Neuromedin U: A Small Peptide in the Big World of Cancer
Abstract
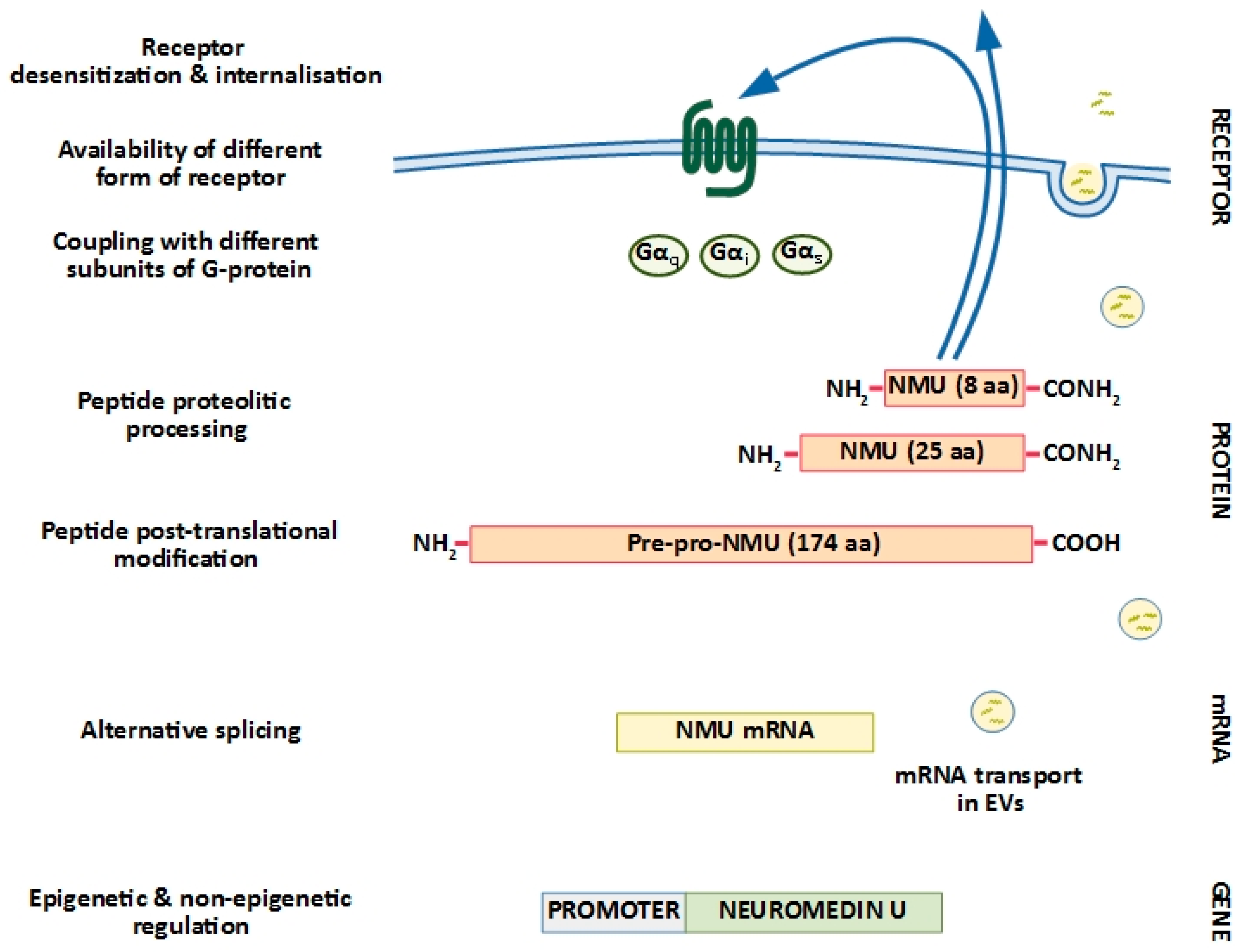
1. Neuromedin U Structure: Implications for Biological Activity
2. Neuromedin U Receptors: Structure and Distribution
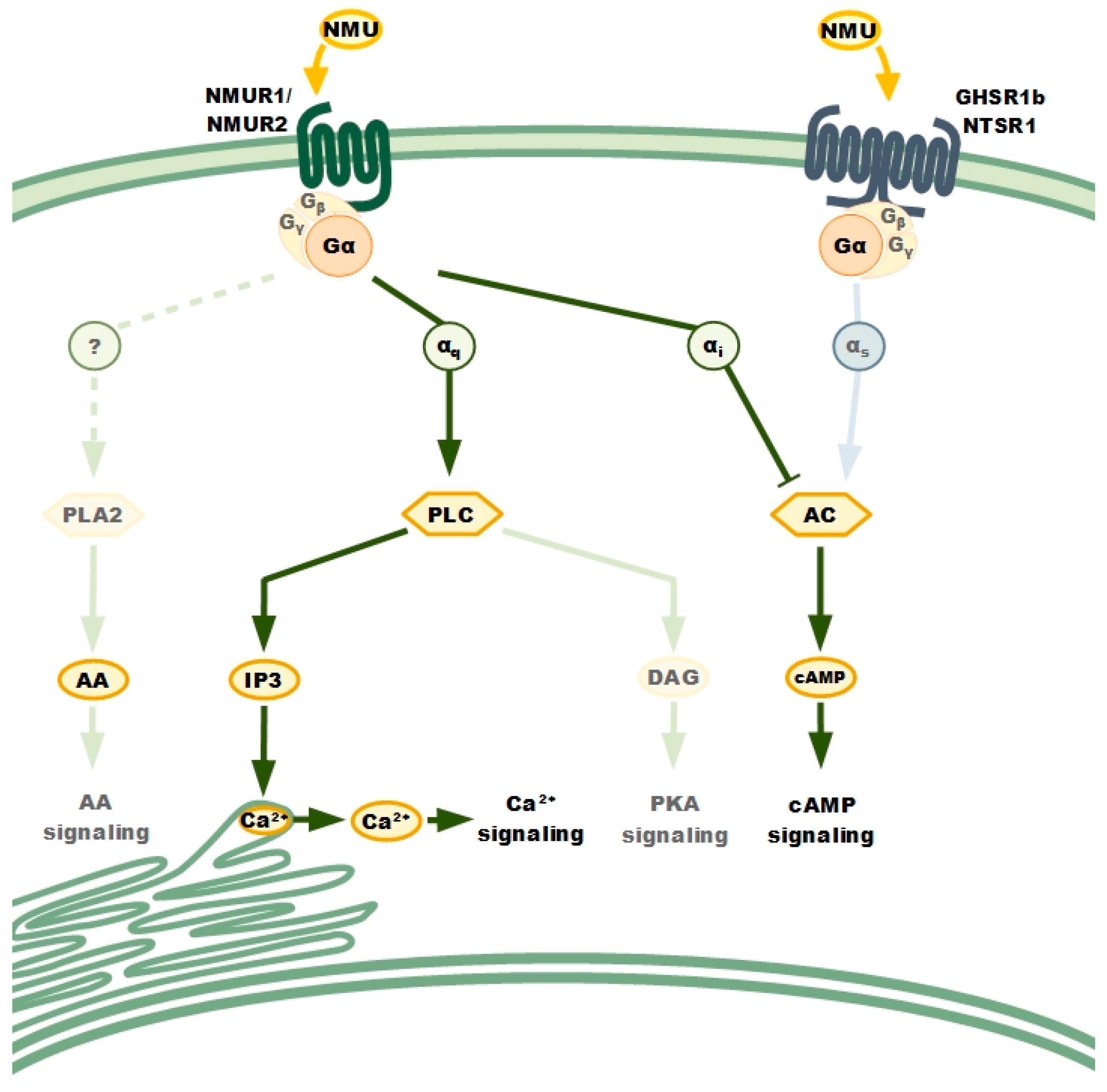
NMURs: Classical GPCRs
3. NMU in Cancer: Knowing the Future Through the Present
3.1. NMU as A Tumour Suppressor
3.2. NMU as A Prognostic Factor
3.3. Effect of NMU on Metastasis Formation
4. NMU Signalling in Cancer Cells Biology
4.1. Cancer Cells Proliferation and Viability
4.2. EMT, Cancer Cell Motility, and Invasiveness
4.3. NMU’s Contribution to Cancer Cell Drug Resistance
5. Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mitchell, J.D.; Maguire, J.J.; Davenport, A.P. Emerging pharmacology and physiology of neuromedin U and the structurally related peptide neuromedin S. Br. J. Pharmacol. 2009, 158, 87–103. [Google Scholar] [CrossRef]
- Brighton, P.J.; Szekeres, P.G.; Willars, G.B. Neuromedin U and its receptors: Structure, function, and physiological roles. Pharmacol. Rev. 2004, 56, 231–248. [Google Scholar] [CrossRef]
- Mori, K.; Ida, T.; Fudetani, M.; Mori, M.; Kaiya, H.; Hino, J.; Nakahara, K.; Murakami, N.; Miyazato, M.; Kangawa, K. Identification of neuromedin U precursor-related peptide and its possible role in the regulation of prolactin release. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Mori, K.; Miyazato, M.; Ida, T.; Murakami, N.; Serino, R.; Ueta, Y.; Kojima, M.; Kangawa, K. Identification of neuromedin S and its possible role in the mammalian circadian oscillator system. EMBO J. 2005, 24, 325–335. [Google Scholar] [CrossRef]
- Mori, K.; Miyazato, M.; Kangawa, K. Neuromedin S: Discovery and functions. Results Probl. Cell Differ. 2008, 46, 201–212. [Google Scholar] [CrossRef]
- Austin, C.; Lo, G.; Nandha, K.A.; Meleagros, L.; Bloom, S.R. Cloning and characterization of the cDNA encoding the human neuromedin U (NmU) precursor: NmU expression in the human gastrointestinal tract. J. Mol. Endocrinol. 1995, 14, 157–169. [Google Scholar] [CrossRef]
- Hosoya, M.; Moriya, T.; Kawamata, Y.; Ohkubo, S.; Fujii, R.; Matsui, H.; Shintani, Y.; Fukusumi, S.; Habata, Y.; Hinuma, S.; et al. Identification and functional characterization of a novel subtype of neuromedin U receptor. J. Biol. Chem. 2000, 275, 29528–29532. [Google Scholar] [CrossRef]
- Raddatz, R.; Wilson, A.E.; Artymyshyn, R.; Bonini, J.A.; Borowsky, B.; Boteju, L.W.; Zhou, S.Q.; Kouranova, E.V.; Nagorny, R.; Guevarra, M.S.; et al. Identification and characterization of two neuromedin U receptors differentially expressed in peripheral tissues and the central nervous system. J. Biol. Chem. 2000, 275, 32452–32459. [Google Scholar] [CrossRef]
