Proteomic Deep Mining the Venom of the Red-Headed Krait, Bungarus flaviceps
Abstract
:1. Introduction
2. Results and Discussion
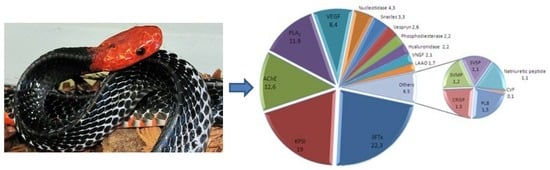
2.1. Major Venom Components
2.2. Minor Venom Components
2.3. Quantification of Venom Components
3. Conclusions
4. Materials and Methods
4.1. Venom Extraction and Ethics Statement
4.2. Tryptic Digestion of Crude Venom
4.3. Chromatography and Mass Spectrometry
4.3.1. One-Dimensional Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE)
4.3.2. Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) Analysis of Excised Gel Bands
4.3.3. Electrospray Mass Spectrometry of Digested Venom
4.3.4. MALDI Mass Spectrometry of Digested Venom
4.4. Data Analysis of Digested Venom
4.5. De Novo Sequencing and Similarity-Driven Analysis of Digested Venom
4.6. Protein Quantitation
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Brahma, R.K.; McCleary, R.J.; Kini, R.M.; Doley, R. Venom gland transcriptomics for identifying, cataloging, and characterizing venom proteins in snakes. Toxicon 2015, 93, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ducancel, F.; Durban, J.; Verdenaud, M. Transcriptomics and venomics: Implications for medicinal chemistry. Future Med. Chem. 2014, 6, 1629–1643. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.J. Snake venomics: From the inventory of toxins to biology. Toxicon 2013, 75, 44–62. [Google Scholar] [CrossRef] [PubMed]
- Zelanis, A.; Tashima, A.K. Unraveling snake venom complexity with ‘omics’ approaches: Challenges and perspectives. Toxicon 2014, 87, 131–134. [Google Scholar] [CrossRef] [PubMed]
- Calvete, J.J.; Sanz, L.; Pla, D.; Lomonte, B.; Gutierrez, J.M. Omics meets biology: Application to the design and preclinical assessment of antivenoms. Toxins 2014, 6, 3388–3405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pla, D.; Petras, D.; Saviola, A.J.; Modahl, C.M.; Sanz, L.; Pérez, A.; Juárez, E.; Frietze, S.; Dorrestein, P.C.; Mackessy, S.P.; et al. Transcriptomics-guided bottom-up and top-down venomics of neonate and adult specimens of the arboreal rear-fanged Brown Treesnake, Boiga irregularis, from Guam. J. Proteom. 2018, 174, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Viala, V.L.; Hildebrand, D.; Trusch, M.; Fucase, T.M.; Sciani, J.M.; Pimenta, D.C.; Arni, R.K.; Schlüter, H.; Betzel, C.; Mirtschin, P.; et al. Venomics of the Australian eastern brown snake (Pseudonaja textilis): Detection of new venom proteins and splicing variants. Toxicon 2015, 107, 252–265. [Google Scholar] [CrossRef] [PubMed]
- Reeks, T.; Lavergne, V.; Sunagar, K.; Jones, A.; Undheim, E.; Dunstan, N.; Fry, B.; Alewood, P.F. Deep venomics of the Pseudonaja genus reveals inter- and intra-specific variation. J. Proteom. 2016, 133, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Modahl, C.M.; Frietze, S.; Mackessy, S.P. Adaptive evolution of distinct prey-specific toxin genes in rear-fanged snake venom. Proc. Biol. Sci. 2018, 285. [Google Scholar] [CrossRef]
- Chatrath, S.T.; Chapeaurouge, A.; Lin, Q.; Lim, T.K.; Dunstan, N.; Mirtschin, P.; Kumar, P.P.; Kini, R.M. Identification of novel proteins from the venom of a cryptic snake Drysdalia coronoides by a combined transcriptomics and proteomics approach. J. Proteome Res. 2011, 10, 739–750. [Google Scholar] [CrossRef] [PubMed]
- Pawlak, J.; Mackessy, S.P.; Sixberry, N.M.; Stura, E.A.; Le Du, M.H.; Menez, R.; Foo, C.S.; Menez, A.; Nirthanan, S.; Kini, R.M. Irditoxin, a novel covalently linked heterodimeric three-finger toxin with high taxon-specific neurotoxicity. FASEB J. 2009, 23, 534–545. [Google Scholar] [CrossRef] [PubMed]
- Heyborne, W.H.; Mackessy, S.P. Identification and characterization of a taxon-specific three-finger toxin from the venom of the Green Vinesnake (Oxybelis fulgidus; family Colubridae). Biochimie 2013, 95, 1923–1932. [Google Scholar] [CrossRef] [PubMed]
- James, J. The Snake Charmer: A Life and Death in Pursuit of Knowledge; Hyperion Press: New York, NY, USA, 2008; p. 288. [Google Scholar]
