Trichothecenes and Fumonisins: Key Players in Fusarium–Cereal Ecosystem Interactions
Abstract
:1. Introduction
2. Role of Trichothecenes and Fumonisins in Fungal Pathogenesis of Plants
3. Cellular Effects of Trichothecenes and Fumonisins on Cereals
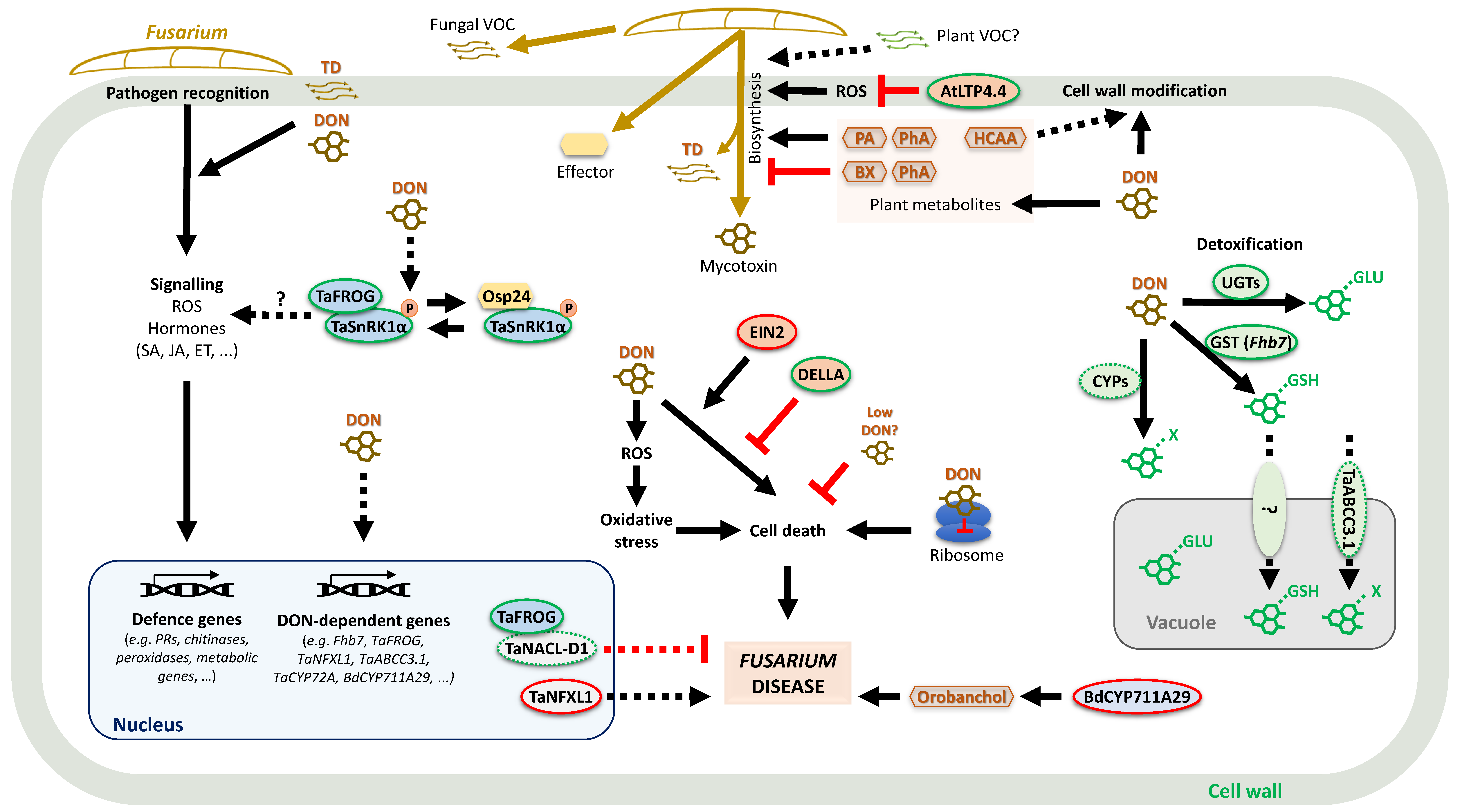
4. Plant Resistance to Trichothecenes and Fumonisins
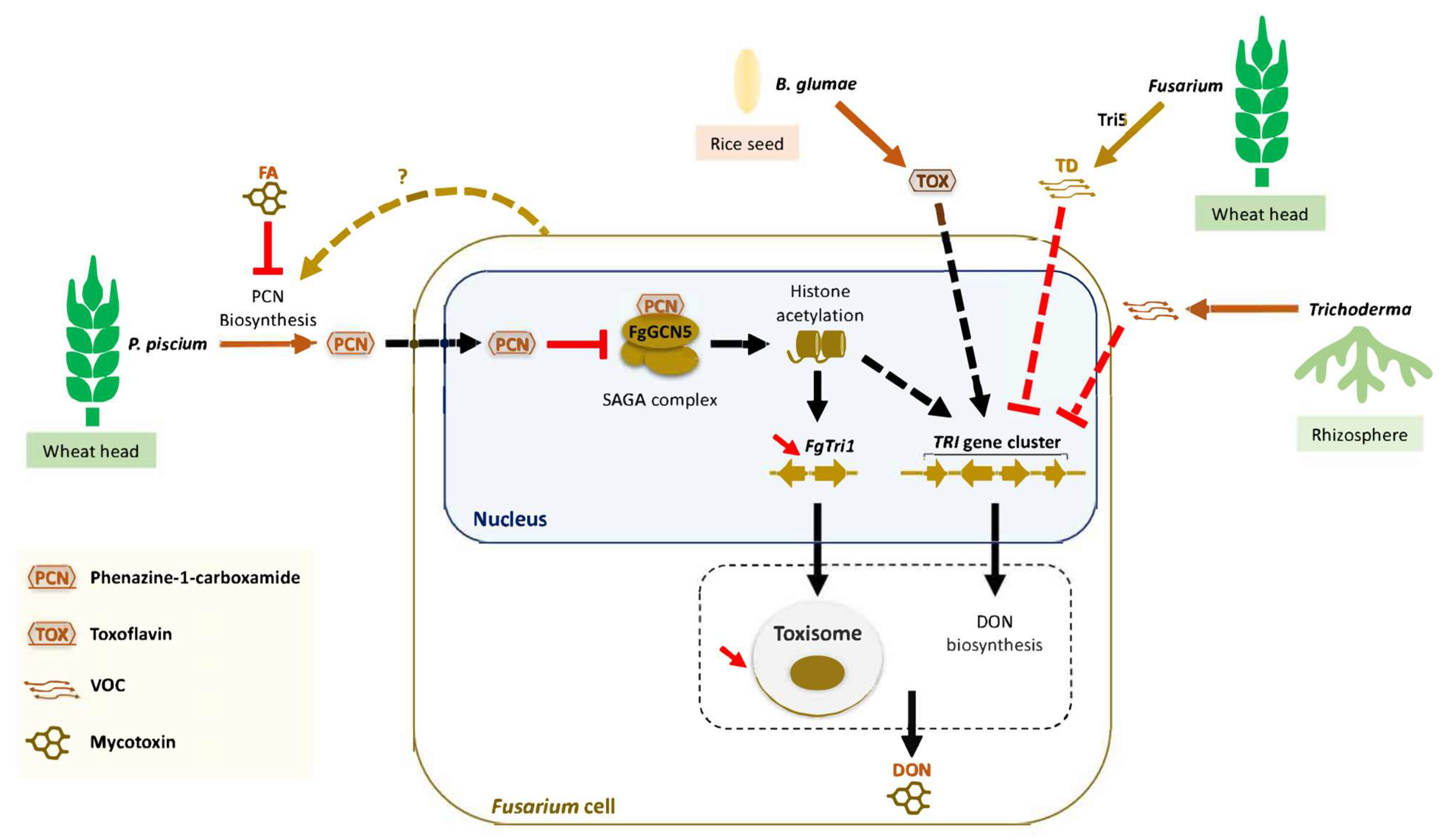
5. Fusarium Mycotoxin–Microbiome and Mycotoxin–Insect Interactions

6. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Foroud, N.A.; Baines, D.; Gagkaeva, T.Y.; Thakor, N.; Badea, A.; Steiner, B.; Bürstmayr, M.; Bürstmayr, H. Trichothecenes in Cereal Grains—An Update. Toxins 2019, 11, 634. [Google Scholar] [CrossRef] [PubMed]
- Munkvold, G.P.; Proctor, R.H.; Moretti, A. Mycotoxin Production in Fusarium According to Contemporary Species Concepts. Annu. Rev. Phytopathol. 2021, 59, 373–402. [Google Scholar] [CrossRef]
- Maier, F.J.; Miedaner, T.; Hadeler, B.; Felk, A.; Salomon, S.; Lemmens, M.; Kassner, H.; Schäfer, W. Involvement of trichothecenes in fusarioses of wheat, barley and maize evaluated by gene disruption of the trichodiene synthase (Tri5) gene in three field isolates of different chemotype and virulence. Mol. Plant Pathol. 2006, 7, 449–461. [Google Scholar] [CrossRef]
- Winter, M.; Samuels, P.; Dong, Y.; Dill-Macky, R. Trichothecene production is detrimental to early root colonization by Fusarium culmorum and F. graminearum in fusarium crown and root rot of wheat. Plant Pathol. 2019, 68, 185–195. [Google Scholar] [CrossRef]
- Zheng, H.; Li, L.; Miao, P.; Wu, C.; Chen, X.; Yuan, M.; Fang, T.; Norvienyeku, J.; Li, G.; Zheng, W. FgSec2A, a guanine nucleotide exchange factor of FgRab8, is important for polarized growth, pathogenicity and deoxynivalenol production in Fusarium graminearum. Environ. Microbiol. 2018, 20, 3378–3392. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; van Diepeningen, A.D.; van der Lee, T.A.; Waalwijk, C.; Xu, J.; Xu, J.; Zhang, H.; Chen, W.; Feng, J. The Fusarium graminearum histone acetyltransferases are important for morphogenesis, DON biosynthesis, and pathogenicity. Front. Microbiol. 2018, 9, 654. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Tong, Q.; Zhang, C.; Ding, K. The transcription factor FgCrz1A is essential for fungal development, virulence, deoxynivalenol biosynthesis and stress responses in Fusarium graminearum. Curr. Genet. 2019, 65, 153–166. [Google Scholar] [CrossRef]
- Lv, W.; Wang, C.; Yang, N.; Que, Y.; Talbot, N.J.; Wang, Z. Genome-wide functional analysis reveals that autophagy is necessary for growth, sporulation, deoxynivalenol production and virulence in Fusarium graminearum. Sci. Rep. 2017, 7, 11062. [Google Scholar] [CrossRef]
