Genetics, Genomics and Evolution of Ergot Alkaloid Diversity
Abstract
:1. Importance of Ergot Alkaloids
2. Structural Content of Ergot Alkaloid Biosynthesis (EAS) Loci Define Alkaloid Potential
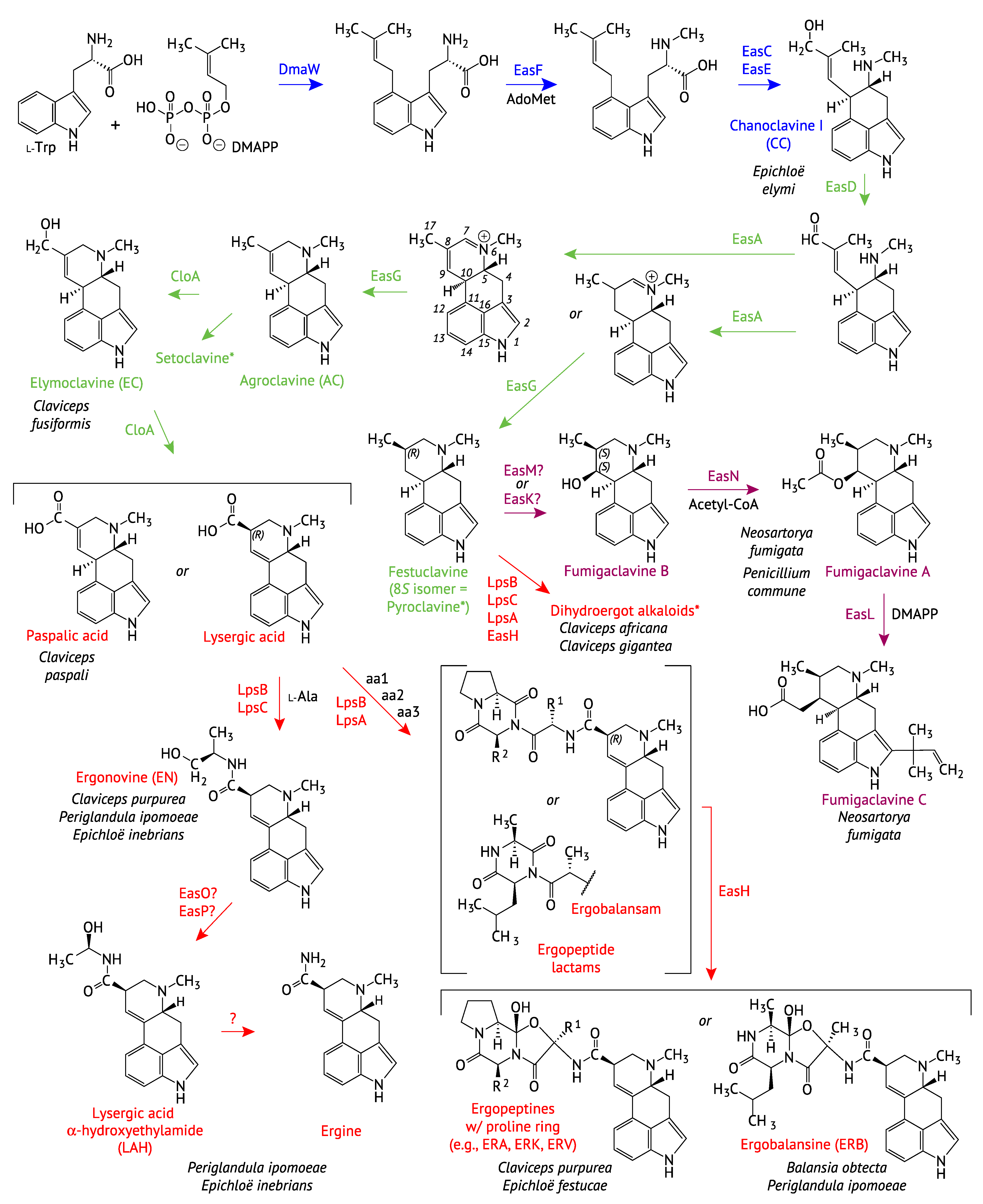
2.1. Genes Encoding the Early Pathway Steps
| Gene name a | Enzyme | EASCC | EASEC | EASERP | EASLAH | EASEN/ERP | EASLAH/ERP | EASFC |
|---|---|---|---|---|---|---|---|---|
| dmaW | Dimethylallyltryptophan synthase | + | + | + | + | + | + | + |
| easF | DimethylallyltryptophanN-methylase | + | + | + | + | + | + | + |
| easC | Catalase | + | + | + | + | + | + | + |
| easE | Chanoclavine-I synthase | + | + | + | + | + | + | + |
| easD | Chanoclavine-I dehydrogenase | + | + | + | + | + | + | |
| easAb | Chanoclavine-1 aldehyde oxidoreductase | + iso | + iso | + iso | + iso | + iso | + red | |
| easG | agroclavine, festuclavine or pyroclavine dehydrogenase | + | + | + | + | + | + | |
| cloAb | agroclavine, festuclavine, or elymoclavine monooxygenase | + | + | + | + | + | ||
| lpsB | lysergyl peptide synthetase subunit 2 | + | + | + | + | |||
| lpsAb | lysergyl peptide synthetase subunit 1 | + | + | + | ||||
| easH | ergopeptide lactam hydroxylase | + | + | + | ||||
| lpsC | lysergyl peptide synthetase and reductase subunit | + | + | + | ||||
| easOc | putative ergonovine oxygenase | + | + | |||||
| easPc | putative LAH synthase | + | + | |||||
| easMc | possible festuclavine 9-monooxygenase | + | ||||||
| easKc | possible festuclavine 9-monooxygenase | + | ||||||
| easN | fumigaclavine acetylase | + | ||||||
| easL | fumigaclavine reverse prenyltransferase | + | ||||||
| Fungal species | Examples of ergot alkaloid-producing fungi | Epichloë elymi | Claviceps fusiformis | Epichloë festucae,Balansia obtecta | Epichloë inebrians | Claviceps purpurea | Periglandula ipomoeae | Neosartorya fumigata |
2.2. Diversification of the EAS Pathways
2.2.1. Completion of the Tetracyclic Ergolene Common Core
2.2.2. Formation of Lysergic Acid, Lysergic Acid Amides and Complex Ergopeptines
2.2.3. Fumigaclavine Production by the Trichocomaceae
| Ergopeptine | AA1 | R1 | AA2 | R2 | AA3 | R3 |
|---|---|---|---|---|---|---|
| Ergotamine (ERA) | Ala | Me | Phe | CH2Ph | Pro | prolyl (CH2)3 |
| Ergovaline (ERV) | Ala | Me | Val | i-Pr | Pro | prolyl (CH2)3 |
| Ergosine | Ala | Me | Leu | i-Bu | Pro | prolyl (CH2)3 |
| Dihydroergosine b | Ala | Me | Leu | i-Bu | Pro | prolyl (CH2)3 |
| β-Ergosine | Ala | Me | Ile | sec-Bu | Pro | prolyl (CH2)3 |
| Ergosedmine | Ile | sec-Bu | Leu | i-Bu | Pro | Prolyl (CH2)3 |
| Ergobine | Ala | Me | ABA | Et | Pro | prolyl (CH2)3 |
| Ergocristine | Val | i-Pr | Phe | CH2Ph | Pro | prolyl (CH2)3 |
| Ergocornine | Val | i-Pr | Val | i-Pr | Pro | prolyl (CH2)3 |
| Ergocryptine c,d (ERK) | Val | i-Pr | Leu | i-Bu | Pro | prolyl (CH2)3 |
| β-Ergocryptine d | Val | i-Pr | Ile | sec-Bu | Pro | prolyl (CH2)3 |
| γ-Ergocryptinine c,e | Val | i-Pr | norLeu | n-Bu | Pro | prolyl (CH2)3 |
