Molecular Characterization of Lys49 and Asp49 Phospholipases A2 from Snake Venom and Their Antiviral Activities against Dengue virus
Abstract
1. Introduction
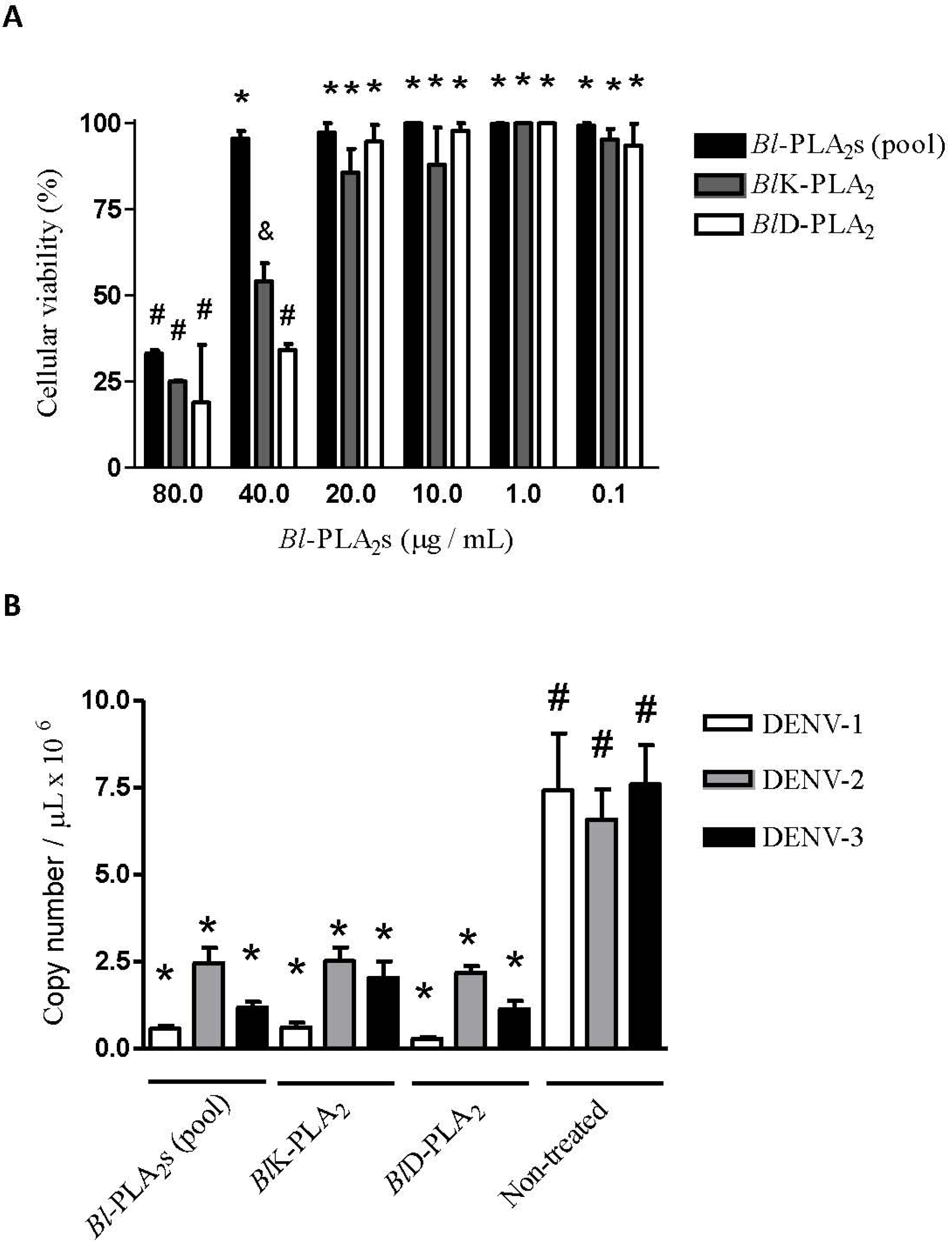
2. Results and Discussion
2.1. Mass Spectrometry Analysis and Sequence Determination

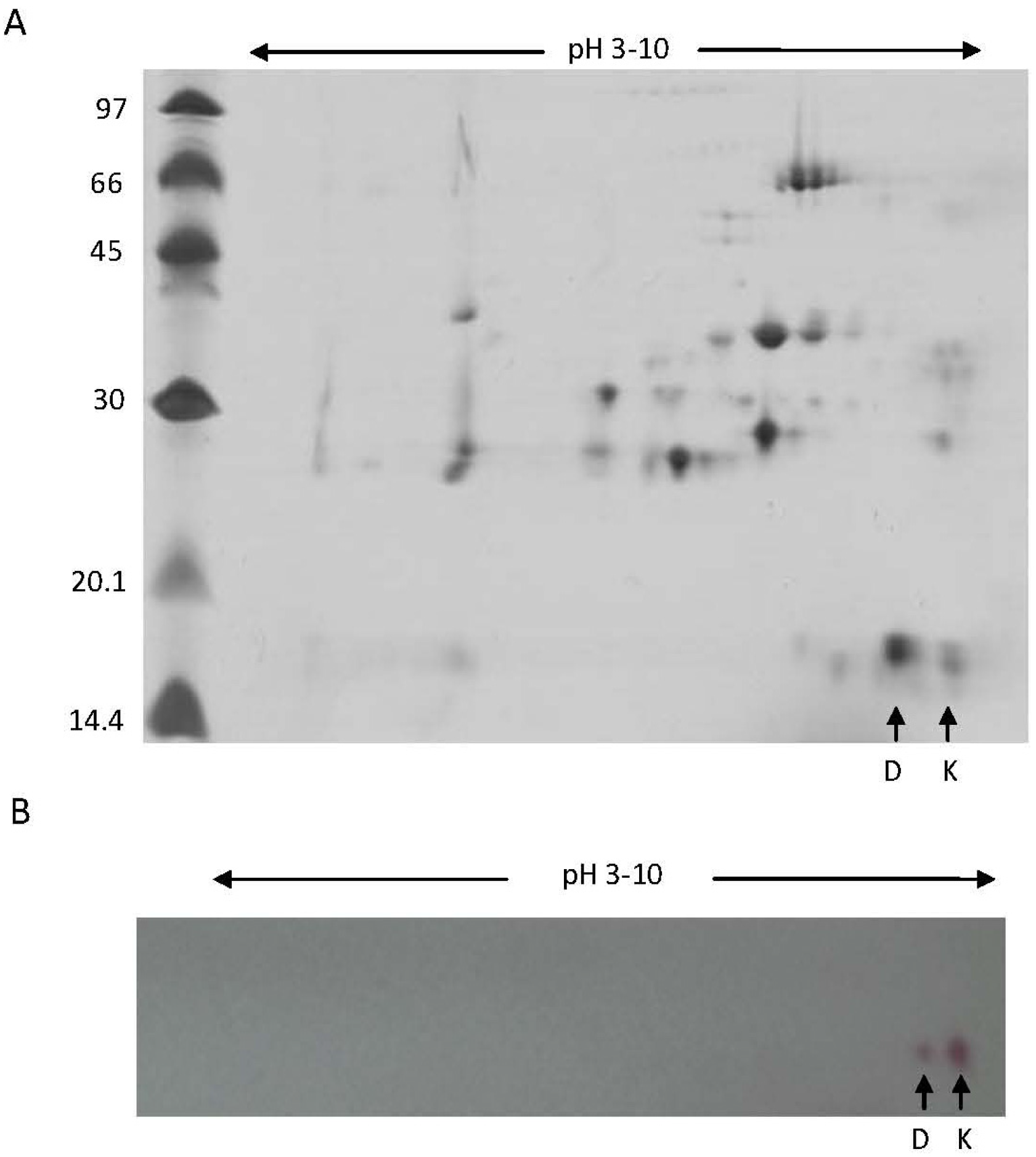
2.2. Biochemical and Immunochemical Properties
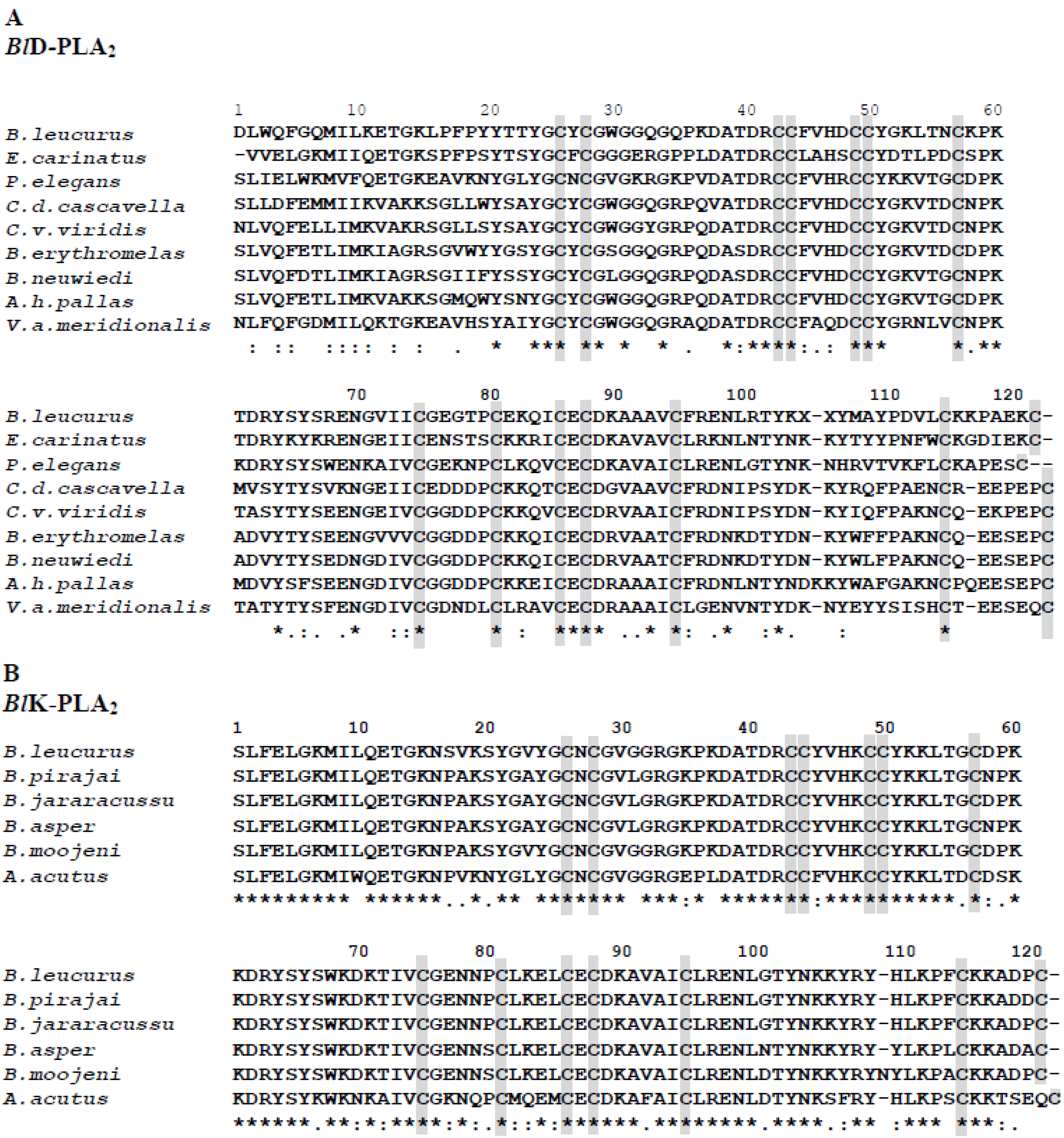

2.3. Phylogenetic Relationships among svPLA2s

3. Materials and Methods
3.1. Purification of BlK-PLA2 and BlD-PLA2
3.2. Protein Characterization
3.3. MALDI-TOF Mass Spectrometry
3.4. 2D-Electrophoresis (2-DE) and Image Analysis
3.5. Protein Sequencing
3.6. Immunoblot and Enzyme-Linked Immunoabsorbent Assay (ELISA)
3.7. Cells and Viruses
3.8. Determination of the Non-Cytotoxic Concentrations
3.10. RNA Extraction
3.11. Generation of RNA Standard
| Target | Sequence (5'-3') | Nucleotide position |
|---|---|---|
| 3'UTR-F | GARAGACCAGAGATCCTGCTGTCT | 10,647–10,670 |
| 3'UTR-R | ACCATTCCATTTTCTGGCGTT | 10,714–10,694 |
| 3'UTR-Probe | VIC-AGCATCATTCCAGGCAC-MGB-NFQ | 10,675–10,691 |
| RnaseP-F | AGATTTGGACCTGCGAGCG | 50–68 |
| RnaseP-R | GAGCGGCTGTCTCCACAAGT | 114–95 |
| RnaseP-Probe | FAM-TTCTGACCTGAAGGCTCTGCGCG-BHQ1 | 71–91 |
3.12. Quantitative Real-Time PCR
3.13. Statistical Analysis
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Koh, D.C.I.; Armugan, A.; Jeyaseelan, K. Snake venom components and their applications in biomedicine. Cell. Mol. Life Sci. 2006, 63, 3030–3041. [Google Scholar] [CrossRef]
- Kini, R.M. Toxins in thrombosis and haemostasis, potential beyond imagination. J. Thromb. Haemost. 2011, 9, 193–208. [Google Scholar]
- Koh, C.Y.; Kini, R.M. From snake venom toxins to therapeutic-cardiovascular examples. Toxicon 2012, 59, 497–506. [Google Scholar] [CrossRef]