- Iqbal, N.; Czékus, Z.; Poór, P.; Ördög, A. Plant defence mechanisms against mycotoxin fumonisin B1. Chem.-Biol. Interact. 2021, 343, 109494. [Google Scholar] [CrossRef]
- Blacutt, A.A.; Gold, S.E.; Voss, K.A.; Gao, M.; Glenn, A.E. Fusarium verticillioides: Advancements in understanding the toxicity, virulence, and niche adaptations of a model mycotoxigenic pathogen of maize. Phytopathology 2018, 108, 312–326. [Google Scholar] [CrossRef]
- Gao, M.; Glenn, A.E.; Gu, X.; Mitchell, T.R.; Satterlee, T.; Duke, M.V.; Scheffler, B.E.; Gold, S.E. Pyrrocidine, a molecular off switch for fumonisin biosynthesis. PLoS Pathog. 2020, 16, e1008595. [Google Scholar] [CrossRef]
- Glenn, A.E.; Zitomer, N.C.; Zimeri, A.M.; Williams, L.D.; Riley, R.T.; Proctor, R.H. Transformation-mediated complementation of a FUM gene cluster deletion in Fusarium verticillioides restores both fumonisin production and pathogenicity on maize seedlings. Mol. Plant-Microbe Interact. 2008, 21, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Garreau de Loubresse, N.; Prokhorova, I.; Holtkamp, W.; Rodnina, M.V.; Yusupova, G.; Yusupov, M. Structural basis for the inhibition of the eukaryotic ribosome. Nature 2014, 513, 517–522. [Google Scholar] [CrossRef]
- Audenaert, K.; Vanheule, A.; Hofte, M.; Haesaert, G. Deoxynivalenol: A Major Player in the Multifaceted Response of Fusarium to Its Environment. Toxins 2014, 6, 1–19. [Google Scholar] [CrossRef]
- Gardiner, D.M.; Kazan, K.; Praud, S.; Torney, F.J.; Rusu, A.; Manners, J.M. Early activation of wheat polyamine biosynthesis during Fusarium head blight implicates putrescine as an inducer of trichothecene mycotoxin production. BMC Plant Biol. 2010, 10, 289. [Google Scholar] [CrossRef]
- Gunupuru, L.; Perochon, A.; Doohan, F. Deoxynivalenol resistance as a component of FHB resistance. Trop. Plant Pathol. 2017, 42, 175–183. [Google Scholar] [CrossRef]
- Bönnighausen, J.; Schauer, N.; Schäfer, W.; Bormann, J. Metabolic profiling of wheat rachis node infection by Fusarium graminearum–decoding deoxynivalenol-dependent susceptibility. New Phytol. 2019, 221, 459–469. [Google Scholar] [CrossRef]
- Yang, J.; Duan, G.; Li, C.; Liu, L.; Han, G.; Zhang, Y.; Wang, C. The Crosstalks Between Jasmonic Acid and Other Plant Hormone Signaling Highlight the Involvement of Jasmonic Acid as a Core Component in Plant Response to Biotic and Abiotic Stresses. Front. Plant Sci. 2019, 10, 1349. [Google Scholar] [CrossRef] [PubMed]
- Brauer, E.K.; Balcerzak, M.; Rocheleau, H.; Leung, W.; Schernthaner, J.; Subramaniam, R.; Ouellet, T. Genome editing of a deoxynivalenol-induced transcription factor confers resistance to Fusarium graminearum in wheat. Mol. Plant-Microbe Interact. 2020, 33, 553–560. [Google Scholar] [CrossRef]
