Circulating MACC1 Transcripts in Glioblastoma Patients Predict Prognosis and Treatment Response
Abstract
:1. Introduction
2. Results
2.1. Patient Cohort
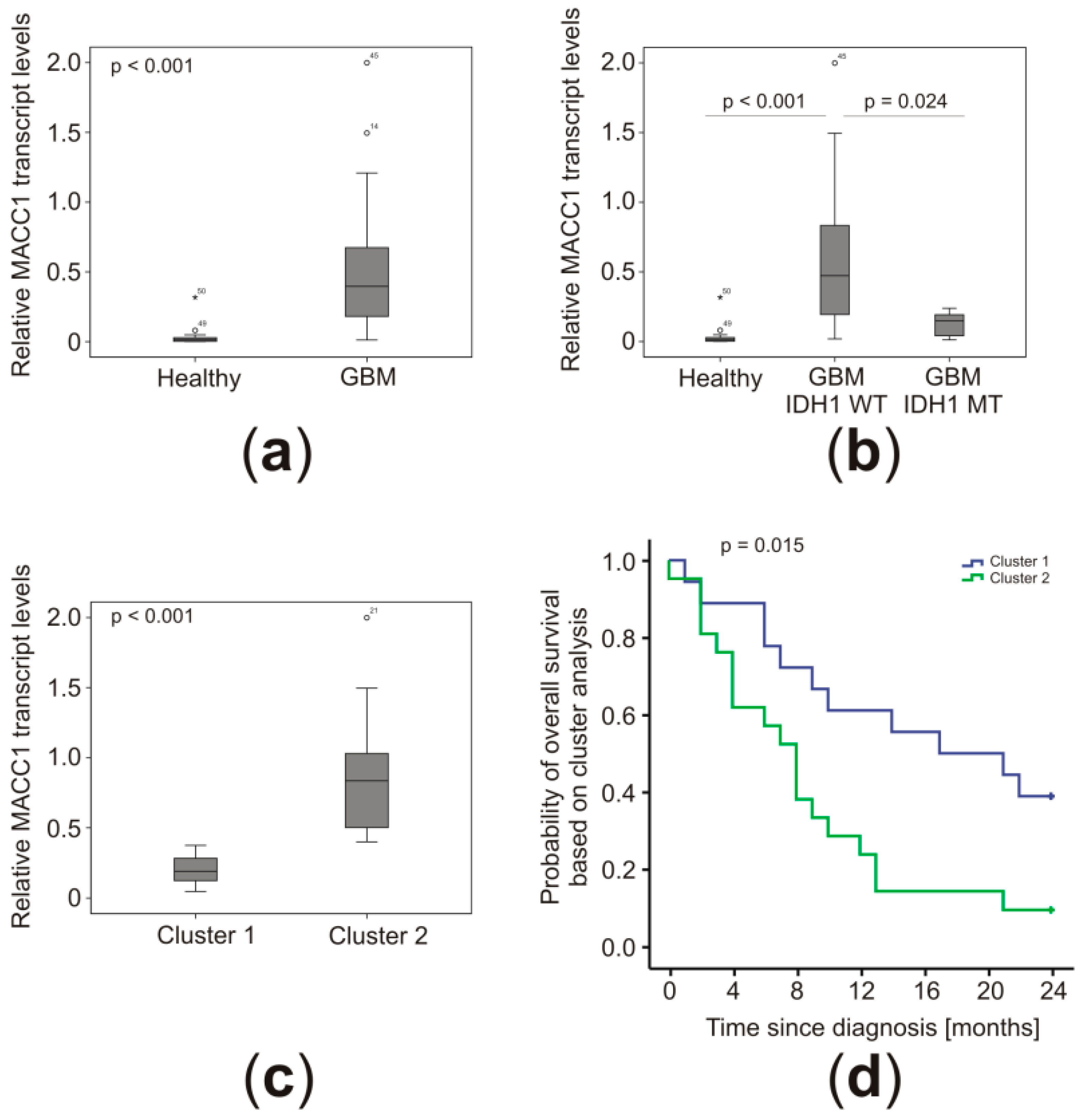
2.2. MACC1 Transcript Concentrations Were Higher in the Plasma of GBM Patients
2.3. Low MACC1 Plasma Levels Clustered together with Other Favorable Markers
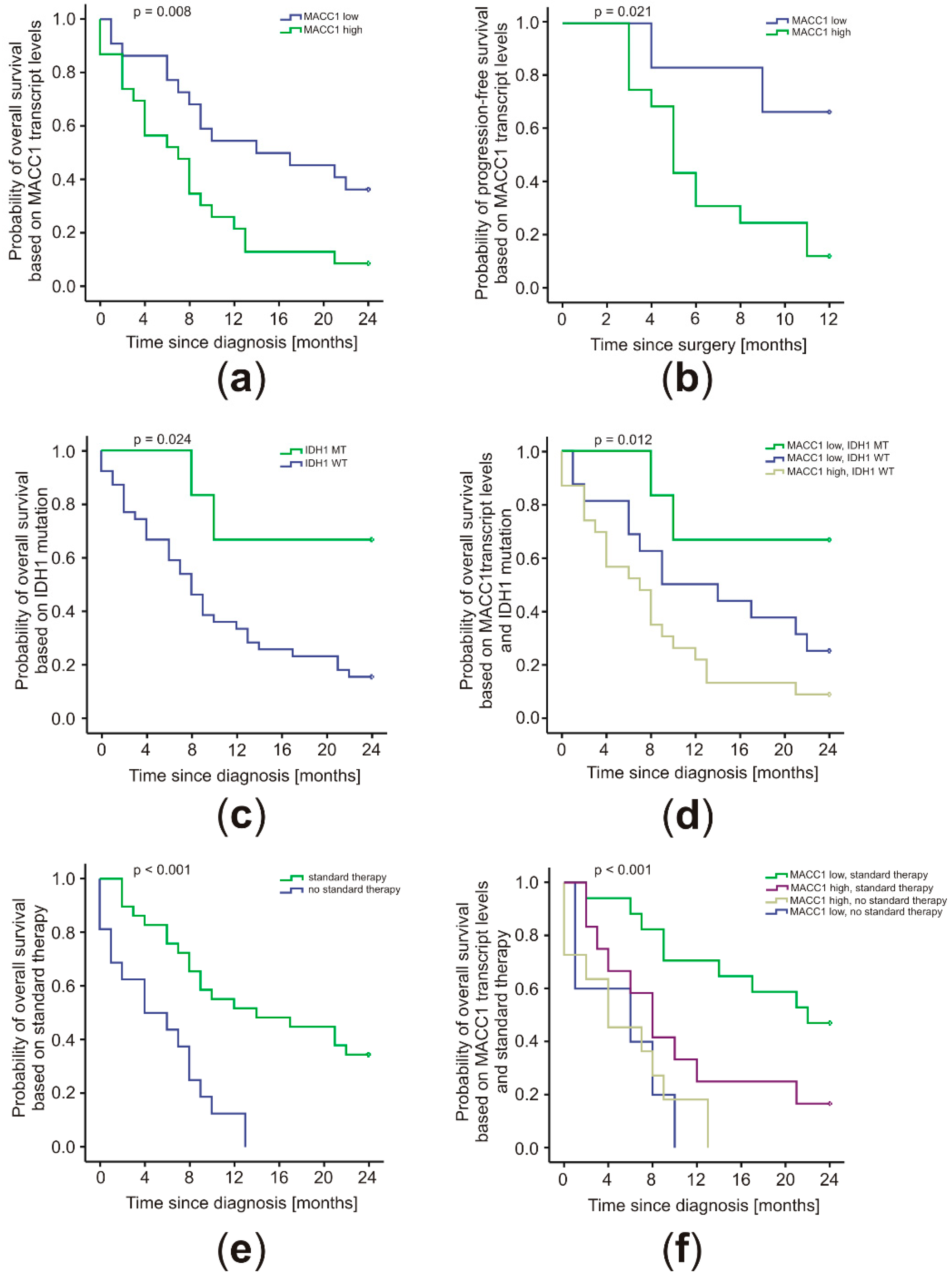
2.4. MACC1 Levels Correlated with Patient Prognosis in Conjunction with the IDH1 Mutation Status and Treatment Regimen
3. Discussion
4. Materials and Methods
4.1. Standard Protocol Approvals and Patient Consents
4.2. Plasma Samples
4.3. Quantitative RT-PCR
4.4. Immunohistochemistry (IHC) and Methylation-Specific High-Resolution Melting (HRM) Analysis
4.5. Statistics
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stupp, R.; Mason, W.P.; Van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; Cloughesy, T.; Perry, J.R.; Wick, W. Standards of care for treatment of recurrent glioblastoma—Are we there yet? Neuro Oncol. 2013, 15, 4–27. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; Van den Bent, M.; Hopkins, K.; Tonn, J.C.; Stupp, R.; Falini, A.; Cohen-Jonathan-Moyal, E.; Frappaz, D.; Henriksson, R.; Balana, C.; et al. EANO guideline for the diagnosis and treatment of anaplastic gliomas and glioblastoma. Lancet Oncol. 2014, 15, 395–403. [Google Scholar] [CrossRef]
- Hegi, M.E.; Diserens, A.C.; Godard, S.; Dietrich, P.Y.; Regli, L.; Ostermann, S.; Otten, P.; Van Melle, G.; De Tribolet, N.; Stupp, R. Clinical trial substantiates the predictive value of O–6–methylguanine–DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin. Cancer Res. 2004, 10, 1871–1874. [Google Scholar] [CrossRef] [PubMed]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; De Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Reifenberger, G.; Von Deimling, A.; Figarella–Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Noushmehr, H.; Weisenberger, D.J.; Diefes, K.; Phillips, H.S.; Pujara, K.; Berman, B.P.; Pan, F.; Pelloski, C.E.; Sulman, E.P.; Bhat, K.P.; et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 2010, 17, 510–522. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi–analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef]
- Zachariah, M.A.; Oliveira–Costa, J.P.; Carter, B.S.; Stott, S.L.; Nahed, B.V. Blood–based biomarkers for the diagnosis and monitoring of gliomas. Neuro. Oncol. 2018, 20, 1155–1161. [Google Scholar] [CrossRef]