- Aiyar, N.; Disa, J.; Foley, J.J.; Buckley, P.T.; Wixted, W.E.; Pullen, M.; Shabon, U.; Dul, E.; Szekeres, P.G. Radioligand binding and functional characterization of recombinant human NmU1 and NmU2 receptors stably expressed in clonal human embryonic kidney-293 cells. Pharmacology 2004, 72, 33–41. [Google Scholar] [CrossRef]
- Tan, C.P.; McKee, K.K.; Liu, Q.Y.; Palyha, O.C.; Feighner, S.D.; Hreniuk, D.L.; Smith, R.G.; Howard, A.D. Cloning and characterization of a human and murine T-cell orphan G-protein-coupled receptor similar to the growth hormone secretagogue and neurotensin receptors. Genomics 1998, 52, 223–229. [Google Scholar] [CrossRef]
- Kojima, M.; Haruno, R.; Nakazato, M.; Date, Y.; Murakami, N.; Hanada, R.; Matsuo, H.; Kangawa, K. Purification and identification of neuromedin U as an endogenous ligand for an orphan receptor GPR66 (FM3). Biochem. Biophys. Res. Commun. 2000, 276, 435–438. [Google Scholar] [CrossRef]
- Szekeres, P.G.; Muir, A.I.; Spinage, L.D.; Miller, J.E.; Butler, S.I.; Smith, A.; Rennie, G.I.; Murdock, P.R.; Fitzgerald, L.R.; Wu, H.L.; et al. Neuromedin U is a potent agonist at the orphan G protein-coupled receptor FM3. J. Biol. Chem. 2000, 275, 20247–20250. [Google Scholar] [CrossRef]
- Shan, L.X.; Qiao, X.D.; Crona, J.H.; Behan, J.; Wang, S.; Laz, T.; Bayne, M.; Gustafson, E.L.; Monsma, F.J.; Hedrick, J.A. Identification of a novel neuromedin U receptor subtype expressed in the central nervous system. J. Biol. Chem. 2000, 275, 39482–39486. [Google Scholar] [CrossRef]
- Fujii, R.; Hosoya, M.; Fukusumi, S.; Kawamata, Y.; Habata, Y.; Hinuma, S.; Onda, H.; Nishimura, O.; Fujino, M. Identification of neuromedin U as the cognate ligand of the orphan G protein-coupled receptor FM-3. J. Biol. Chem. 2000, 275, 21068–21074. [Google Scholar] [CrossRef]
- Lin, T.Y.; Huang, W.L.; Lee, W.Y.; Luo, C.W. Identifying a Neuromedin U Receptor 2 Splice Variant and Determining its Roles in the Regulation of Signaling and Tumorigenesis in vitro. PLoS ONE 2015, 10. [Google Scholar] [CrossRef]
- Takahashi, K.; Furukawa, C.; Takano, A.; Ishikawa, N.; Kato, T.; Hayama, S.; Suzuki, C.; Yasui, W.; Inai, K.; Sone, S.; et al. The neuromedin U-growth hormone secretagogue receptor 1b/neurotensin receptor 1 oncogenic signaling pathway as a therapeutic target for lung cancer. Cancer Res. 2006, 66, 9408–9419. [Google Scholar] [CrossRef]