- Asano, T.; Masuda, D.; Yasuda, M.; Nakashita, H.; Kudo, T.; Kimura, M.; Yamaguchi, K.; Nishiuchi, T. AtNFXL1, an Arabidopsis homologue of the human transcription factor NF-X1, functions as a negative regulator of the trichothecene phytotoxin-induced defense response. Plant J. 2008, 53, 450–464. [Google Scholar] [CrossRef]
- Qi, P.-F.; Balcerzak, M.; Rocheleau, H.; Leung, W.; Wei, Y.-M.; Zheng, Y.-L.; Ouellet, T. Jasmonic acid and abscisic acid play important roles in host–pathogen interaction between Fusarium graminearum and wheat during the early stages of fusarium head blight. Physiol. Mol. Plant Pathol. 2016, 93, 39–48. [Google Scholar] [CrossRef]
- Gutiérrez-Nájera, N.A.; Saucedo-García, M.; Noyola-Martínez, L.; Vázquez-Vázquez, C.; Palacios-Bahena, S.; Carmona-Salazar, L.; Plasencia, J.; El-Hafidi, M.; Gavilanes-Ruiz, M. Sphingolipid Effects on the Plasma Membrane Produced by Addition of Fumonisin B1 to Maize Embryos. Plants 2020, 9, 150. [Google Scholar] [CrossRef]
- Otaiza-González, S.N.; Mary, V.S.; Arias, S.L.; Bertrand, L.; Velez, P.A.; Rodriguez, M.G.; Rubinstein, H.R.; Theumer, M.G. Cell death induced by fumonisin B1 in two maize hybrids: Correlation with oxidative status biomarkers and salicylic and jasmonic acids imbalances. Eur. J. Plant Pathol. 2022, 163, 203–221. [Google Scholar] [CrossRef]
- Yanagawa, D.; Ishikawa, T.; Imai, H. Synthesis and degradation of long-chain base phosphates affect fumonisin B1-induced cell death in Arabidopsis thaliana. J. Plant Res. 2017, 130, 571–585. [Google Scholar] [CrossRef]
- Glenz, R.; Schmalhaus, D.; Krischke, M.; Mueller, M.J.; Waller, F. Elevated levels of phosphorylated sphingobases do not antagonize sphingobase-or Fumonisin B1-induced plant cell death. Plant Cell Physiol. 2019, 60, 1109–1119. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, Q.; Cui, S.; Ren, J.; Qian, W.; Yang, Y.; He, S.; Chu, J.; Sun, X.; Yan, C.; et al. Hijacking of the jasmonate pathway by the mycotoxin fumonisin B1 (FB1) to initiate programmed cell death in Arabidopsis is modulated by RGLG3 and RGLG4. J. Exp. Bot. 2015, 66, 2709–2721. [Google Scholar] [CrossRef] [PubMed]
- Lanubile, A.; De Michele, R.; Loi, M.; Fakhari, S.; Marocco, A.; Paciolla, C. Cell death induced by mycotoxin fumonisin B1 is accompanied by oxidative stress and transcriptional modulation in Arabidopsis cell culture. Plant Cell Rep. 2022, 41, 1733–1750. [Google Scholar] [CrossRef]
- Iqbal, N.; Czékus, Z.; Angeli, C.; Bartók, T.; Poór, P.; Ördög, A. Fumonisin B1-Induced Oxidative Burst Perturbed Photosynthetic Activity and Affected Antioxidant Enzymatic Response in Tomato Plants in Ethylene-Dependent Manner. J. Plant Growth Regul. 2023, 42, 1865–1878. [Google Scholar] [CrossRef]