- Jiang, Y.; Li, Y.; Lee, W.; Xu, X.; Zhang, Y.; Zhao, R.; Zhang, Y.; Wang, W. Venom gland transcriptomes of two elapid snakes (Bungarus multicinctus and Naja atra) and evolution of toxin genes. BMC Genom. 2011, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- Shan, L.L.; Gao, J.F.; Zhang, Y.X.; Shen, S.S.; He, Y.; Wang, J.; Ma, X.M.; Ji, X. Proteomic characterization and comparison of venoms from two elapid snakes (Bungarus multicinctus and Naja atra) from China. J. Proteom. 2016, 138, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Oh, A.M.F.; Tan, C.H.; Ariaranee, G.C.; Quraishi, N.; Tan, N.H. Venomics of Bungarus caeruleus (Indian krait): Comparable venom profiles, variable immunoreactivities among specimens from Sri Lanka, India and Pakistan. J. Proteom. 2017, 164, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Das, I. A Field Guide to the Reptiles of South-East Asia; New Holland Publishers: London, UK, 2010; p. 376. [Google Scholar]
- Chang, C.C.; Lee, C.Y. Isolation of neurotoxins from the venom of Bungarus multicinctus and their modes of neuromuscular blocking action. Arch. Int. Pharmacodyn. Ther. 1963, 144, 241–257. [Google Scholar] [PubMed]
- Chu, C.C.; Li, S.H.; Chen, Y.H. Resolution of isotoxins in the beta-bungarotoxin family. J. Chromatogr. A 1995, 694, 492–497. [Google Scholar] [CrossRef]
- Su, M.J.; Chang, C.C. Presynaptic effects of snake venom toxins which have phospholipase A2 activity (beta-bungarotoxin, taipoxin, crotoxin). Toxicon 1984, 22, 631–640. [Google Scholar] [CrossRef]
- Rusmili, M.R.A.; Yee, T.T.; Mustafa, M.R.; Hodgson, W.C.; Othman, I. Proteomic characterization and comparison of Malaysian Bungarus candidus and Bungarus fasciatus venoms. J. Proteom. 2014, 110, 129–144. [Google Scholar] [CrossRef] [PubMed]
- McDowell, R.S.; Dennis, M.S.; Louie, A.; Shuster, M.; Mulkerrin, M.G.; Lazarus, R.A.; Ma, Z.Q. Mambin, a potent glycoprotein IIb-IIIa antagonist and platelet aggregation inhibitor structurally related to the short neurotoxins. Biochemistry 1992, 31, 4766–4772. [Google Scholar] [CrossRef] [PubMed]
- Chaki, S.; Muramatsu, M.; Ushiyama, Y.; Otomo, S. Purification and partial characterization of K+ channel blockers from the venom of Dendroaspis angusticeps. Neurochem. Int. 1992, 20, 553–558. [Google Scholar] [CrossRef]
- Kini, R.M.; Doley, R.S. Structure, function and evolution of three-finger toxins: Mini proteins with multiple targets. Toxicon 2010, 56, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Siang, A.S.; Doley, R.; Vonk, F.J.; Kini, R.M. Transcriptomic analysis of the venom gland of the red-headed krait (Bungarus flaviceps) using expressed sequence tags. BMC Mol. Biol. 2010, 11, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Ziganshin, R.H.; Kovalchuk, S.I.; Arapidi, G.P.; Starkov, V.; Hoang, A.N.; Thi Nguyen, T.T.; Nguyen, K.C.; Shoibonov, B.B.; Tsetlin, V.I.; Utkin, Y.N. Quantitative proteomic analysis of Vietnamese krait venoms: Neurotoxins are the major components in Bungarus multicinctus and phospholipases A2 in Bungarus fasciatus. Toxicon 2015, 107, 197–209. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, M.J.; Kato, I. Protein inhibitors of proteinases. Annu. Rev. Biochem. 1980, 49, 593–626. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, M.J. Protein inhibitors of serine proteinases—Mechanism and classification. Adv. Exp. Med. Biol. 1986, 199, 1–17. [Google Scholar] [PubMed]
- Chang, L.S.; Chung, C.; Huang, H.B.; Lin, S.R. Purification and characterization of a chymotrypsin inhibitor from the venom of Ophiophagus hannah (King Cobra). Biochem. Biophys. Res. Commun. 2001, 283, 862–867. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.C.; Yan, F.R.; Chang, L.S. Taiwan cobra chymotrypsin inhibitor: Cloning, functional expression and gene organization. Biochim. Biophys. Acta 2005, 1747, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Kini, R.M. Structure-function relationships and mechanism of anticoagulant phospholipase A2 enzymes from snake venoms. Toxicon 2005, 45, 1147–1161. [Google Scholar] [CrossRef] [PubMed]