- Lomonte, B.; Sasa, A.Y.; Gutierrez, J.M. The phospholipase A2 homologues of snake venoms, biological activities and their possible adaptative roles. Protein Pept. Lett. 2009, 16, 860–876. [Google Scholar] [CrossRef]
- Samy, R.P.; Gopalakrishnakone, P.; Kesturu, G.; Swamy, S.N.; Hemshekhar, M.; Tan, K.S.; Rowan, G.E.G.; Stiles, B.G.; Chow, V.T.K. Snake venom phospholipases A2: A novel tool against bacterial diseases. Curr. Med. Chem. 2012, 19, 6150–6162. [Google Scholar] [CrossRef]
- Doley, R.; Zhou, X.; Kini, R.M. Snake Venom Phospholipase A2 Enzymes. In Handbook of Venoms and Toxins of Reptiles; Mackessy, S.P., Ed.; CRC Press: New York, NY, USA, 2010; pp. 173–198. [Google Scholar]
- Huang, P.; Mackessy, S.P. Biochemical characterization of phospholipase A2 (trimorphin) from the venom of the Sonoran lyre snake Trimorphodon biscutatus lambda (family Colubridae). Toxicon 2004, 44, 25–43. [Google Scholar]
- Six, D.A.; Dennis, E.A. The expanding of superfamily of phospholipase A2 enzymes, classification and characterization. Biochim. Biophys. Acta 2000, 1488, 1–19. [Google Scholar]
- Bazaa, A.; Luis, J.; Srairi-Abid, N.; Kallech-Ziri, O.; Kessentini-Zouari, R. MVL-PLA2; a phospholipase A2 from Microvipera lebetine transmediterranea venom; inhibits tumor cells adhesion and migration. Matrix Biol. 2009, 28, 188–193. [Google Scholar] [CrossRef]
- Bazaa, A.; Pasquier, E.; Defilles, C.; Limam, I.; Kessentini-Zouari, R.; Kallech-Ziri, O.; el Battari, A.; Braguer, D.; el Ayeb, M.; Marrakchi, N.; et al. MVL-PLA2; a snake venom phospholipase A2; inhibits angiogenesis through an increase in microtubule dynamics and disorganization of focal adhesions. PLoS One 2010, 5, e10124. [Google Scholar] [CrossRef]
- Khunsap, S.; Pakmanee, N.; Khow, O.; Chanhome, L.; Sitprija, V.; Suntravat, M.; Lucena, S.E.; Perez, J.C.; Sánchez, E.E. Purification of a phospholipase A2 from Daboia russelli siamensis venom with anticancer effects. J. Venom. Res. 2011, 2, 42–51. [Google Scholar]