- Xie, L.H.; Yang, Q.X.; Wu, Y.F.; Xiao, J.B.; Qu, H.X.; Jiang, Y.M.; Li, T.T. Fumonisin B1 Biosynthesis Is Associated with Oxidative Stress and Plays an Important Role in Infection on Banana Fruit. J. Agric. Food Chem. 2023, 71, 5372–5381. [Google Scholar] [CrossRef]
- Xie, L.; Wu, Y.; Wang, Y.; Jiang, Y.; Yang, B.; Duan, X.; Li, T. Fumonisin B1 induced aggressiveness and infection mechanism of Fusarium proliferatum on banana fruit. Environ. Pollut. 2021, 288, 117793. [Google Scholar] [CrossRef]
- Wang, H.; Sun, S.; Ge, W.; Zhao, L.; Hou, B.; Wang, K.; Lyu, Z.; Chen, L.; Xu, S.; Guo, J.; et al. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 2020, 368, eaba5435. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Hei, R.; Yang, Y.; Zhang, S.; Wang, Q.; Wang, W.; Zhang, Q.; Yan, M.; Zhu, G.; Huang, P.; et al. An orphan protein of Fusarium graminearum modulates host immunity by mediating proteasomal degradation of TaSnRK1α. Nat. Commun. 2020, 11, 4382. [Google Scholar] [CrossRef] [PubMed]
- Perochon, A.; Jianguang, J.; Kahla, A.; Arunachalam, C.; Scofield, S.R.; Bowden, S.; Wallington, E.; Doohan, F.M. TaFROG Encodes a Pooideae Orphan Protein That Interacts with SnRK1 and Enhances Resistance to the Mycotoxigenic Fungus Fusarium graminearum. Plant Physiol. 2015, 169, 2895–2906. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Steed, A.; Travella, S.; Keller, B.; Nicholson, P. Fusarium graminearum exploits ethylene signalling to colonize dicotyledonous and monocotyledonous plants. New Phytol. 2009, 182, 975–983. [Google Scholar] [CrossRef]
- Saville, R.J.; Gosman, N.; Burt, C.J.; Makepeace, J.; Steed, A.; Corbitt, M.; Chandler, E.; Brown, J.K.; Boulton, M.I.; Nicholson, P. The ‘Green Revolution’ dwarfing genes play a role in disease resistance in Triticum aestivum and Hordeum vulgare. J. Exp. Bot. 2011, 63, 1271–1283. [Google Scholar] [CrossRef] [PubMed]
- Atanasova-Penichon, V.; Barreau, C.; Richard-Forget, F. Antioxidant Secondary Metabolites in Cereals: Potential Involvement in Resistance to Fusarium and Mycotoxin Accumulation. Front. Microbiol. 2016, 7, 566. [Google Scholar] [CrossRef]
- Baldwin, T.; Baldwin, S.; Klos, K.; Bregitzer, P.; Marshall, J. Deletion of the benzoxazinoid detoxification gene NAT1 in Fusarium graminearum reduces deoxynivalenol in spring wheat. PLoS ONE 2019, 14, e0214230. [Google Scholar] [CrossRef]
- Doppler, M.; Kluger, B.; Bueschl, C.; Steiner, B.; Buerstmayr, H.; Lemmens, M.; Krska, R.; Adam, G.; Schuhmacher, R. Stable Isotope-Assisted Plant Metabolomics: Investigation of Phenylalanine-Related Metabolic Response in Wheat Upon Treatment With the Fusarium Virulence Factor Deoxynivalenol. Front. Plant Sci. 2019, 10, 1137. [Google Scholar] [CrossRef]