- Brighton, P.J.; Szekeres, P.G.; Wise, A.; Willars, G.B. Signaling and ligand binding by recombinant neuromedin U receptors: Evidence for dual coupling to G alpha(q/11) and G alpha(i) and an irreversible ligand-receptor interaction. Mol. Pharmacol. 2004, 66, 1544–1556. [Google Scholar] [CrossRef]
- Cabrera-Vera, T.M.; Vanhauwe, J.; Thomas, T.O.; Medkova, M.; Preininger, A.; Mazzoni, M.R.; Hamm, H.E. Insights into G protein structure, function, and regulation. Endocr. Rev. 2003, 24, 765–781. [Google Scholar] [CrossRef]
- Alhosaini, K.; Bahattab, O.; Qassam, H.; Challiss, R.A.J.; Willars, G.B. Ligand-Specific Signaling Profiles and Resensitization Mechanisms of the Neuromedin U2 Receptor. Mol. Pharmacol. 2018, 94, 674–688. [Google Scholar] [CrossRef]
- Martinez, V.G.; O’Driscoll, L. Neuromedin U: A Multifunctional Neuropeptide with Pleiotropic Roles. Clin. Chem. 2015, 61, 471–482. [Google Scholar] [CrossRef]
- Hanada, R.; Teranishi, H.; Pearson, J.T.; Kurokawa, M.; Hosoda, H.; Fukushima, N.; Fukue, Y.; Serino, R.; Fujihara, H.; Ueta, Y.; et al. Neuromedin U has a novel anorexigenic effect independent of the leptin signaling pathway. Nat. Med. 2004, 10, 1067–1073. [Google Scholar] [CrossRef]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Yamashita, K.; Upadhyay, S.; Osada, M.; Hoque, M.O.; Xiao, Y.; Mori, M.; Sato, F.; Meltzer, S.J.; Sidransky, D. Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. Cancer Cell 2002, 2, 485–495. [Google Scholar] [CrossRef]
- Tokumaru, Y.; Yamashita, K.; Osada, M.; Nomoto, S.; Sun, D.I.; Xiao, Y.; Hoque, M.O.; Westra, W.H.; Califano, J.A.; Sidransky, D. Inverse correlation between cyclin A1 hypermethylation and p53 mutation in head and neck cancer identified by reversal of epigenetic silencing. Cancer Res. 2004, 64, 5982–5987. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, C.; Li, F.; Hua, Q.Q.; Chen, S.M.; Xiao, B.K.; Dai, M.Y.; Li, M.; Zheng, A.Y.; Yu, D.; et al. Overexpression of neuromedin U is correlated with regional metastasis of head and neck squamous cell carcinoma. Mol. Med. Rep. 2016, 14, 1075–1082. [Google Scholar] [CrossRef] [PubMed]
- Ketterer, K.; Kong, B.; Frank, D.; Giese, N.A.; Bauer, A.; Hoheisel, J.; Korc, M.; Kleeff, J.; Michalski, C.W.; Friess, H. Neuromedin U is overexpressed in pancreatic cancer and increases invasiveness via the hepatocyte growth factor c-Met pathway. Cancer Lett. 2009, 277, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Shetzline, S.E.; Rallapalli, R.; Dowd, K.J.; Zou, S.M.; Nakata, Y.; Swider, C.R.; Kalota, A.; Choi, J.K.; Gewirtz, A.M. Neuromedin U: A Myb-regulated autocrine growth factor for human myeloid leukemias. Blood 2004, 104, 1833–1840. [Google Scholar] [CrossRef]