- Samy, R.P.; Pachiappan, A.; Gopalakrishnakone, P.; Thwin, M.M.; Hian, Y.E.; Chow, V.T.K.; Bow, H.; Weng, J.T. In vitro antimicrobial activity of natural toxins and animal venoms tested against Burkholderia pseudomallei. BMC Infect. Dis. 2006, 6, 1–16. [Google Scholar] [CrossRef]
- Vargas, L.J.; Londoño, M.; Quintana, J.C.; Rua, C.; Segura, C.; Lomonte, B.; Nuñez, V. An acidic phospholipase A2 with antibacterial activity from Porthidium nasutum venom. Comp. Biochem. Physiol. B 2012, 161, 341–347. [Google Scholar] [CrossRef]
- Gunther, A.G.; Stegmann, T. How lysophosphatidylcholine inhibits cell-cell fusion mediated by the envelope glycoprotein of human immunodeficiency virus. Virology 1997, 235, 201–208. [Google Scholar] [CrossRef]
- Fenard, D.; Lambeau, G.; Valentin, E.; Lefebvre, J.C.; Lazdunski, M.; Doglio, A. Secreted phospholipases A2; a new class of HIV inhibitors that block virus entry into host cells. J. Clin. Invest. 1999, 104, 611–618. [Google Scholar] [CrossRef]
- Ooi, E.E.; Gubler, D.J. Global spread of epidemic dengue, the influence of environmental change. Future Virol. 2009, 4, 571–580. [Google Scholar] [CrossRef]
- Hotez, P.J.; Bottazzi, M.E.; Franco-Paredes, C.; Ault, S.K.; Periago, M.R. The neglected tropical diseases of Latin America and the Caribbean, a review of disease burden and distribution of a roadmap for control and elimination. PLoS Negl. Trop. Dis. 2008, 2, e300. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Impact of Dengue. Available online: http//www.who.int/csr/disease/dengue/impact/em (accessed on 31 May 2013).
- Thomas, S.J.; Strickman, D.; Vuaghan, D.W. Dengue epidemiology; ecology; resurgence. Adv. Virus Res. 2003, 61, 235–289. [Google Scholar] [CrossRef]
- Shepard, D.S.; Coudeville, L.; Halasa, Y.A.; Zambrano, B.; Dayan, G.H. Economic impact of dengue illness in the Americas. Am. J. Trop. Med. Hyg. 2011, 84, 200–207. [Google Scholar] [CrossRef]