| Ergobutyrine | Val | i-Pr | ABA | Et | Pro | prolyl (CH2)3 |
| Ergoladinine e | Val | i-Pr | Met | EtSCH3 | Pro | prolyl (CH2)3 |
| Ergogaline | Val | i-Pr | homoIle | 2-Me- n-Bu | Pro | prolyl (CH2)3 |
| Ergostine | ABA | Et | Phe | CH2Ph | Pro | prolyl (CH2)3 |
| Ergonine | ABA | Et | Val | i-Pr | Pro | prolyl (CH2)3 |
| Ergoptine c | ABA | Et | Leu | i-Bu | Pro | prolyl (CH2)3 |
| β-ergoptine | ABA | Et | Ile | sec-Bu | Pro | prolyl (CH2)3 |
| Ergobutine | ABA | Et | ABA | Et | Pro | prolyl (CH2)3 |
| Ergobalansine (ERB) | Ala | Me | Leu | i-Bu | Ala | Me |
| Unnamed, from Dicyma sp. | Ala | Me | Leu | i-Bu | Phe | CH2Ph |
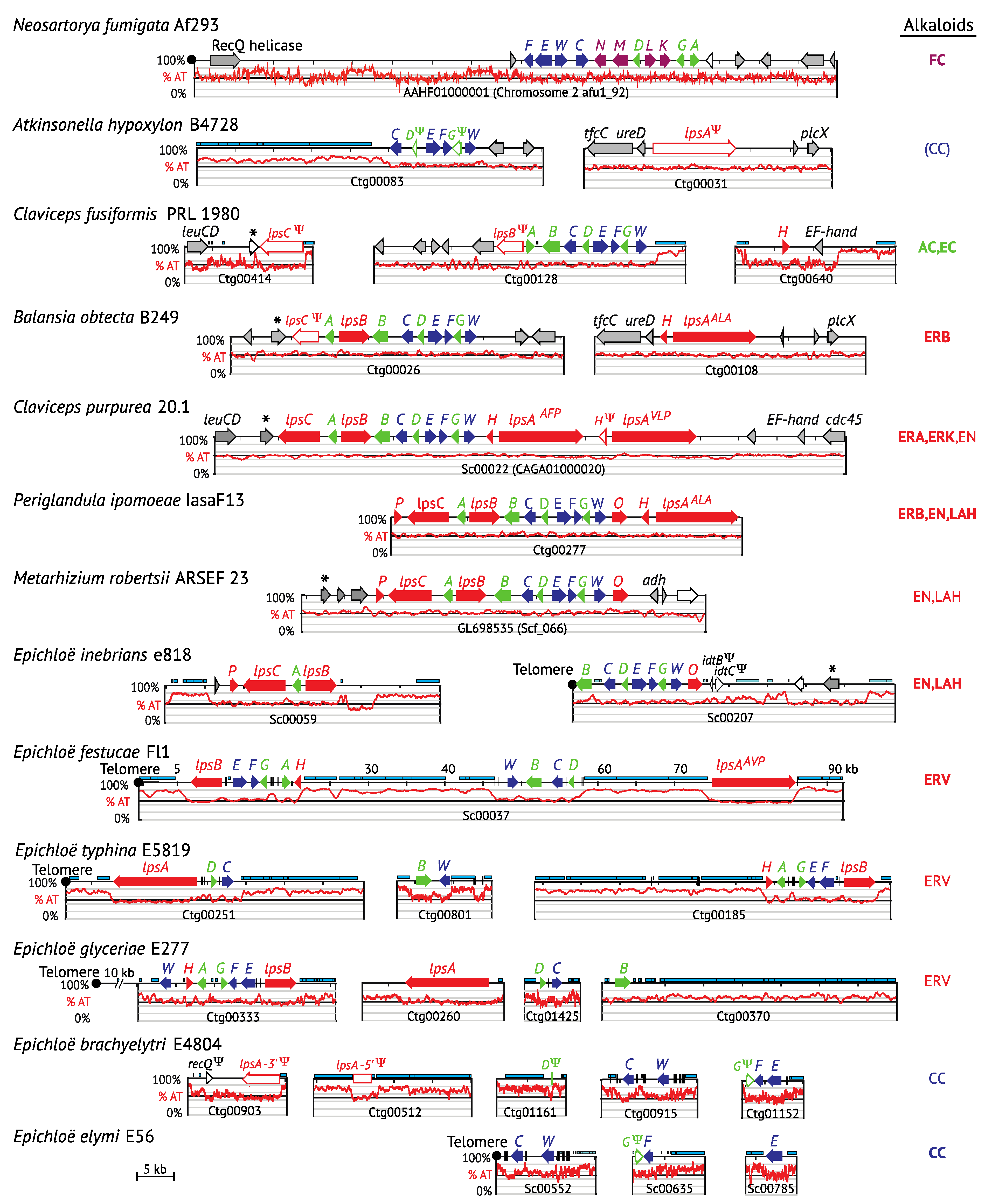
2.3. Contents of the EAS Loci
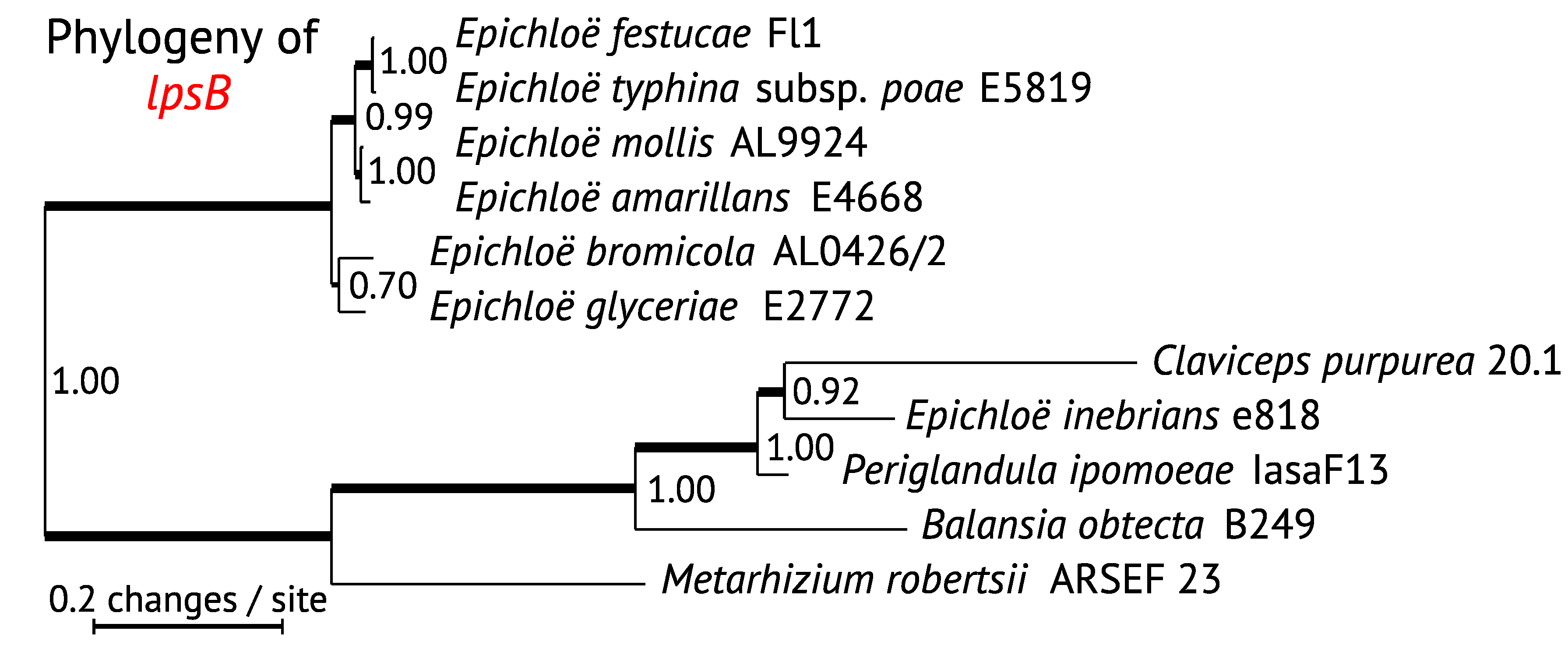
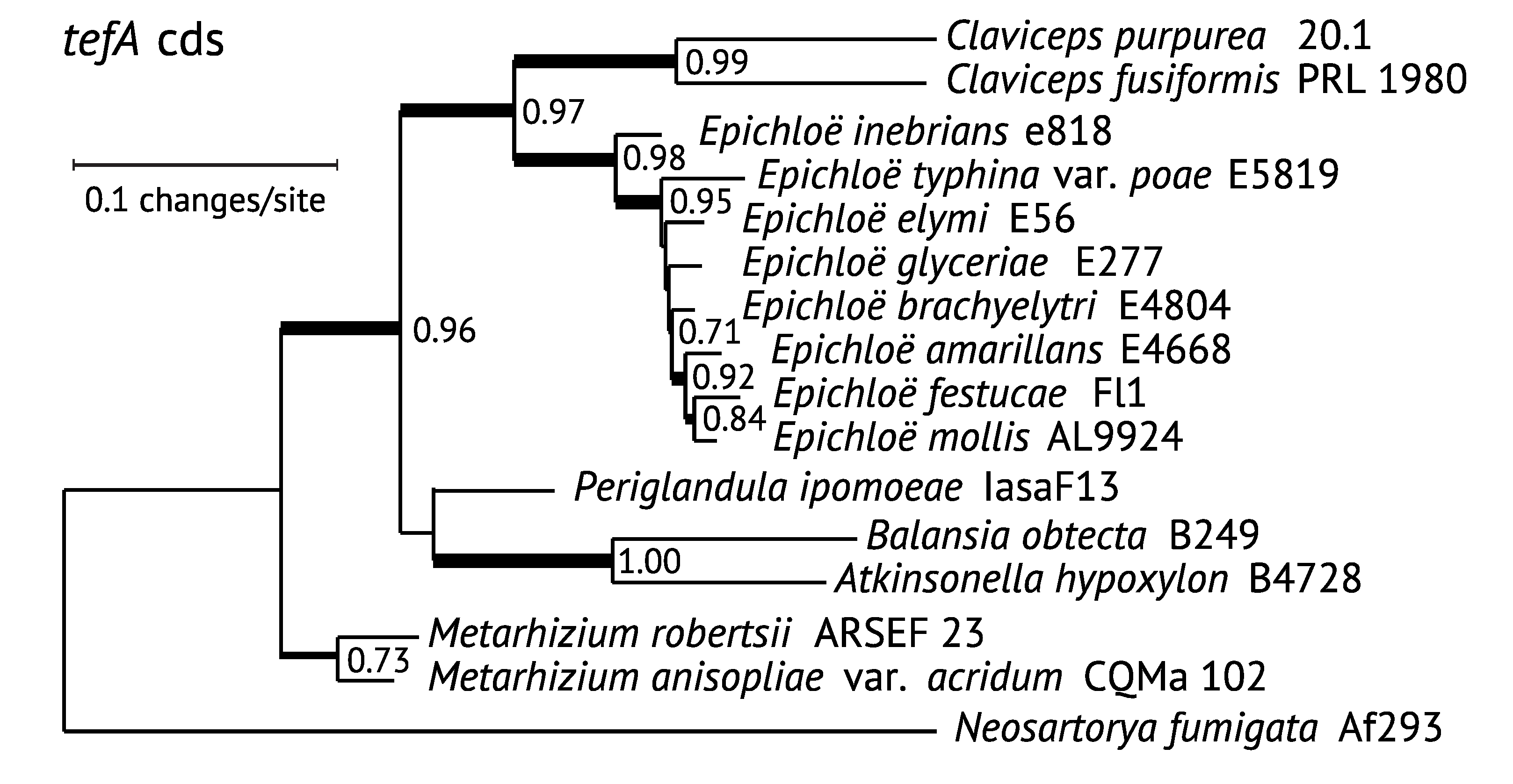
3. Phylogenetic Relationships of EAS Genes
3.1. Comparison of EAS Gene Phylogenies

3.2. Mapping EAS Gene Gains and Losses
3.3. Positional Changes of Clusters with Respect to Telomeres
3.4. Evolution of LpsC
3.5. Evolution of Module Specificity in LpsA
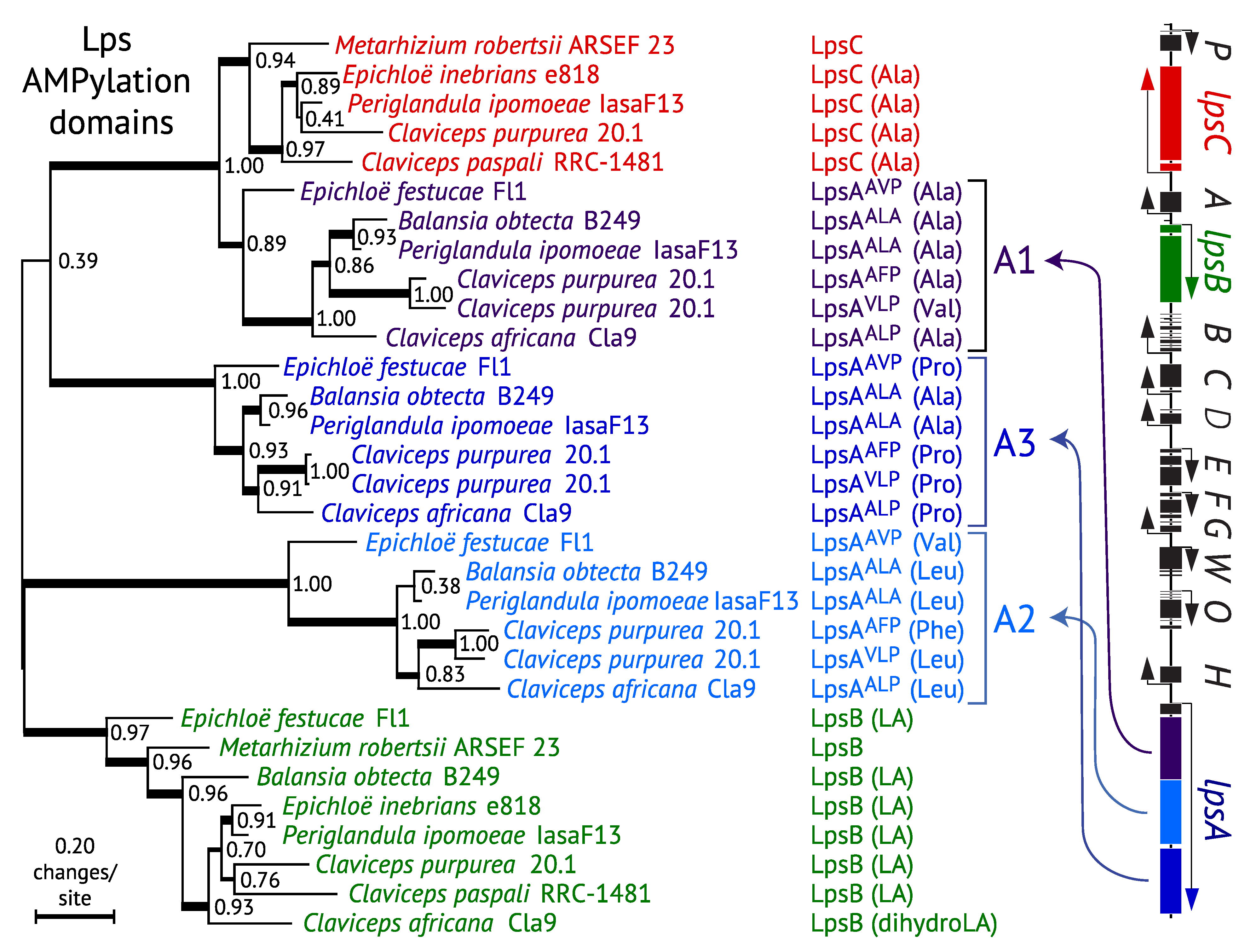
4. Ergot Alkaloid Diversity within Epichloë Species
4.1. Distribution of EAS Genes across Epichloë Species
| Epichloë species a | Host species | Detection method b | EAS gene variations (strains observed) c | Reference |
|---|---|---|---|---|
| Epichloë amarillans | Agrostis hyemalis | GT, DG, G | 0* (4), (ERV) (1) | [24] |
| E. aotearoae | Echinopogon ovatus | G | 0 (1) | [24,43] |
| E. baconii | Agrostis tenuis,Calamagrostis villosa | GT, G | 0* (3) | [43] |
| E. brachyelytri | Brachyelytrum erectum | GT, G | 0 (1), CC (3) | [24] |
| E. bromicola | Bromus erectus,Bromus benekenii,Bromus tomentellus,Agropyron hispidus | GT, DG, G | 0* (5) | [43] |
| E. cabralii (H) | Phleum alpinum Bromus laevipes | G, GT | 0 (1), (ERV) (2) | [50] |
| E. canadensis (H) | Elymus canadensis | GT, DG | CC (1), ERV (1) | [43,47] |
| E. chisosa (H) | Achnatherum eminens | DG | 0 (1) | [43] |
| E. coenophiala (H) | Lolium arundinaceum | GT, DG | 0* (11), ERV (12), ERV (39) | [43,49,51] |
| E. elymi | Elymus virginicus | GT, G | 0 (1), CC (1) | [24] |
| E. festucae | Festuca trachyphylla,Festuca rubra subsp.rubra,Lolium giganteum | GT, G | 0 (1), ERV (1), (ERV) (2) | [24] |
| E. festucae var. lolii | Lolium perenne | GT, G | ERV (2), (ERV) (1) | [56,57] |
| E. festucae var. lolii x E. typhina (H) | Lolium perenne | DG | ERV (1) | [43,58] |
| E. funkii (H) | Achnatherum robustum | GT, DG | CC (1) | [43] |
| E. gansuensis | Achnatherum inebrians | G | 0 (1) | [24] |
| E. inebrians | Achnatherum inebrians | G | EN, LAH (1) | [24] |
| E. glyceriae | Glyceria striata | GT | (ERV) (2) | [24] |
| E. mollis | Holcus mollis | G | ERV (1) | [43] |
| E. occultans (H) | Lolium sp. (2x) | GT | 0 (3) | [43] |
| E. schardlii (H) | Cinna arundinacea | GT | 0 (1) | [59] |
| E. siegelii (H) | Lolium pratense | DG | 0 (1) | [43] |
| E. sylvatica | Brachypodium sylvaticum | GT | 0 (2) | [34] |
| E. typhina | Lolium perenne,Dactylis glomerata | G, GT | 0 (3) | [24,43] |