- Stein, U.; Walther, W.; Arlt, F.; Schwabe, H.; Smith, J.; Fichtner, I.; Birchmeier, W.; Schlag, P.M. MACC1, a newly identified key regulator of HGF–MET signaling, predicts colon cancer metastasis. Nat. Med. 2009, 15, 59–67. [Google Scholar] [CrossRef]
- Radhakrishnan, H.; Walther, W.; Zincke, F.; Kobelt, D.; Imbastari, F.; Erdem, M.; Kortum, B.; Dahlmann, M.; Stein, U. MACC1–the first decade of a key metastasis molecule from gene discovery to clinical translation. Cancer Metastasis Rev. 2018, 37, 805–820. [Google Scholar] [CrossRef] [PubMed]
- Stein, U. MACC1–a novel target for solid cancers. Expert Opin. Ther. Targets 2013, 17, 1039–1052. [Google Scholar] [CrossRef] [PubMed]
- Burock, S.; Herrmann, P.; Wendler, I.; Niederstrasser, M.; Wernecke, K.D.; Stein, U. Circulating metastasis associated in colon cancer 1 transcripts in gastric cancer patient plasma as diagnostic and prognostic biomarker. World J. Gastroenterol. 2015, 21, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Stein, U.; Burock, S.; Herrmann, P.; Wendler, I.; Niederstrasser, M.; Wernecke, K.D.; Schlag, P.M. Circulating MACC1 transcripts in colorectal cancer patient plasma predict metastasis and prognosis. PLoS ONE 2012, 7, e49249. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Cai, M.; Weng, Y.; Zhang, F.; Meng, D.; Song, J.; Zhou, H.; Xie, Z. Circulating MACC1 as a novel diagnostic and prognostic biomarker for nonsmall cell lung cancer. J. Cancer Res. Clin. Oncol. 2015, 141, 1353–1361. [Google Scholar] [CrossRef] [PubMed]
- Hagemann, C.; Fuchs, S.; Monoranu, C.M.; Herrmann, P.; Smith, J.; Hohmann, T.; Grabiec, U.; Kessler, A.F.; Dehghani, F.; Löhr, M.; et al. Impact of MACC1 on human malignant glioma progression and patients’ unfavorable prognosis. Neuro Oncol. 2013, 15, 1696–1709. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Moore, L.M.; Li, X.; Yung, W.K.; Zhang, W. IDH1/2 mutations target a key hallmark of cancer by deregulating cellular metabolism in glioma. Neuro Oncol. 2013, 15, 1114–1126. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Lin, Y.; Xu, W.; Jiang, W.; Zha, Z.; Wang, P.; Yu, W.; Li, Z.; Gong, L.; Peng, Y.; et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF–1alpha. Science 2009, 324, 261–265. [Google Scholar] [CrossRef] [PubMed]