- Whitehead, S.S.; Blaney, J.E.; Durbin, A.P.; Murphy, B.R. Prospects for a dengue virus vaccine. Nat. Rev. Microbiol. 2007, 5, 518–528. [Google Scholar] [CrossRef]
- Porto, M.; Teixeira, D.M. Bothrops leucurus (White-tailed-lancehead). Herpetol. Rev. 1995, 26, 156. [Google Scholar]
- Sanchez, E.F.; Eble, J.A. P-III Metalloproteinase (Leucurolysin-B) from Bothrops Leucurus Venom, Isolation and Possible Inhibition. In Drug Design of Zinc-Enzyme Inhibitors; Supuran, C.T., Winum, J.-Y., Eds.; John Wiley & Sons Inc: Hoboken, NJ, USA, 2009; pp. 789–812. [Google Scholar]
- Higuchi, D.A.; Barbosa, C.M.V.; Bincoleto, C.; Chagas, J.R.; Magalhaes, A.; Richardson, M.; Sanchez, E.F.; Pesquero, J.B.; Araujo, R.C.; Pesquero, J.L. Purification and partial characterization of two phospholipases A2 from Bothrops leucurus (white-tailed-jararaca) snake venom. Biochimie 2007, 89, 319–328. [Google Scholar] [CrossRef]
- Muller, V.D.M.; Russo, R.R.; Cintra, A.D.O.; Sartim, M.A.; Alves-Paiva, R.M.; Figueiredo, L.T.M.; Sampaio, S.V.; Aquino, V.H. Crotoxin and phospholipases A2 from Crotalus. durissus terrificus showed antiviral activity against dengue and yellow fever viruses. Toxicon 2012, 59, 507–515. [Google Scholar] [CrossRef]
- Julander, J.G.; Perry, S.T.; Shresta, S. Important advances in the field of anti-dengue virus research. Antivir. Chem. Chemother. 2011, 21, 105–116. [Google Scholar] [CrossRef]
- Mattiazzi, M.; Sun, Y.; Wolinski, H.; Bavdek, A.; Petan, T.; Anderluh, G.; Kohlwein, S.D.; Drubin, D.G.; Krizaj, I.; Petrovic, U. A neurotoxic phospholipase A2 impairs yeast amphiphysin activity and reduces endocytosis. PLoS One 2012, 7, e40931. [Google Scholar] [CrossRef]