- Taylor, L.; Gutierrez, S.; McCormick, S.P.; Bakker, M.G.; Proctor, R.H.; Teresi, J.; Kurtzman, B.; Hao, G.; Vaughan, M.M. Use of the volatile trichodiene to reduce Fusarium head blight and trichothecene contamination in wheat. Microb. Biotechnol. 2021, 15, 513–527. [Google Scholar] [CrossRef]
- Blumke, A.; Sode, B.; Ellinger, D.; Voigt, C.A. Reduced susceptibility to Fusarium head blight in Brachypodium distachyon through priming with the Fusarium mycotoxin deoxynivalenol. Mol. Plant Pathol. 2015, 16, 472–483. [Google Scholar] [CrossRef]
- Kirana, R.P.; Gaurav, K.; Arora, S.; Wiesenberger, G.; Doppler, M.; Michel, S.; Zimmerl, S.; Matic, M.; Eze, C.E.; Kumar, M. Identification of a UDP-glucosyltransferase conferring deoxynivalenol resistance in Aegilops tauschii and wheat. Plant Biotechnol. J. 2023, 21, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Gunupuru, L.R.; Arunachalam, C.; Malla, K.B.; Kahla, A.; Perochon, A.; Jia, J.; Thapa, G.; Doohan, F.M. A wheat cytochrome P450 enhances both resistance to deoxynivalenol and grain yield. PLoS ONE 2018, 13, e0204992. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, J.; Song, B.; Li, H.; Xu, H.; Qu, B.; Dang, F.; Liao, Y. Resistance to Fusarium head blight and seedling blight in wheat is associated with activation of a cytochrome P450 gene. Phytopathology 2010, 100, 183–191. [Google Scholar] [CrossRef]
- Walter, S.; Kahla, A.; Arunachalam, C.; Perochon, A.; Khan, M.R.; Scofield, S.R.; Doohan, F.M. A wheat ABC transporter contributes to both grain formation and mycotoxin tolerance. J. Exp. Bot. 2015, 66, 2583–2593. [Google Scholar] [CrossRef]
- Wang, G.-P.; Hou, W.-Q.; Zhang, L.; Wu, H.-Y.; Zhao, L.-F.; Du, X.-Y.; Xin, M.; Li, A.-F.; Wang, H.-W.; Kong, L.-R. Functional analysis of a wheat pleiotropic drug resistance gene involved in Fusarium head blight resistance. J. Integr. Agric. 2016, 15, 2215–2227. [Google Scholar] [CrossRef]
- McLaughlin, J.E.; Darwish, N.I.; Garcia-Sanchez, J.; Tyagi, N.; Trick, H.N.; McCormick, S.; Dill-Macky, R.; Tumer, N.E. A lipid transfer protein has antifungal and antioxidant activity and suppresses Fusarium head blight disease and DON accumulation in transgenic wheat. Phytopatholy 2021, 111, 671–683. [Google Scholar] [CrossRef] [PubMed]
- Perochon, A.; Kahla, A.; Vranić, M.; Jia, J.; Malla, K.B.; Craze, M.; Wallington, E.; Doohan, F.M. A wheat NAC interacts with an orphan protein and enhances resistance to Fusarium head blight disease. Plant Biotechnol. J. 2019, 17, 1892–1904. [Google Scholar] [CrossRef]