| E. typhina ssp. clarkii | Holcus lanatus | GT | ERV (1) | unpublished |
| E. typhina ssp. poae | Poa nemoralis,Bromus laevipes | GT, G | 0 (3), ERV (1) | [24,50] |
| E. uncinata (H) | Lolium pratense | DG | 0 (1) | [43] |
| E. sp. AroTG-2(H) | Achnatherum robustum | GT | EN (1) | [10] |
| E. sp. BlaTG-3(H) | Bromus laevipes | GT | 0* (1), CC (2) | [50] |
| E. sp. FaTG-2(H) | Lolium sp. (6x) | GT, DG | ERV (10), ERV (33) | [43,49,51,60] |
| E. sp. FaTG-3(H) | Lolium sp. (6x), (8X) | GT, DG | 0 (11) | [43,51,60] |
| E. sp. FaTG-4(H) | Lolium sp. (10x) | GT, DG | ERV (1), ERV (11) | [43,51] |
| E. sp. FcaTG-1(H) | Festuca campestris | GT | 0 (3) | unpublished |
| E. sp. FveTG-1(H) | Festuca versuta | GT | 0 (2) | unpublished |
| E. sp. PalTG-1(H) | Poa alsodes | GT | 0* (1) | unpublished |
| E. sp. PauTG-1(H) | Poa autumnalis | GT | 0 (1) | unpublished |
4.2. Pseudogenes and Gene Remnants within the EAS Locus
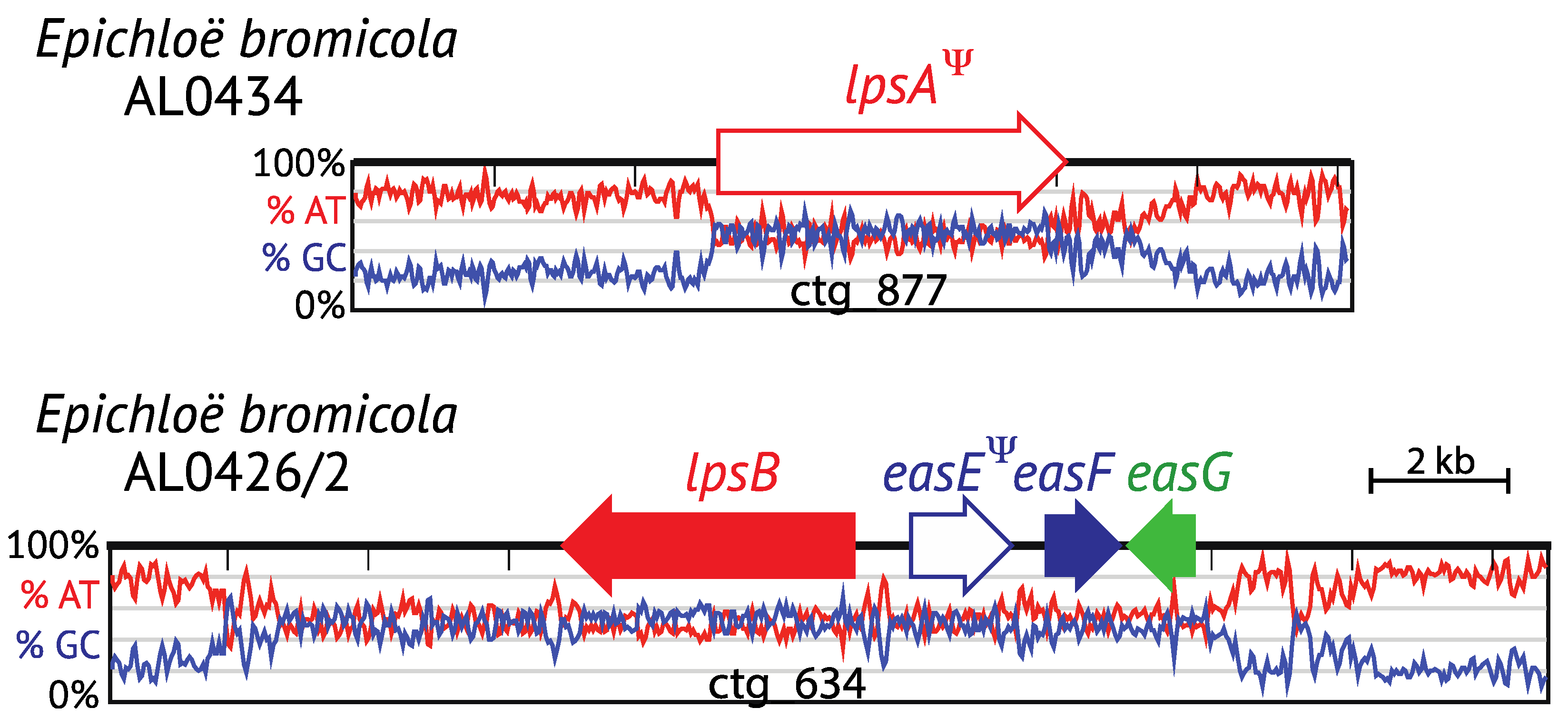
4.3. Hybrids: EAS Gene Cluster Variations

4.4. Gene Losses in Hybrids with Multiple Copies
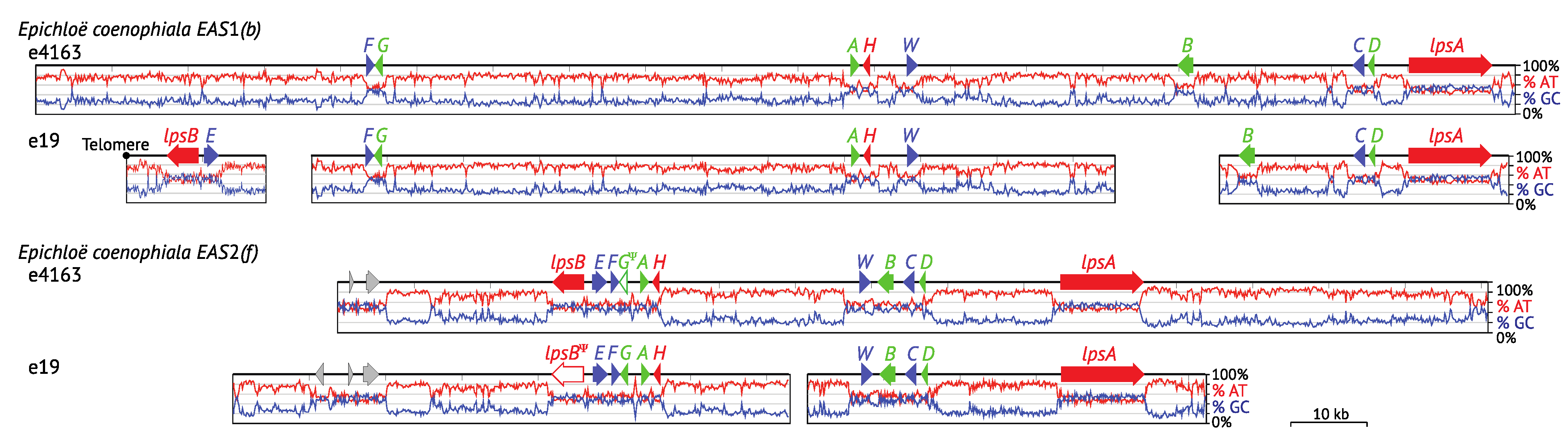
4.5. Endophyte Genetic Variation within a Single Host Species
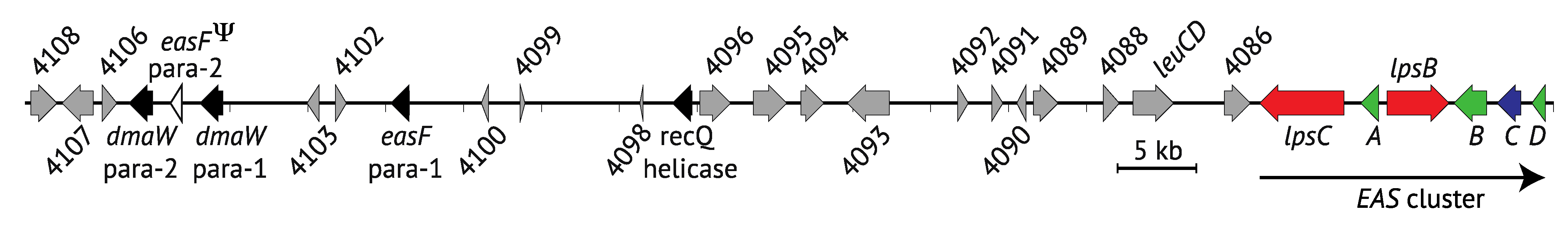
5. Synteny and Rearrangements in the EAS Loci
5.1. Syntenic Regions of the EAS Loci

5.2. Epichloë Species Have More EAS Loci Rearrangements
5.3. The Complex History of EAS Loci
6. Conclusions
Acknowledgments
Conflicts of Interest
References
- Billings, M. The Crusades: Five Centuries of Holy Wars; Sterling Publishing Company: New York, NY, USA, 1996. [Google Scholar]
- Alm, T. The witch trials of Finnmark, northern Norway, during the 17th century: Evidence for ergotism as a contributing factor. Econ. Bot. 2003, 57, 403–416. [Google Scholar] [CrossRef]
- Caporael, L.R. Ergotism: the Satan loosed in Salem? Science 1976, 192, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Woolf, A. Witchcraft or mycotoxin? The Salem witch trials. J. Toxicol.: Clin. Toxicol. 2000, 38, 457–460. [Google Scholar] [CrossRef]
- Zadoks, J.C. On the Political Economy of Plant Disease Epidemics: Capita Selecta in Historical Epidemiology; Wageningen Academic Publishers: Wageningen, The Netherlands, 2008. [Google Scholar]
- Tribble, M.A.; Gregg, C.R.; Margolis, D.M.; Amirkhan, R.; Smith, J.W. Fatal ergotism induced by an HIV protease inhibitor. Headache 2002, 42, 694–695. [Google Scholar] [CrossRef] [PubMed]
- Urga, K.; Debella, A.; W’Medihn, Y.; Bayu, A.; Zewdie, W. Laboratory studies on the outbreak of gangrenous ergotism associated with consumption of contaminated barley in Arsi, Ethiopia. Ethiop. J. Health Dev. 2002, 16, 317–323. [Google Scholar]
- Scott, P. Ergot alkaloids: Extent of human and animal exposure. World Mycotoxin J. 2009, 2, 141–149. [Google Scholar] [CrossRef]
- Lyons, P.C.; Plattner, R.D.; Bacon, C.W. Occurrence of peptide and clavine ergot alkaloids in tall fescue grass. Science 1986, 232, 487–489. [Google Scholar] [CrossRef] [PubMed]