- Petricevich, V.L.; Mendonça, R.Z. Inhibitory potential of Crotalus durissus terrificus venom on measles virus growth. Toxicon 2003, 42, 143–153. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshilds, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Scott, D.L. Phospholipase A2 Structure and Catalytic Properties. In Venom Phospholipase A2 Enzymes, Structure; Function and Mechanism; Kini, R.M., Ed.; John Wiley & Sons: Chichester, UK, 1997; pp. 97–128. [Google Scholar]
- Páramo, L.; Lomonte, B.; Pizarro-Cerda, J.; Bengoechea, J.A.; Gorvel, J.P.; Moreno, E. Bactericidal activity of Lys49 and Asp49 myotoxic phaspholipases A2 from Bothrops asper snake venom. Eur. J. Biochem. 1998, 253, 452–561. [Google Scholar]
- Watanabe, L.; Fontes, M.R.; Soares, A.M.; Giglio, J.R.; Arni, R.K. Initiating structural studies of Lys49-PLA2 homologues complexed with an anionic detergent; a fatty acid and a natural lipid. Protein Pep. Lett. 2003, 10, 525–530. [Google Scholar] [CrossRef]
- Freedman, J.E.; Snyder, S.H. Vipoxin. A protein from Russell’s viper venom with high affinity for biogenic amine receptors. J. Biol. Chem. 1981, 256, 13172–13179. [Google Scholar]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Polgar, J.; Magnenat, E.M.; Peitsch, M.C.; Wells, T.N.; Clemetson, K.J. Asp-49 is not an absolute prerequisite for the enzymic activity of low-Mr phospholipase A2, Purification; characterization and computer modelling of an enzymically active Ser-49 phospholipase A2; ecarpholin S; from the venom of Echis carinatus sochureki (saw-scaled viper). Biochem. J. 1996, 319, 965–968. [Google Scholar]
- Chijiwa, T.; Tokunaga, E.; Ikeda, R.; Tereda, K.; Ogawa, T.; Oda-Ueda, N.; Hattori, S.; Nozaki, M.; Ohno, M. Discovery of novel Arg49 phospholipase A2 isozyme from Phrotobothrops elegans venom and regional evolution of Crotalinae snake venom phospholipase A2 isozymes in the southwestern islands of Japan and Taiwan. Toxicon 2006, 48, 672–682. [Google Scholar] [CrossRef]
- Fry, B.G.; Wüster, W. Assembling an arsenal, Origin and evolution of the snake venom proteome inferred from Phylogenetic analysis of toxin sequences. Mol. Biol. Evol. 2004, 21, 870–883. [Google Scholar] [CrossRef]