- Harding, M.A.; Theodorescu, D. RhoGDI2: A new metastasis suppressor gene: Discovery and clinical translation. Urol. Oncol.-Semin. Ori. 2007, 25, 401–406. [Google Scholar] [CrossRef]
- Wu, Y.; McRoberts, K.; Berr, S.S.; Frierson, H.F.; Conaway, M.; Theodorescu, D. Neuromedin U is regulated by the metastasis suppressor RhoGDI2 and is a novel promoter of tumor formation, lung metastasis and cancer cachexia. Oncogene 2007, 26, 765–773. [Google Scholar] [CrossRef]
- Przygodzka, P.; Papiewska-Pajak, I.; Bogusz, H.; Kryczka, J.; Sobierajska, K.; Kowalska, M.A.; Boncela, J. Neuromedin U is upregulated by Snail at early stages of EMT in HT29 colon cancer cells. BBA-Gen. Subj. 2016, 1860, 2445–2453. [Google Scholar] [CrossRef]
- You, S.J.; Gao, L. Identification of NMU as a potential gene conferring alectinib resistance in non-small cell lung cancer based on bioinformatics analyses. Gene 2018, 678, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.Y.; Wu, F.J.; Chang, C.L.; Li, Z.Y.; Luo, C.W. NMU signaling promotes endometrial cancer cell progression by modulating adhesion signaling. Oncotarget 2016, 7, 10228–10242. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.Y.; Wang, C.C.; Lee, W.Y.W.; Trovik, J.; Chung, T.K.H.; Kwong, J. Long non-coding RNA HAND2-AS1 inhibits invasion and metastasis in endometrioid endometrial carcinoma through inactivating neuromedin U. Cancer Lett. 2018, 413, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Garczyk, S.; Klotz, N.; Szczepanski, S.; Denecke, B.; Antonopoulos, W.; Von Stillfried, S.; Knchel, R.; Rose, M.; Dahl, E. Oncogenic features of neuromedin U in breast cancer are associated with NMUR2 expression involving crosstalk with members of the WNT signaling pathway. Oncotarget 2017, 8, 36246–36265. [Google Scholar] [CrossRef] [PubMed]
- Martinez, V.G.; Crown, J.; Porter, R.K.; O’Driscoll, L. Neuromedin U alters bioenergetics and expands the cancer stem cell phenotype in HER2-positive breast cancer. Int. J. Cancer 2017, 140, 2771–2784. [Google Scholar] [CrossRef] [PubMed]
- Rani, S.; Corcoran, C.; Shiels, L.; Germano, S.; Breslin, S.; Madden, S.; McDermott, M.S.; Browne, B.C.; O'Donovan, N.; Crown, J.; et al. Neuromedin U: A Candidate Biomarker and Therapeutic Target to Predict and Overcome Resistance to HER-Tyrosine Kinase Inhibitors. Cancer Res. 2014, 74, 3821–3833. [Google Scholar] [CrossRef]