- Sunderkotter, C.; Steinbrink, K.; Goebeler, M.; Bhardwaj, R.; Sorg, C. Macrophages and angiogenesis. J. Leukocyte Biol. 1994, 55, 410–422. [Google Scholar] [CrossRef] [PubMed]
- Verheul, H.M.W.; Hoekman, K.; Bakker, S.L.D.; Eekman, C.A.; Folman, C.C.; Broxterman, H.J.; Pinedo, H.M. Platelet: Transporter of vascular endothelial growth factor. Clin. Cancer Res. 1997, 3, 2187–2190. [Google Scholar] [PubMed]
- Itakura, J.; Ishiwata, T.; Shen, B.; Kornmann, M.; Korc, M. Concomitant over-expression of vascular endothelial growth factor and its receptors in pancreatic cancer. Int. J. Cancer 2000, 85, 27–34. [Google Scholar] [CrossRef] [Green Version]
- Yamazaki, Y.; Takani, K.; Atoda, H.; Morita, T. Snake venom vascular endothelial growth factors (VEGFs) exhibit potent activity through their specific recognition of KDR (VEGF receptor 2). J. Biol. Chem. 2003, 278, 51985–51988. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, Y.; Matsunaga, Y.; Tokunaga, Y.; Obayashi, S.; Saito, M.; Morita, T. Snake venom vascular endothelial growth factors (VEGF-Fs) exclusively vary their structures and functions among species. J. Biol. Chem. 2009, 284, 9885–9891. [Google Scholar] [CrossRef] [PubMed]
- Aird, S.D. Ophidian envenomation strategies and the role of purines. Toxicon 2002, 40, 335–393. [Google Scholar] [CrossRef]
- Margres, M.J.; Aronow, K.; Loyacano, J.; Rokyta, D.R. The venom-gland transcriptome of the eastern coral snake (Micrurus fulvius) reveals high venom complexity in the intragenomic evolution of venoms. BMC Genom. 2013, 14, 531. [Google Scholar] [CrossRef] [PubMed]
- McGivern, J.J.; Wray, K.P.; Margres, M.J.; Couch, M.E.; Mackessy, S.P.; Rokyta, D.R. RNA-seq and high-definition mass spectrometry reveal the complex and divergent venoms of two rear-fanged colubrid snakes. BMC Genom. 2014, 15, 1061. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.M.; Clemetson, J.M.; Clemetson, K.J. Snake venom C-type lectins interacting with platelet receptors. Toxin Rev. 2007, 26, 77–93. [Google Scholar] [CrossRef]
- Zha, H.G.; Lee, W.H.; Zhang, Y. Cloning of cDNAs encoding C-type lectins from Elapidae snakes Bungarus fasciatus and Bungarus multicinctus. Toxicon 2001, 39, 1887–1892. [Google Scholar] [CrossRef]
- Pung, Y.F.; Wong, P.T.; Kumar, P.P.; Hodgson, W.C.; Kini, R.M. Ohanin, a novel protein from king cobra venom, induces hypolocomotion and hyperalgesia in mice. J. Biol. Chem. 2005, 280, 13137–13147. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.W. A brief review of the scientific history of several lesser-known snake venom proteins: L-amino acid oxidases, hyaluronidases and phosphodiesterases. Toxicon 2013, 62, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Trummal, K.; Aaspollu, A.; Tonismagi, K.; Samel, M.; Subbi, J.; Siigur, J.; Siigur, E. Phosphodiesterase from Vipera lebetina venom—Structure and characterization. Biochimie 2014, 106, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Lavin, M.F.; Earl, S.; Birrell, G.; St. Pierre, L.; Guddat, L.; de Jersey, J.; Masci, P. Snake venom nerve growth factors. In Handbook of Venoms and Toxins of Reptiles; Mackessy, S.P., Ed.; CRC Press: Boca Raton, FL, USA, 2009; pp. 377–391. [Google Scholar]
- Kostiza, T.; Meier, J. Nerve growth factors from snake venoms: Chemical properties, mode of action and biological significance. Toxicon 1996, 34, 787–806. [Google Scholar] [CrossRef]
- Chen, H.S.; Wang, Y.M.; Huang, W.T.; Huang, K.F.; Tsai, I.H. Cloning, characterization and mutagenesis of Russell’s viper venom L-amino acid oxidase: Insights into its catalytic mechanism. Biochimie 2012, 94, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Zuliani, J.P.; Kayano, A.M.; Zaqueo, K.D.; Neto, A.C.; Sampaio, S.V.; Soares, A.M.; Stabeli, R.G. Snake venom L-amino acid oxidases: Some consideration about their functional characterization. Protein Pept. Lett. 2009, 16, 908–912. [Google Scholar] [CrossRef] [PubMed]