- Perochon, A.; Váry, Z.; Malla, K.B.; Halford, N.G.; Paul, M.J.; Doohan, F.M. The wheat SnRK1α family and its contribution to Fusarium toxin tolerance. Plant Sci. 2019, 288, 110217. [Google Scholar] [CrossRef]
- Sun, Z.; Zhou, Y.; Hu, Y.; Jiang, N.; Hu, S.; Li, L.; Li, T. Identification of wheat LACCASEs in response to Fusarium graminearum as potential deoxynivalenol trappers. Front. Plant Sci. 2022, 13, 832800. [Google Scholar] [CrossRef]
- He, W.-J.; Shi, M.-M.; Yang, P.; Huang, T.; Zhao, Y.; Wu, A.-B.; Dong, W.-B.; Li, H.-P.; Zhang, J.-B.; Liao, Y.-C. A quinone-dependent dehydrogenase and two NADPH-dependent aldo/keto reductases detoxify deoxynivalenol in wheat via epimerization in a Devosia strain. Food Chem. 2020, 321, 126703. [Google Scholar] [CrossRef]
- Sgarbi, C.; Malbrán, I.; Saldúa, L.; Lori, G.A.; Lohwasser, U.; Arif, M.A.R.; Börner, A.; Yanniccari, M.; Castro, A.M. Mapping resistance to argentinean fusarium (Graminearum) head blight isolates in wheat. Int. J. Mol. Sci. 2021, 22, 13653. [Google Scholar] [CrossRef]
- Wu, J.-x.; Wu, J.-l.; Yin, J.; Zheng, P.; Yao, N. Ethylene Modulates Sphingolipid Synthesis in Arabidopsis. Front. Plant Sci. 2015, 6, 1122. [Google Scholar] [CrossRef]
- Santiago, R.; Cao, A.; Malvar, R.A.; Butrón, A. Genomics of maize resistance to Fusarium ear rot and fumonisin contamination. Toxins 2020, 12, 431. [Google Scholar] [CrossRef]
- Ye, J.; Zhong, T.; Zhang, D.; Ma, C.; Wang, L.; Yao, L.; Zhang, Q.; Zhu, M.; Xu, M. The Auxin-Regulated Protein ZmAuxRP1 Coordinates the Balance between Root Growth and Stalk Rot Disease Resistance in Maize. Mol. Plant 2019, 12, 360–373. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.-Y.; Li, C.-Y.; Yao, N. Fumonisin B1: A tool for exploring the multiple functions of sphingolipids in plants. Front. Plant Sci. 2020, 11, 600458. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.; Zhao, Y.; Liu, L.; Chen, S.; Li, C.; Meng, F.; Liu, H.; Hu, S.; Wang, J.; Wang, Q. Overexpression of FBR 41 enhances resistance to sphinganine analog mycotoxin-induced cell death and Alternaria stem canker in tomato. Plant Biotechnol. J. 2020, 18, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Kimberlin, A.N.; Majumder, S.; Han, G.; Chen, M.; Cahoon, R.E.; Stone, J.M.; Dunn, T.M.; Cahoon, E.B. Arabidopsis 56-amino acid serine palmitoyltransferase-interacting proteins stimulate sphingolipid synthesis, are essential, and affect mycotoxin sensitivity. Plant Cell 2013, 25, 4627–4639. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, N.; Keller, N.P. Mycotoxins in Conversation With Bacteria and Fungi. Front. Microbiol. 2019, 10, 403. [Google Scholar] [CrossRef] [PubMed]