- Harten, S.K.; Esteban, M.A.; Shukla, D.; Ashcroft, M.; Maxwell, P.H. Inactivation of the von Hippel-Lindau tumour suppressor gene induces Neuromedin U expression in renal cancer cells. Mol. Cancer 2011, 10, 89. [Google Scholar] [CrossRef]
- Zhang, S.L.; Wang, Q.; Han, Q.; Han, H.Z.; Lu, P.X. Identification and analysis of genes associated with papillary thyroid carcinoma by bioinformatics methods. Bioscience Rep. 2019, 39. [Google Scholar] [CrossRef]
- Alevizos, I.; Mahadevappa, M.; Zhang, X.; Ohyama, H.; Kohno, Y.; Posner, M.; Gallagher, G.T.; Varvares, M.; Cohen, D.; Kim, D.; et al. Oral cancer in vivo gene expression profiling assisted by laser capture microdissection and microarray analysis. Oncogene 2001, 20, 6196–6204. [Google Scholar] [CrossRef]
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The Different Mechanisms of Cancer Drug Resistance: A Brief Review. Adv. Pharm. Bull. 2017, 7, 339–348. [Google Scholar] [CrossRef]
| Cancer Type | Expression of NMU in Tissues (Cancer/Healthy) | NMU Receptors | Signal Contributors | Observed Biological Effects | |
|---|---|---|---|---|---|
| Expression | Signal Transduction Research | ||||
| Oesophageal [23] | ↓ | no data | no data | no data | NMU silencing in cells and cancer tissue is a result of NMU-promoter region hypermethylation. NMU treatment caused diminished colony formation efficacy of cancer cells. |
| Head and neck [24,25] | ↑ [24] | no data | no data | no data | NMU-promoter region hypermethylation [25]. NMU upregulation in the advanced stage of cancer [24]. |
| Pancreatic [26] | ↑ | NMUR2 | no data | c-Met | NMU and NMUR2 upregulation in the cancer tissues and cancer cell lines correlates with increased invasiveness and metastatic potential of cells. NMU and NMUR2 expression upregulation in metastatic tissues of the liver and lymph nodes. |
| Leukaemia [27] | no data | NMUR1 | NMUR1 | c-Myb | NMU treatment resulted in the increased leukaemia cells proliferation and increase in colony formation ability. NMU silencing resulted in decrease in leukaemia cells viability. |
| Bladder [28,29] | no data | no data | no data | RhoGDI2 | NMU expression in cells with metastatic features enhanced pulmonary metastasis |
| Colorectal [30] | ↑ | NMUR2 | no data | Snail | NMU upregulation in cancer cells at the early stage of EMT. NMU mRNA detected in microvesicle fraction released from invasive cells. |
| Lung [16,31] | ↑ [16] | GHSR1b/NSTR1 (heterodimer) | GSHR1b/NSTR1 | FOXM1 [31] CD80 [31] CHEK1 [31] IL1RN [31] MYCN [31] PIM1 [31] | NMU upregulation in the cancer tissues and cell lines led to increase in cancer cells growth and invasion [16]. NMU is potentially involved in cancer cells resistance to alectinib [31]. NMU silencing resulted in decreased cells viability and ability to form colonies [16]. |