- Shymanovich, T.; Saari, S.; Lovin, M.; Jarmusch, A.; Jarmusch, S.; Musso, A.; Charlton, N.; Young, C.; Cech, N.; Faeth, S. Alkaloid variation among epichloid endophytes of sleepygrass (Achnatherum robustum) and consequences for resistance to insect herbivores. J. Chem. Ecol. 2015, 41, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Miles, C.O.; Lane, G.A.; di Menna, M.E.; Garthwaite, I.; Piper, E.L.; Ball, O.J.P.; Latch, G.C.M.; Allen, J.M.; Hunt, M.B.; Bush, L.P.; et al. High levels of ergonovine and lysergic acid amide in toxic Achnatherum inebrians accompany infection by an Acremonium-like endophytic fungus. J. Agric. Food Chem. 1996, 44, 1285–1290. [Google Scholar] [CrossRef]
- De Groot, A.N.; van Dongen, P.W.; Vree, T.B.; Hekster, Y.A.; van Roosmalen, J. Ergot alkaloids. Current status and review of clinical pharmacology and therapeutic use compared with other oxytocics in obstetrics and gynaecology. Drugs 1998, 56, 523–535. [Google Scholar] [CrossRef] [PubMed]
- Burn, D. Neurology (2) Parkinson’s disease: Treatment. Pharm. J. 2000, 264, 476–479. [Google Scholar]
- Crosignani, P.G. Current treatment issues in female hyperprolactinaemia. Eur. J. Obstet. Gynecol. Reprod. Biol. 2006, 125, 152–164. [Google Scholar] [CrossRef] [PubMed]
- Micale, V.; Incognito, T.; Ignoto, A.; Rampello, L.; Sparta, M.; Drago, F. Dopaminergic drugs may counteract behavioral and biochemical changes induced by models of brain injury. Eur. Neuropsychopharmacol. 2006, 16, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, A. LSD: Mein Sorgenkind. Die Entdeckung einer “Wunderdroge”; Deutscher Taschenbuch Verlag: München, Germany, 2006. [Google Scholar]
- Robinson, S.L.; Panaccione, D.G. Diversification of ergot alkaloids in natural and modified fungi. Toxins 2015, 7, 201–218. [Google Scholar] [CrossRef] [PubMed]
- Wallwey, C.; Heddergott, C.; Xie, X.; Brakhage, A.A.; Li, S.-M. Genome mining reveals the presence of a conserved biosynthetic gene cluster for the biosynthesis of ergot alkaloid precursors in the fungal family Arthrodermataceae. Microbiology 2012, 158, 1634–1644. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, M.J.; Roa, A.M.; Reyes, F.; Vega, A.; Rivera-Sagredo, A.; Thomas, D.R.; Diez, E.; Hueso-Rodriguez, J.A. A novel ergot alkaloid as a 5-HT(1A) inhibitor produced by Dicyma sp. J. Med. Chem. 2003, 46, 5117–5120. [Google Scholar] [CrossRef] [PubMed]
- Gerhards, N.; Neubauer, L.; Tudzynski, P.; Li, S.-M. Biosynthetic pathways of ergot alkaloids. Toxins 2014, 6, 3281–3295. [Google Scholar] [CrossRef] [PubMed]
- Panaccione, D.G. Origins and significance of ergot alkaloid diversity in fungi. FEMS Microbiol. Lett. 2005, 251, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Panaccione, D.G.; Schardl, C.L.; Coyle, C.M. Pathways to diverse ergot alkaloid profiles in fungi. In Recent Advances in Phytochemistry; Romeo, J.T., Ed.; Elsevier: Amsterdam, The Netherlands, 2006; Volume 40, pp. 23–52. [Google Scholar]