- Lutz, M.P.; Feichtinger, G.; Defago, G.; Duffy, B. Mycotoxigenic Fusarium and deoxynivalenol production repress chitinase gene expression in the biocontrol agent Trichoderma atroviride P1. Appl. Environ. Microbiol. 2003, 69, 3077–3084. [Google Scholar] [CrossRef]
- van Rij, E.T.; Girard, G.; Lugtenberg, B.J.J.; Bloemberg, G.V. Influence of fusaric acid on phenazine-1-carboxamide synthesis and gene expression of Pseudomonas chlororaphis strain PCL1391. Microbiology 2005, 151, 2805–2814. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, J.; Yang, N.; Wen, Z.; Sun, X.; Chai, Y.; Ma, Z. Wheat microbiome bacteria can reduce virulence of a plant pathogenic fungus by altering histone acetylation. Nat. Commun. 2018, 9, 3429. [Google Scholar] [CrossRef]
- Abdallah, M.F.; De Boevre, M.; Landschoot, S.; De Saeger, S.; Haesaert, G.; Audenaert, K. Fungal Endophytes Control Fusarium graminearum and Reduce Trichothecenes and Zearalenone in Maize. Toxins 2018, 10, 493. [Google Scholar] [CrossRef]
- Tian, Y.; Yu, D.; Liu, N.; Tang, Y.; Yan, Z.; Wu, A. Confrontation assays and mycotoxin treatment reveal antagonistic activities of Trichoderma and the fate of Fusarium mycotoxins in microbial interaction. Environ. Pollut. 2020, 267, 115559. [Google Scholar] [CrossRef] [PubMed]
- Jung, B.; Park, J.; Kim, N.; Li, T.; Kim, S.; Bartley, L.E.; Kim, J.; Kim, I.; Kang, Y.; Yun, K. Cooperative interactions between seed-borne bacterial and air-borne fungal pathogens on rice. Nat. Commun. 2018, 9, 31. [Google Scholar] [CrossRef] [PubMed]
- Vanhoutte, I.; Audenaert, K.; De Gelder, L. Biodegradation of Mycotoxins: Tales from Known and Unexplored Worlds. Front. Microbiol. 2016, 7, 561. [Google Scholar] [CrossRef]
- Tian, Y.; Tan, Y.; Liu, N.; Yan, Z.; Liao, Y.; Chen, J.; De Saeger, S.; Yang, H.; Zhang, Q.; Wu, A. Detoxification of Deoxynivalenol via Glycosylation Represents Novel Insights on Antagonistic Activities of Trichoderma when Confronted with Fusarium graminearum. Toxins 2016, 8, 335. [Google Scholar] [CrossRef]
- De Zutter, N.; Audenaert, K.; Arroyo-Manzanares, N.; De Boevre, M.; Van Poucke, C.; De Saeger, S.; Haesaert, G.; Smagghe, G. Aphids transform and detoxify the mycotoxin deoxynivalenol via a type II biotransformation mechanism yet unknown in animals. Sci. Rep. 2016, 6, 38640. [Google Scholar] [CrossRef]
- Tian, Y.; Tan, Y.; Yan, Z.; Liao, Y.; Chen, J.; De Boevre, M.; De Saeger, S.; Wu, A. Antagonistic and Detoxification Potentials of Trichoderma Isolates for Control of Zearalenone (ZEN) Producing Fusarium graminearum. Front. Microbiol. 2018, 8, 2710. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Guo, Z.; Yang, Z.; Han, H.; Wang, S.; Xu, H.; Yang, X.; Yang, F.; Wu, Q.; Xie, W.; et al. Whitefly hijacks a plant detoxification gene that neutralizes plant toxins. Cell 2021, 184, 1693–1705.e17. [Google Scholar] [CrossRef]
