engineering of Breast Cancer Cells Interferes with Adhesion and Migration. Molecules 2020, 25, 2632. [Google Scholar] [CrossRef]
- Kamiguchi, H.; Lemmon, V. A neuronal form of the cell adhesion molecule L1 contains a tyrosine-based signal required for sorting to the axonal growth cone. J. Neurosci. 1998, 18, 3749–3756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, H.J.; Galileo, D.S. Small-molecule inhibitors of FGFR, integrins and FAK selectively decrease L1CAM-stimulated glioblastoma cell motility and proliferation. Cell Oncol. 2016, 39, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Sytnyk, V.; Leshchyns’ka, I.; Schachner, M. Neural Cell Adhesion Molecules of the Immunoglobulin Superfamily Regulate Synapse Formation, Maintenance, and Function. Trends Neurosci. 2017, 40, 295–308. [Google Scholar] [CrossRef] [PubMed]
- Angiolini, F.; Belloni, E.; Giordano, M.; Campioni, M.; Forneris, F.; Paronetto, M.P.; Lupia, M.; Brandas, C.; Pradella, D.; Di Matteo, A.; et al. A novel L1CAM isoform with angiogenic activity generated by NOVA2-mediated alternative splicing. eLife 2019, 8, e44305. [Google Scholar] [CrossRef]
- He, Z. Crossed wires: L1 and neuropilin interactions. Neuron 2000, 27, 191–193. [Google Scholar] [CrossRef] [Green Version]
- McNutt, M. Cancer immunotherapy. Science 2013, 342, 1417. [Google Scholar] [CrossRef]
- Rosenberg, S.A. Progress in human tumour immunology and immunotherapy. Nature 2001, 411, 380–384. [Google Scholar] [CrossRef]
- Morise, J.; Takematsu, H.; Oka, S. The role of human natural killer-1 (HNK-1) carbohydrate in neuronal plasticity and disease. Biochim. Et Biophys. Acta-Gen. Subj. 2017, 1861, 2455–2461. [Google Scholar] [CrossRef] [PubMed]
- Suzuki-Anekoji, M.; Suzuki, M.; Kobayashi, T.; Sato, Y.; Nakayama, J.; Suzuki, A.; Bao, X.; Angata, K.; Fukuda, M. HNK-1 glycan functions as a tumor suppressor for astrocytic tumor. J. Biol. Chem. 2011, 286, 32824–32833. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Lectin Family | Saccharide Ligands | Subcellular Location | Examples of Functions | Example-Proteins |
|---|---|---|---|---|
| C-Type-Lectin Calcium-dependent | Man6Ps (a), Gal (b), N-acetylgalactosamine (Sialic acid), Fuc (c), etc. | Cell membrane, extracellular | Cell adhesion, (selectins), Glycoprotein clearance, Innate immunity (Collectins) | DC-SIGN (d), Dectin1, NKG2D (e), REG3 (f) proteins, L-Selectin, MMR (g), MBL (h)-protein, Tetranectin, PKD1 (i), Thrombomodulin, Attractin (DPPT-L), Human Macrophage Galactose-Type Lectin (MGL) (j) |
| Siglecs (k) (I-Type-Lectin) | Sialic acid | Cell membrane | Molecular and cell recognition | Siglec1, Siglec2, Siglec4 & Siglecs 3-16 |
| F-Type Lectin (fucose binding lectins) | Fuc termini | Extracellular | Innate immunity | L-fucose binding proteins |
| F-box Lectin | GlcNAc2 | Cytoplasm | Degradation of misfolded proteins | β-TRCP1 and β-TRCP2 (l) proteins |
| L-Type-Lectin | Man6Ps, Gal, Sialic acid, Fuc, etc. | ER (m), ERGIC (n), Golgi | Protein sorting in the ER | ERGIC-53, ERGL (o), VIP36 (p), and VIPL (q) |
| M-Type-Lectin | Man8Ps | ER | ER-associated degradation | EDEM1 (r), EDEM2, EDEM3 |
| P-Type-Lectin | Man6Ps and others | Secretory pathway | Post-Golgi glycoprotein trafficking | Mannose 6-phosphate receptor (M6Ps) |
| R-Type-Lectin | Man6Ps, Gal, Sialic acid, Xyl (s) | Golgi, Cell membrane | Enzyme targeting, hormone turnover | Mannose receptor family e.g., DEC-205 |
| S-Type-Lectin (Galectins) | β-Galactosides | Cytoplasm, extracellular | Cell surface crosslinking | Galectin-1, -2, -5, -7, -10, -11, -14 and -15 |
| X-type-Lectin (Intelectins) | Gal, galactofuranose, pentose | Cell membrane, extracellular | Innate immunity, fertilization and embryogenesis | Intelectin-1 and 2 |
| Ficolins | Sialic acid, GlcNAc, GalNAc | Cell membrane, extracellular | Innate immunity | H-ficolin and M-ficolin |
| Calnexin family | Glc1Man9 | ER | Protein sorting in the ER | Calnexin, calmegin, calreticulin |
| Chitinase like-Lectin | Chito (t)-oligosaccharides | Extracellular | Collagen metabolism | Chitinase 3-like 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jarahian, M.; Marofi, F.; Maashi, M.S.; Ghaebi, M.; Khezri, A.; Berger, M.R. Re-Expression of Poly/Oligo-Sialylated Adhesion Molecules on the Surface of Tumor Cells Disrupts Their Interaction with Immune-Effector Cells and Contributes to Pathophysiological Immune Escape. Cancers 2021, 13, 5203. https://doi.org/10.3390/cancers13205203
Jarahian M, Marofi F, Maashi MS, Ghaebi M, Khezri A, Berger MR. Re-Expression of Poly/Oligo-Sialylated Adhesion Molecules on the Surface of Tumor Cells Disrupts Their Interaction with Immune-Effector Cells and Contributes to Pathophysiological Immune Escape. Cancers. 2021; 13(20):5203. https://doi.org/10.3390/cancers13205203
Chicago/Turabian StyleJarahian, Mostafa, Faroogh Marofi, Marwah Suliman Maashi, Mahnaz Ghaebi, Abdolrahman Khezri, and Martin R. Berger. 2021. "Re-Expression of Poly/Oligo-Sialylated Adhesion Molecules on the Surface of Tumor Cells Disrupts Their Interaction with Immune-Effector Cells and Contributes to Pathophysiological Immune Escape" Cancers 13, no. 20: 5203. https://doi.org/10.3390/cancers13205203