- Dang, L.; White, D.W.; Gross, S.; Bennett, B.D.; Bittinger, M.A.; Driggers, E.M.; Fantin, V.R.; Jang, H.G.; Jin, S.; Keenan, M.C.; et al. Cancer–associated IDH1 mutations produce 2–hydroxyglutarate. Nature 2009, 462, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Turcan, S.; Rohle, D.; Goenka, A.; Walsh, L.A.; Fang, F.; Yilmaz, E.; Campos, C.; Fabius, A.W.; Lu, C.; Ward, P.S.; et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 2012, 483, 479–483. [Google Scholar] [CrossRef]
- Reifenberger, G.; Collins, V.P. Pathology and molecular genetics of astrocytic gliomas. J. Mol. Med. 2004, 82, 656–670. [Google Scholar] [CrossRef] [PubMed]
- Batash, R.; Asna, N.; Schaffer, P.; Francis, N.; Schaffer, M. Glioblastoma multiforme, diagnosis and treatment; recent literature review. Curr. Med. Chem. 2017, 24, 3002–3009. [Google Scholar] [CrossRef] [PubMed]
- Tokarz, P.; Pawlowska, E.; Bialkowska–Warzecha, J.; Blasiak, J. The significance of DNA methylation profile in metastasis–related genes for the progression of colorectal cancer. Cell. Mol. Biol. 2017, 63, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Ilm, K.; Fuchs, S.; Mudduluru, G.; Stein, U. MACC1 is post–transcriptionally regulated by miR–218 in colorectal cancer. Oncotarget 2016, 7, 53443–53458. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Chen, X.; Su, L.; Li, C.; Zhi, Q.; Yu, B.; Sheng, H.; Wang, J.; Feng, R.; Cai, Q.; et al. Epigenetic silencing of miR–338–3p contributes to tumorigenicity in gastric cancer by targeting SSX2IP. PLoS ONE 2013, 8, e66782. [Google Scholar] [CrossRef] [PubMed]
- Shang, C.; Hong, Y.; Guo, Y.; Xue, Y.X. Mir–338–3p inhibits malignant biological behaviors of glioma cells by targeting MACC1 gene. Med. Sci. Monit. 2016, 22, 710–716. [Google Scholar] [PubMed]
- Sanson, M.; Marie, Y.; Paris, S.; Idbaih, A.; Laffaire, J.; Ducray, F.; El Hallani, S.; Boisselier, B.; Mokhtari, K.; Hoang–Xuan, K.; et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J. Clin. Oncol. 2009, 27, 4150–4154. [Google Scholar] [CrossRef]
- Yang, P.; Zhang, W.; Wang, Y.; Peng, X.; Chen, B.; Qiu, X.; Li, G.; Li, S.; Wu, C.; Yao, K.; et al. IDH mutation and MGMT promoter methylation in glioblastoma: Results of a prospective registry. Oncotarget 2015, 6, 40896–40906. [Google Scholar] [CrossRef]
- Li, H.; Li, J.; Cheng, G.; Zhang, J.; Li, X. IDH mutation and MGMT promoter methylation are associated with the pseudoprogression and improved prognosis of glioblastoma multiforme patients who have undergone concurrent and adjuvant temozolomide–based chemoradiotherapy. Clin. Neurol. Neurosurg. 2016, 151, 31–36. [Google Scholar] [CrossRef]