| Endometrial [32,33] | ↑ [32,33] | NMUR1 (low) NMUR2 (high) | NMUR2 | ITGA1 [32] CD44 [32] MMP-2,3,9 [32] COLA4A1 [32] COLA4A2 [32] HAS3 [32] HYAL1 [32] HYAL2 [32] HYAL3 [32] c-SRC [32] RAC1 [32] RHOA [32] TGFB [32] EGF [32] HAND2-AS1 [33] | NMU upregulation in the cancer tissues correlated with poor outcome. NMU upregulation in cell lines increased cell proliferation and motility of cells isolated from grade II tumours [32]. NMU silencing resulted in decreased cells migration, invasion, proliferation, and adhesion [32,33]. Cancer cells with decreased NMU expression formed smaller tumours in mice models [32]. |
| Breast [34,35,36] | ↑ [34] | NMUR1 NMUR2 NSTR1 | NMUR1 [36] NMUR2 [34,36] | WNT (Myc, RAC1) β-catenin [35] E-cadherin vimentin [35] TGFB Ephrin receptor | NMU upregulation in the cancer tissues was proposed as prognostic biomarker for poor outcome [34,36]. NMU upregulation on the cellular level caused [34,35,36]:
|
| Renal [37] | ↑ | NMU1R (low) | no data | VHL HIF-1α HIF-2α | NMU upregulation in the cancer tissues was proposed as prognostic biomarker for poor outcome. NMU upregulation on the cellular level caused increased migration and invasion ability. |
| Ovary [15] | ↑ | NMUR1 NMUR2 NMUR2S | NMUR1 NMUR2 | no data | NMU and NMUR2 expression was increased in cancer tissues and on the cellular level significantly enhanced migration and invasion abilities. |
| Thyroid [38] | ↑ | no data | no data | no data | NMU upregulation correlated with patients decreased disease-free survival time. |
| Oral [39] | ↓ | no data | no data | no data | no data |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Przygodzka, P.; Soboska, K.; Sochacka, E.; Boncela, J. Neuromedin U: A Small Peptide in the Big World of Cancer. Cancers 2019, 11, 1312. https://doi.org/10.3390/cancers11091312
Przygodzka P, Soboska K, Sochacka E, Boncela J. Neuromedin U: A Small Peptide in the Big World of Cancer. Cancers. 2019; 11(9):1312. https://doi.org/10.3390/cancers11091312
Chicago/Turabian StylePrzygodzka, Patrycja, Kamila Soboska, Ewelina Sochacka, and Joanna Boncela. 2019. "Neuromedin U: A Small Peptide in the Big World of Cancer" Cancers 11, no. 9: 1312. https://doi.org/10.3390/cancers11091312
APA StylePrzygodzka, P., Soboska, K., Sochacka, E., & Boncela, J. (2019). Neuromedin U: A Small Peptide in the Big World of Cancer. Cancers, 11(9), 1312. https://doi.org/10.3390/cancers11091312