- Flieger, M.; Sedmera, P.; Vokoun, J.; R̆ic̄icovā, A.; R̆ehác̆ek, Z. Separation of four isomers of lysergic acid α-hydroxyethylamide by liquid chromatography and their spectroscopic identification. J. Chromatogr. A 1982, 236, 441–452. [Google Scholar] [CrossRef]
- Schardl, C.L.; Young, C.A.; Hesse, U.; Amyotte, S.G.; Andreeva, K.; Calie, P.J.; Fleetwood, D.J.; Haws, D.C.; Moore, N.; Oeser, B.; et al. Plant-symbiotic fungi as chemical engineers: Multi-genome analysis of the Clavicipitaceae reveals dynamics of alkaloid loci. PLoS Genet. 2013, 9, e1003323. [Google Scholar] [CrossRef] [PubMed]
- Coyle, C.M.; Cheng, J.Z.; O’Connor, S.E.; Panaccione, D.G. An old yellow enzyme gene controls the branch point between Aspergillus fumigatus and Claviceps purpurea ergot alkaloid pathways. Appl. Environ. Microbiol. 2010, 76, 3898–3903. [Google Scholar] [CrossRef] [PubMed]
- Ortel, I.; Keller, U. Combinatorial assembly of simple and complex d-lysergic acid alkaloid peptide classes in the ergot fungus Claviceps purpurea. J. Biol. Chem. 2009, 284, 6650–6660. [Google Scholar] [CrossRef] [PubMed]
- Havemann, J.; Vogel, D.; Loll, B.; Keller, U. Cyclolization of D-lysergic acid alkaloid peptides. Chem. Biol. 2014, 21, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Haarmann, T.; Lorenz, N.; Tudzynski, P. Use of a nonhomologous end joining deficient strain (∆ku70) of the ergot fungus Claviceps purpurea for identification of a nonribosomal peptide synthetase gene involved in ergotamine biosynthesis. Fungal Genet. Biol. 2008, 45, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Uhlig, S.; Petersen, D.; Rolèn, E.; Egge-Jacobsen, W.; Vrålstad, T. Ergosedmine, a new peptide ergot alkaloid (ergopeptine) from the ergot fungus, Claviceps purpurea parasitizing Calamagrostis arundinacea. Phytochem. Lett. 2011, 4, 79–85. [Google Scholar] [CrossRef]
- Schardl, C.L.; Panaccione, D.G.; Tudzynski, P. Ergot alkaloids—Biology and molecular biology. Alkaloids 2006, 63, 45–86. [Google Scholar] [PubMed]
- Matuschek, M.; Wallwey, C.; Wollinsky, B.; Xie, X.; Li, S.-M. In vitro conversion of chanoclavine-I aldehyde to the stereoisomers festuclavine and pyroclavine controlled by the second reduction step. RSC Adv. 2012, 2, 3662. [Google Scholar] [CrossRef]
- Unsöld, I.A.; Li, S.M. Overproduction, purification and characterization of FgaPT2, a dimethylallyltryptophan synthase from Aspergillus fumigatus. Microbiol. SGM 2005, 151, 1499–1505. [Google Scholar] [CrossRef]
- Coyle, C.M.; Panaccione, D.G. An ergot alkaloid biosynthesis gene and clustered hypothetical genes from Aspergillus fumigatus. Appl. Environ. Microbiol. 2005, 71, 3112–3118. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, X.Z.; Li, C.J.; Swoboda, G.A.; Young, C.A.; Sugawara, K.; Leuchtmann, A.; Schardl, C.L. Two distinct Epichloë species symbiotic with Achnatherum inebrians, drunken horse grass. Mycologia 2015, in press. [Google Scholar]