- Millward, C.P.; Brodbelt, A.R.; Haylock, B.; Zakaria, R.; Baborie, A.; Crooks, D.; Husband, D.; Shenoy, A.; Wong, H.; Jenkinson, M.D. The impact of MGMT methylation and IDH-1 mutation on long–term outcome for glioblastoma treated with chemoradiotherapy. Acta Neurochir. 2016, 158, 1943–1953. [Google Scholar] [CrossRef]
- Galimi, F.; Torti, D.; Sassi, F.; Isella, C.; Cora, D.; Gastaldi, S.; Ribero, D.; Muratore, A.; Massucco, P.; Siatis, D.; et al. Genetic and expression analysis of MET, MACC1, and HGF in metastatic colorectal cancer: Response to met inhibition in patient xenografts and pathologic correlations. Clin. Cancer Res. 2011, 17, 3146–3156. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Li, G.; Dai, B.; Tan, W.; Zhao, H.; Li, X.; Wang, A. Silence of MACC1 expression by RNA interference inhibits proliferation, invasion and metastasis, and promotes apoptosis in U251 human malignant glioma cells. Mol. Med. Rep. 2015, 12, 3423–3431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shang, C.; Hong, Y.; Guo, Y.; Liu, Y.H.; Xue, Y.X. Influence of the MACC1 gene on sensitivity to chemotherapy in human U251 glioblastoma cells. Asian Pac. J. Cancer Prev. 2015, 16, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Zhang, Y.; Liang, H. MicroRNA–598 inhibits cell proliferation and invasion of glioblastoma by directly targeting metastasis associated in colon cancer–1 (MACC1). Oncol. Res. 2018, 26, 1275–1283. [Google Scholar] [CrossRef] [PubMed]
- Bush, N.A.; Chang, S.M.; Berger, M.S. Current and future strategies for treatment of glioma. Neurosurg. Rev. 2017, 40, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Blumenthal, D.T.; Rankin, C.; Stelzer, K.J.; Spence, A.M.; Sloan, A.E.; Moore, D.F., Jr.; Padula, G.D.; Schulman, S.B.; Wade, M.L.; Rushing, E.J. A Phase III study of radiation therapy (RT) and O(6)–benzylguanine + BCNU versus RT and BCNU alone and methylation status in newly diagnosed glioblastoma and gliosarcoma: Southwest Oncology Group (SWOG) study S0001. Int. J. Clin. Oncol. 2015, 20, 650–658. [Google Scholar] [CrossRef] [PubMed]
- Andronesi, O.C.; Arrillaga–Romany, I.C.; Ly, K.I.; Bogner, W.; Ratai, E.M.; Reitz, K.; Iafrate, A.J.; Dietrich, J.; Gerstner, E.R.; Chi, A.S.; et al. Pharmacodynamics of mutant–IDH1 inhibitors in glioma patients probed by in vivo 3D MRS imaging of 2–hydroxyglutarate. Nat. Commun. 2018, 9, 1474. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, X.B.; Liu, Y.H.; Xue, Y.X.; Wang, P.; Liu, L.B.; Liu, J.; Yao, Y.L.; Ma, J. Roles of serine/threonine phosphatases in low–dose endothelial monocyte–activating polypeptide–II–induced opening of blood–tumor barrier. J. Mol. Neurosci. 2015, 57, 11–20. [Google Scholar] [CrossRef]