- Fry, B.G.; Scheib, H.; van der Weer, L.; Young, B.; McNaughtan, J.; Ramjan, S.F.R.; Vidal, N.; Poelmann, R.E.; Norman, J.A. Evolution of an arsenal. Mol. Cell. Proteomics 2008, 7, 215–246. [Google Scholar]
- Cummings, K.L.; Tarleton, R.L. Rapid quantitation of Trypanosoma cruzi in host tissue by real-time PCR. Mol. Biochem. Parasitol. 2003, 129, 53–59. [Google Scholar] [CrossRef]
- Trane, L.D.; Bedini, B.; Donatelli, I.; Campitelli, L.; Chiappini, B.; de Marco, M.A.; Delogu, M.; Buonaboglia, C.; Vaccari, G. A sensitive one-step real time PCR for detection of avian influence viruses using a MGB probe and an internal positive control. BMC Infect. Dis. 2006, 6, 87. [Google Scholar] [CrossRef]
- Caldas, S.; Caldas, I.S.; Diniz, L.F.; Lima, W.G.; Oliveira, R.P.; Cecílio, A.B.; Ribeiro, I.; Talvania, A.; Bahia, M.T. Real-Time PCR strategy for parasite quantification in blood and tissue samples of experimental Trypanosoma cruzi infection. Acta Trop. 2012, 123, 170–177. [Google Scholar] [CrossRef]
- Naumann, G.B.; Silva, L.F.; Silva, L.; Faria, G.; Richardson, M.; Evangelista, K.; Kohlhoff, M.; Gontijo, C.M.F.; Navdaev, A.; Rezende, F.F.; et al. Cytotoxicity and inhibition of platelet aggregation caused by an L-amino acid oxidase from Bopthrops leucurus venom. Biochim. Biophys. Acta 2011, 1810, 683–694. [Google Scholar]
- Wilson, K.J.; Yuan, P.M. Protein and Peptide Purification. In Protein Sequencing a Practical Approach; Findlay, J.B.C., Geisow, M.J., Eds.; IRL Press: Oxford, UK, 1989; pp. 3–41. [Google Scholar]
- Dulbecco, R. Microbiology. In Evolution of Microbiology and Microbes; Dulbecco, R., Ed.; Lippincott Company: Philadelphia, PA, USA, 1994; pp. 769–795. [Google Scholar]
- Gurukumar, K.R.; Priyadarshini, D.; Patil, J.A.; Bhagat, A.; Singh, A.; Shah, S.; Cecilia, D. Development of real time PCR for detection and quantitation of Dengue Viruses. Virol. J. 2009, 6, 10. [Google Scholar] [CrossRef]