- Blin, K.; Medema, M.H.; Kazempour, D.; Fischbach, M.A.; Breitling, R.; Takano, E.; Weber, T. antiSMASH 2.0—A versatile platform for genome mining of secondary metabolite producers. Nucl. Acids Res. 2013, 41, W204–W212. [Google Scholar] [CrossRef] [PubMed]
- Khaldi, N.; Seifuddin, F.T.; Turner, G.; Haft, D.; Nierman, W.C.; Wolfe, K.H.; Fedorova, N.D. SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet. Biol. 2010, 47, 736–741. [Google Scholar] [CrossRef] [PubMed]
- Keller, N.P.; Turner, G.; Bennett, J.W. Fungal secondary metabolism—From biochemistry to genomics. Nat. Rev. Microbiol. 2005, 3, 937–947. [Google Scholar] [CrossRef] [PubMed]
- Schardl, C.L.; Young, C.A.; Moore, N.; Krom, N.; Dupont, P.-Y.; Pan, J.; Florea, S.; Webb, J.S.; Jaromczyk, J.; Jaromczyk, J.W.; et al. Genomes of plant-associated Clavicipitaceae. Adv. Bot. Res. 2014, 70, 291–327. [Google Scholar]
- Gao, Q.; Jin, K.; Ying, S.-H.; Zhang, Y.; Xiao, G.; Shang, Y.; Duan, Z.; Hu, X.; Xie, X.-Q.; Zhou, G.; et al. Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum. PLoS Genet. 2011, 7, e1001264. [Google Scholar] [CrossRef] [PubMed]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.-F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucl. Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl. Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Anisimova, M.; Gascuel, O. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 2006, 55, 539–552. [Google Scholar] [CrossRef] [PubMed]
- Schardl, C.L.; Young, C.A.; Pan, J.; Florea, S.; Takach, J.E.; Panaccione, D.G.; Farman, M.L.; Webb, J.S.; Jaromczyk, J.; Charlton, N.D.; et al. Currencies of mutualisms: Sources of alkaloid genes in vertically transmitted epichloae. Toxins 2013, 5, 1064–1088. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Bhardwaj, M.; Faulkner, J.R.; Nagabhyru, P.; Charlton, N.D.; Higashi, R.M.; Miller, A.-F.; Young, C.A.; Grossman, R.B.; Schardl, C.L. Ether bridge formation in loline alkaloid biosynthesis. Phytochemistry 2014, 98, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Bhardwaj, M.; Nagabhyru, P.; Grossman, R.B.; Schardl, C.L. Enzymes from fungal and plant origin required for chemical diversification of insecticidal loline alkaloids in grass-Epichloë symbiota. PLoS One 2014, 9, e115590. [Google Scholar] [CrossRef] [PubMed]
- Schardl, C.L.; Farman, M.L.; Jaromczyk, J.W.; Young, C.A.; Webb, J.S.; Jaromczyk, J.; Moore, N.; Bec, S.; Calie, P.J.; Charlton, N.; et al. Genome sequences of 56 plant-associated Clavicipitaceae. 2015, unpublished. [Google Scholar]
- Charlton, N.D.; Craven, K.D.; Mittal, S.; Hopkins, A.A.; Young, C.A. Epichloë canadensis, a new interspecific epichloid hybrid symbiotic with Canada wildrye (Elymus canadensis). Mycologia 2012, 104, 1187–1199. [Google Scholar] [CrossRef] [PubMed]
- Schardl, C.L.; Young, C.A.; Faulkner, J.R.; Florea, S.; Pan, J. Chemotypic diversity of epichloae, fungal symbionts of grasses. Fungal Ecol. 2012, 5, 331–344. [Google Scholar] [CrossRef]