- Reznikov, A.G.; Chaykovskaya, L.V.; Polyakova, L.I.; Kornelyuk, A.I. Antitumor effect of endothelial monocyte–activating polypeptide–II on human prostate adenocarcinoma in mouse xenograft model. Exp. Oncol. 2007, 29, 267–271. [Google Scholar]
- Schwarz, R.E.; Awasthi, N.; Konduri, S.; Caldwell, L.; Cafasso, D.; Schwarz, M.A. Antitumor effects of EMAP II against pancreatic cancer through inhibition of fibronectin–dependent proliferation. Cancer Biol. Ther. 2010, 9, 632–639. [Google Scholar] [CrossRef]
- Zhou, W.; Liu, L.; Xue, Y.; Zheng, J.; Liu, X.; Ma, J.; Li, Z.; Liu, Y. Combination of endothelial–Monocyte–Activating Polypeptide–II with Temozolomide Suppress Malignant Biological Behaviors of human glioblastoma stem cells via miR-590–3p/MACC1 inhibiting PI3K/AKT/mTOR signal pathway. Front. Mol. Neurosci. 2017, 10, 68. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Song, J.; Wang, W.; Zhang, Y.; Zheng, S. MACC1 antibody target therapy suppresses growth and migration of nonsmall cell lung cancer. Mol. Med. Rep. 2017, 16, 7329–7336. [Google Scholar] [CrossRef] [PubMed]
- Endo, A. The origin of the statins. Atheroscler. Suppl. 2004, 5, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Juneja, M.; Kobelt, D.; Walther, W.; Voss, C.; Smith, J.; Specker, E.; Neuenschwander, M.; Gohlke, B.O.; Dahlmann, M.; Radetzki, S.; et al. Statin and rottlerin small–molecule inhibitors restrict colon cancer progression and metastasis via MACC1. PLoS Biol. 2017, 15, e2000784. [Google Scholar] [CrossRef] [PubMed]
- Guillot, F.; Misslin, P.; Lemaire, M. Comparison of fluvastatin and lovastatin blood–brain barrier transfer using in vitro and in vivo methods. J. Cardiovasc. Pharmacol. 1993, 21, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Endo, A. A historical perspective on the discovery of statins. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2010, 86, 484–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, A.; Brenneman, B.; Floyd, D.; Comeau, L.; Spurio, K.; Olmez, I.; Lee, J.; Nakano, I.; Godlewski, J.; Bronisz, A.; et al. Statins affect human glioblastoma and other cancers through TGF–β inhibition. Oncotarget 2019, 10, 1716–1728. [Google Scholar] [CrossRef] [PubMed]