- World Health Organization. CDC protocol of real time RT PCR for influenza A (H1N1). Available online: http://www.who.int/csr/resources/publications/swineflu/CDCrealtimeRTPCRprotocol20090428.pdf (accessed on 27 June 2013).
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Cecilio, A.B.; Caldas, S.; Oliveira, R.A.D.; Santos, A.S.B.; Richardson, M.; Naumann, G.B.; Schneider, F.S.; Alvarenga, V.G.; Estevão-Costa, M.I.; Fuly, A.L.; et al. Molecular Characterization of Lys49 and Asp49 Phospholipases A2 from Snake Venom and Their Antiviral Activities against Dengue virus. Toxins 2013, 5, 1780-1798. https://doi.org/10.3390/toxins5101780
Cecilio AB, Caldas S, Oliveira RAD, Santos ASB, Richardson M, Naumann GB, Schneider FS, Alvarenga VG, Estevão-Costa MI, Fuly AL, et al. Molecular Characterization of Lys49 and Asp49 Phospholipases A2 from Snake Venom and Their Antiviral Activities against Dengue virus. Toxins. 2013; 5(10):1780-1798. https://doi.org/10.3390/toxins5101780
Chicago/Turabian StyleCecilio, Alzira B., Sergio Caldas, Raiana A. De Oliveira, Arthur S. B. Santos, Michael Richardson, Gustavo B. Naumann, Francisco S. Schneider, Valeria G. Alvarenga, Maria I. Estevão-Costa, Andre L. Fuly, and et al. 2013. "Molecular Characterization of Lys49 and Asp49 Phospholipases A2 from Snake Venom and Their Antiviral Activities against Dengue virus" Toxins 5, no. 10: 1780-1798. https://doi.org/10.3390/toxins5101780
APA StyleCecilio, A. B., Caldas, S., Oliveira, R. A. D., Santos, A. S. B., Richardson, M., Naumann, G. B., Schneider, F. S., Alvarenga, V. G., Estevão-Costa, M. I., Fuly, A. L., Eble, J. A., & Sanchez, E. F. (2013). Molecular Characterization of Lys49 and Asp49 Phospholipases A2 from Snake Venom and Their Antiviral Activities against Dengue virus. Toxins, 5(10), 1780-1798. https://doi.org/10.3390/toxins5101780