- Takach, J.E.; Mittal, S.; Swoboda, G.A.; Bright, S.K.; Trammell, M.A.; Hopkins, A.A.; Young, C.A. Genotypic and chemotypic diversity of Neotyphodium endophytes in tall fescue from Greece. Appl. Environ. Microbiol. 2012, 78, 5501–5510. [Google Scholar] [CrossRef] [PubMed]
- Charlton, N.D.; Craven, K.D.; Afkhami, M.E.; Hall, B.A.; Ghimire, S.R.; Young, C.A. Interspecific hybridization and bioactive alkaloid variation increases diversity in endophytic Epichloë species of Bromus laevipes. FEMS Microbiol. Ecol. 2014, 90, 276–289. [Google Scholar] [CrossRef] [PubMed]
- Takach, J.E.; Young, C.A. Alkaloid genotype diversity of tall fescue endophytes. Crop. Sci. 2014, 54, 667–678. [Google Scholar] [CrossRef]
- Young, C.A.; Charlton, N.D.; Takach, J.E.; Swoboda, G.A.; Trammell, M.A.; Huhman, D.V.; Hopkins, A.A. Characterization of Epichloë coenophiala within the US: Are all tall fescue endophytes created equal? Front. Chem. 2014, 2, 95. [Google Scholar] [CrossRef] [PubMed]
- Bacon, C.W. Procedure for isolating the endophyte from tall fescue and screening isolates for ergot alkaloids. Appl. Env. Microbiol. 1988, 54, 2615–2618. [Google Scholar]
- Blankenship, J.D.; Spiering, M.J.; Wilkinson, H.H.; Fannin, F.F.; Bush, L.P.; Schardl, C.L. Production of loline alkaloids by the grass endophyte, Neotyphodium uncinatum, in defined media. Phytochemistry 2001, 58, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Chujo, T.; Scott, B. Histone H3K9 and H3K27 methylation regulates fungal alkaloid biosynthesis in a fungal endophyte–plant symbiosis. Mol. Microbiol. 2014, 92, 413–434. [Google Scholar] [CrossRef] [PubMed]
- Fleetwood, D.J.; Scott, B.; Lane, G.A.; Tanaka, A.; Johnson, R.D. A complex ergovaline gene cluster in epichloë endophytes of grasses. Appl. Environ. Microbiol. 2007, 73, 2571–2579. [Google Scholar] [CrossRef] [PubMed]
- Fleetwood, D.J.; Khan, A.K.; Johnson, R.D.; Young, C.A.; Mittal, S.; Wrenn, R.E.; Hesse, U.; Foster, S.J.; Schardl, C.L.; Scott, B. Abundant degenerate miniature inverted-repeat transposable elements in genomes of epichloid fungal endophytes of grasses. Genome Biol. Evol. 2011, 3, 1253–1264. [Google Scholar] [CrossRef] [PubMed]
- Panaccione, D.G.; Johnson, R.D.; Wang, J.H.; Young, C.A.; Damrongkool, P.; Scott, B.; Schardl, C.L. Elimination of ergovaline from a grass-Neotyphodium endophyte symbiosis by genetic modification of the endophyte. Proc. Natl. Acad. Sci. USA 2001, 98, 12820–12825. [Google Scholar] [CrossRef] [PubMed]
- Ghimire, S.R.; Rudgers, J.A.; Charlton, N.D.; Young, C.; Craven, K.D. Prevalence of an intraspecific Neotyphodium hybrid in natural populations of stout wood reed (Cinna arundinacea L.) from eastern North America. Mycologia 2011, 103, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Christensen, M.J.; Leuchtmann, A.; Rowan, D.D.; Tapper, B.A. Taxonomy of Acremonium endophytes of tall fescue (Festuca arundinacea), meadow fescue (F. pratensis), and perennial rye-grass (Lolium perenne). Mycol. Res. 1993, 97, 1083–1092. [Google Scholar] [CrossRef]