- Shojaei, S.; Alizadeh, J.; Thliveris, J.; Koleini, N.; Kardami, E.; Hatch, G.M.; Xu, F.; Hombach–Klonisch, S.; Klonisch, T.; Ghavami, S. Statins: a new approach to combat temozolomide chemoresistance in glioblastoma. J. Investig. Med. 2018, 66, 1083–1087. [Google Scholar] [CrossRef]
- Bababeygy, S.R.; Polevaya, N.V.; Youssef, S.; Sun, A.; Xiong, A.; Pruqpichailers, T.; Veeravagu, A.; Hou, L.C.; Steinman, L.; Tse, V. HMG–CoA reductase inhibition causes increased necrosis and apoptosis in an in vivo mouse glioblastoma multiforme model. Anticancer Res. 2009, 29, 4901–4908. [Google Scholar]
- Cemeus, C.; Zhao, T.T.; Barrett, G.M.; Lorimer, I.A.; Dimitroulakos, J. Lovastatin enhances gefiutinib activity in glioblastoma cells irrespective of EGFvIII and PTEN status. J. Neurooncol. 2008, 90, 9–17. [Google Scholar] [CrossRef]
- Larner, J.; Jane, J.; Laws, E.; Packer, R.; Myers, C.; Shaffrey, M. A phase I-II trial of lovastatin for anaplastic astrocytoma and glioblastoma multiforme. Am. J. Clin. Oncol. 1998, 21, 579–583. [Google Scholar] [CrossRef] [PubMed]
- Gaist, D.; Hallas, J.; Friis, S.; Hansen, S.; Sørensen, H.T. Statin use and survival following glioblastoma multiforme. Cancer Epidemiol. 2014, 38, 722–777. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.; Burger, P.C.; Jouvet, A.; Scheithauer, B.W.; Kleihues, P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007, 114, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Fleischhacker, M.; Beinert, T.; Ermitsch, M.; Seferi, D.; Possinger, K.; Engelmann, C.; Jandrig, B. Detection of amplifiable messenger RNA in the serum of patients with lung cancer. Ann. N. Y. Acad. Sci. 2001, 945, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Stein, U.; Burock, S.; Herrmann, P.; Wendler, I.; Niederstrasser, M.; Wernecke, K.D.; Schlag, P.M. Diagnostic and prognostic value of metastasis inducer S100A4 transcripts in plasma of colon, rectal, and gastric cancer patients. J. Mol. Diagn. 2011, 13, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Feldheim, J.; Kessler, A.F.; Schmitt, D.; Wilczek, L.; Linsenmann, T.; Dahlmann, M.; Monoranu, C.M.; Ernestus, R.I.; Hagemann, C.; Löhr, M. Expression of activating transcription factor 5 (ATF5) is increased in astrocytomas of different WHO grades and correlates with survival of glioblastoma patients. OncoTargets Ther. 2018, 11, 8673–8684. [Google Scholar] [CrossRef] [PubMed]
| Patients’ characteristics | |||