- Santos, A.C.d.S.; Diniz, A.G.; Tiago, P.V.; Oliveira, N.T.d. Entomopathogenic Fusarium species: A review of their potential for the biological control of insects, implications and prospects. Fungal Biol. Rev. 2020, 34, 41–57. [Google Scholar] [CrossRef]
- Drakulic, J.; Bruce, T.J.A.; Ray, R.V. Direct and host-mediated interactions between Fusarium pathogens and herbivorous arthropods in cereals. Plant Pathol. 2017, 66, 3–13. [Google Scholar] [CrossRef]
- Guo, Z.; Döll, K.; Dastjerdi, R.; Karlovsky, P.; Dehne, H.-W.; Altincicek, B. Effect of Fungal Colonization of Wheat Grains with Fusarium spp. on Food Choice, Weight Gain and Mortality of Meal Beetle Larvae (Tenebrio molitor). PLoS ONE 2014, 9, e100112. [Google Scholar] [CrossRef]
- Drakulic, J.; Caulfield, J.; Woodcock, C.; Jones, S.P.; Linforth, R.; Bruce, T.J.; Ray, R.V. Sharing a Host Plant (Wheat [Triticum aestivum]) Increases the Fitness of Fusarium graminearum and the Severity of Fusarium Head Blight but Reduces the Fitness of Grain Aphids (Sitobion avenae). Appl. Environ. Microbiol. 2015, 81, 3492–3501. [Google Scholar] [CrossRef]
- Drakulic, J.; Ajigboye, O.; Swarup, R.; Bruce, T.; Ray, R.V. Aphid Infestation Increases Fusarium langsethiae and T-2 and HT-2 Mycotoxins in Wheat. Appl. Environ. Microbiol. 2016, 82, 6548–6556. [Google Scholar] [CrossRef]
- Guo, Z.; Pfohl, K.; Karlovsky, P.; Dehne, H.-W.; Altincicek, B. Dissemination of Fusarium proliferatum by mealworm beetle Tenebrio molitor. PLoS ONE 2018, 13, e0204602. [Google Scholar] [CrossRef]
- Kang, Z.; Buchenauer, H. Studies on the infection process of Fusarium culmorum in wheat spikes: Degradation of host cell wall components and localization of trichothecene toxins in infected tissue. Eur. J. Plant Pathol. 2002, 108, 653–660. [Google Scholar] [CrossRef]
- Wipfler, R.; McCormick, S.P.; Proctor, R.; Teresi, J.; Hao, G.; Ward, T.; Alexander, N.; Vaughan, M.M. Synergistic Phytotoxic Effects of Culmorin and Trichothecene Mycotoxins. Toxins 2019, 11, 555. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perochon, A.; Doohan, F.M. Trichothecenes and Fumonisins: Key Players in Fusarium–Cereal Ecosystem Interactions. Toxins 2024, 16, 90. https://doi.org/10.3390/toxins16020090
Perochon A, Doohan FM. Trichothecenes and Fumonisins: Key Players in Fusarium–Cereal Ecosystem Interactions. Toxins. 2024; 16(2):90. https://doi.org/10.3390/toxins16020090
Chicago/Turabian StylePerochon, Alexandre, and Fiona M. Doohan. 2024. "Trichothecenes and Fumonisins: Key Players in Fusarium–Cereal Ecosystem Interactions" Toxins 16, no. 2: 90. https://doi.org/10.3390/toxins16020090