- Leuchtmann, A.; Schmidt, D.; Bush, L.P. Different levels of protective alkaloids in grasses with stroma-forming and seed-transmitted Epichloë/Neotyphodium endophytes. J. Chem. Ecol. 2000, 26, 1025–1036. [Google Scholar] [CrossRef]
- Schardl, C.L.; Craven, K.D.; Speakman, S.; Stromberg, A.; Lindstrom, A.; Yoshida, R. A novel test for host-symbiont codivergence indicates ancient origin of fungal endophytes in grasses. Syst. Biol. 2008, 57, 483–498. [Google Scholar] [CrossRef] [PubMed]
- Petroski, R.; Powell, R.G.; Clay, K. Alkaloids of Stipa robusta (sleepygrass) infected with an Acremonium endophyte. Nat. Toxins 1992, 1, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Moon, C.D.; Guillaumin, J.-J.; Ravel, C.; Li, C.; Craven, K.D.; Schardl, C.L. New Neotyphodium endophyte species from the grass tribes Stipeae and Meliceae. Mycologia 2007, 99, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Farman, M.L. Telomeres in the rice blast fungus Magnaporthe oryzae: The world of the end as we know it. FEMS Microbiol. Lett. 2007, 273, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Starnes, J.H.; Thornbury, D.W.; Novikova, O.S.; Rehmeyer, C.J.; Farman, M.L. Telomere-targeted retrotransposons in the rice blast fungus Magnaporthe oryzae: Agents of telomere instability. Genetics 2012, 191, 389–406. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Kim, Y.-S.; Smith, K.M.; Li, W.; Hood, H.M.; Staben, C.; Selker, E.U.; Sachs, M.S.; Farman, M.L. Characterization of chromosome ends in the filamentous fungus Neurospora crassa. Genetics 2009, 181, 1129–1145. [Google Scholar] [CrossRef] [PubMed]
- Stajich, J.E.; Wilke, S.K.; Ahren, D.; Au, C.H.; Birren, B.W.; Borodovsky, M.; Burns, C.; Canback, B.; Casselton, L.A.; Cheng, C.K.; et al. Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus). Proc. Natl. Acad. Sci. USA 2010, 107, 11889–11894. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Young, C.A.; Schardl, C.L.; Panaccione, D.G.; Florea, S.; Takach, J.E.; Charlton, N.D.; Moore, N.; Webb, J.S.; Jaromczyk, J. Genetics, Genomics and Evolution of Ergot Alkaloid Diversity. Toxins 2015, 7, 1273-1302. https://doi.org/10.3390/toxins7041273
Young CA, Schardl CL, Panaccione DG, Florea S, Takach JE, Charlton ND, Moore N, Webb JS, Jaromczyk J. Genetics, Genomics and Evolution of Ergot Alkaloid Diversity. Toxins. 2015; 7(4):1273-1302. https://doi.org/10.3390/toxins7041273
Chicago/Turabian StyleYoung, Carolyn A., Christopher L. Schardl, Daniel G. Panaccione, Simona Florea, Johanna E. Takach, Nikki D. Charlton, Neil Moore, Jennifer S. Webb, and Jolanta Jaromczyk. 2015. "Genetics, Genomics and Evolution of Ergot Alkaloid Diversity" Toxins 7, no. 4: 1273-1302. https://doi.org/10.3390/toxins7041273
APA StyleYoung, C. A., Schardl, C. L., Panaccione, D. G., Florea, S., Takach, J. E., Charlton, N. D., Moore, N., Webb, J. S., & Jaromczyk, J. (2015). Genetics, Genomics and Evolution of Ergot Alkaloid Diversity. Toxins, 7(4), 1273-1302. https://doi.org/10.3390/toxins7041273