| Sex | female: 11/24% | male: 34/76% | |
| Median age at diagnosis | 65 years | ||
| ECOG at diagnosis | 0: 16/36% | 1: 21/47% | >1: 8/17% |
| Tumor characteristics | |||
| Median tumor volume | 36.0 cm3 (1.8–97.8 cm3) | ||
| IDH1 R132H mutation | absent: 39/87% | present: 6/13% | |
| MGMT promoter methylation 1 | unmethylated: 20/50% | methylated: 20/50% | |
| Therapy | |||
| Radiation therapy | yes: 40/89% | no: 5/11% | |
| Chemotherapy with TMZ | yes: 29/64% | no: 16/36% | |
| Outcome | |||
| OS | 0–6 m: 16/36% | >6 m: 29/64% | |
| PFS 2 | 0–6 m: 13/57% | >6 m: 10/43% | |
| Patients | Female | Male | Age * | OS | |||||||
| n | % | n | % | n | % | years | SD | months/days | SD | ||
| Cluster 1 | 18 | 46 | 3 | 30 | 15 | 52 | 57.0 | 11.9 | 16.0/488 | 8.5/259 | |
| Cluster 2 | 21 | 54 | 7 | 70 | 14 | 48 | 69.1 | 9.3 | 9.3/283 | 6.9/209 | |
| Combined | 39 | 100 | 10 | 100 | 29 | 100 | 63.5 | 12.1 | 12.4/377 | 8.3/252 | |
| MACC1 status | MACC1 * | MGMT status | |||||||||
| low | high | %calibrator | SD | not methylated | methylated | ||||||
| Cluster 1 | 18 | 0 | 0.20 | 0.10 | 9 | 9 | |||||
| Cluster 2 | 0 | 21 | 0.84 | 0.41 | 11 | 10 | |||||
| Combined | 18 | 21 | 0.54 | 0.44 | 20 | 19 | |||||
| IDH1 R132H mutation | tumor volume | ||||||||||
| absent | present | cm3 | SD | ||||||||
| Cluster 1 | 14 | 4 | 33 | 24 | |||||||
| Cluster 2 | 21 | 0 | 44 | 28 | |||||||
| Combined | 35 | 4 | 39 | 26 | |||||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hagemann, C.; Neuhaus, N.; Dahlmann, M.; Kessler, A.F.; Kobelt, D.; Herrmann, P.; Eyrich, M.; Freitag, B.; Linsenmann, T.; Monoranu, C.M.; et al. Circulating MACC1 Transcripts in Glioblastoma Patients Predict Prognosis and Treatment Response. Cancers 2019, 11, 825. https://doi.org/10.3390/cancers11060825
Hagemann C, Neuhaus N, Dahlmann M, Kessler AF, Kobelt D, Herrmann P, Eyrich M, Freitag B, Linsenmann T, Monoranu CM, et al. Circulating MACC1 Transcripts in Glioblastoma Patients Predict Prognosis and Treatment Response. Cancers. 2019; 11(6):825. https://doi.org/10.3390/cancers11060825
Chicago/Turabian StyleHagemann, Carsten, Nikolas Neuhaus, Mathias Dahlmann, Almuth F. Kessler, Dennis Kobelt, Pia Herrmann, Matthias Eyrich, Benjamin Freitag, Thomas Linsenmann, Camelia M. Monoranu, and et al. 2019. "Circulating MACC1 Transcripts in Glioblastoma Patients Predict Prognosis and Treatment Response" Cancers 11, no. 6: 825. https://doi.org/10.3390/cancers11060825