- Mackessy, S.P.; Baxter, L.M. Bioweapons synthesis and storage: The venom gland of front-fanged snakes. Zool. Anz. 2006, 245, 147–159. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Brown, R.L.; Morita, T. Purification and cloning of toxins from elapid venoms that target cyclic nucleotide-gated ion channels. Biochemistry 2002, 41, 11331–11337. [Google Scholar] [CrossRef] [PubMed]
- Chapeaurouge, A.; Abu Reza, M.D.; Mackessy, S.P.; Carvalho, P.C.; Valente, R.H.; Teixeira-Ferreira, A.; Jonas Perales, J.; Lin, Q.; Kini, R.M. Interrogating the venom of the viperid snake Sistrurus catenatus edwardsii by a combined approach of electrospray and MALDI mass spectrometry. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Markland, F.S.; Swenson, S. Snake venom metalloproteinases. Toxicon 2013, 62, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.W.; Serrano, S.M. Insights into and speculations about snake venom metalloproteinase (SVMP) synthesis, folding and disulfide bond formation and their contribution to venom complexity. FEBS J. 2008, 275, 3016–3030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doley, R.; Mackessy, S.P.; Kini, R.M. Role of accelerated segment switch in exons to alter targeting (ASSET) in the molecular evolution of snake venom proteins. BMC Evol. Biol. 2009, 9, 146. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xiong, Y.L.; Bon, C. An activator of blood-coagulation factor-X from the venom of Bungarus fasciatus. Toxicon 1995, 33, 1277–1288. [Google Scholar] [CrossRef]
- Johns, D.G.; Ao, Z.H.; Heidrich, B.J.; Hunsberger, G.E.; Graham, T.; Payne, L.; Elshourbagy, N.; Lu, Q.; Aiyar, N.; Douglas, S.A. Dendroaspis natriuretic peptide binds to the natriuretic peptide clearance receptor. Biochem. Biophys. Res. Commun. 2007, 358, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Trinh, K.X.; Khac, Q.L.; Trinh, L.X.; Warren, D.A. Hyponatraemia, rhabdomyolysis, alterations in blood pressure and persistent mydriasis in patients envenomed by Malayan kraits (Bungarus candidus) in southern Viet Nam. Toxicon 2010, 56, 1070–1075. [Google Scholar] [CrossRef] [PubMed]
- Sridharan, S.; Kini, R.M. Snake venom natriuretic peptides: Potential molecular probes. BMC Pharmacol. Toxicol. 2015, 16. [Google Scholar] [CrossRef] [Green Version]
- Vink, S.; Jin, A.H.; Poth, K.J.; Head, G.A.; Alewood, P.F. Natriuretic peptide drug leads from snake venom. Toxicon 2012, 59, 434–445. [Google Scholar] [CrossRef] [PubMed]
- Rehana, S.; Kini, R.M. Molecular isoforms of cobra venom factor-like proteins in the venom of Austrelaps superbus. Toxicon 2007, 50, 32–52. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.W.; Bredehorst, R.; Fritzinger, D.C.; Grunwald, T.; Ziegelmuller, P.; Kock, M.A. Structure and function of cobra venom factor, the complement-activating protein in cobra venom. Adv. Exp. Med. Biol. 1996, 391, 97–114. [Google Scholar] [PubMed]
- Guan, H.H.; Goh, K.S.; Davamani, F.; Wu, P.L.; Huang, Y.W.; Jeyakanthan, J.; Wu, W.; Chen, C.J. Structures of two elapid snake venom metalloproteases with distinct activities highlight the disulfide patterns in the D domain of ADAMalysin family proteins. J. Struct. Biol. 2010, 169, 294–303. [Google Scholar] [CrossRef] [PubMed]
- Pla, D.; Sanz, L.; Sasa, M.; Acevedo, M.E.; Dwyer, Q.; Durban, J.; Pérez, A.; Rodriguez, Y.; Lomonte, B.; Calvete, J.J. Proteomic analysis of venom variability and ontogeny across the arboreal palm-pitvipers (genus Bothriechis). J. Proteom. 2017, 152, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Leprevost, F.V.; Valente, R.H.; Lima, D.B.; Perales, J.; Melani, R.; Yates, J.R., III; Barbosa, V.C.; Junqueira, M.; Carvalho, P.C. PepExplorer: A similarity-driven tool for analyzing de novo sequencing results. Mol. Cell. Proteom. 2014, 13, 2480–2489. [Google Scholar] [CrossRef] [PubMed]
- Florens, L.; Carozza, M.J.; Swanson, S.K.; Fournier, M.; Coleman, M.K.; Workman, J.L.; Washburn, M.P. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods 2006, 40, 303–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
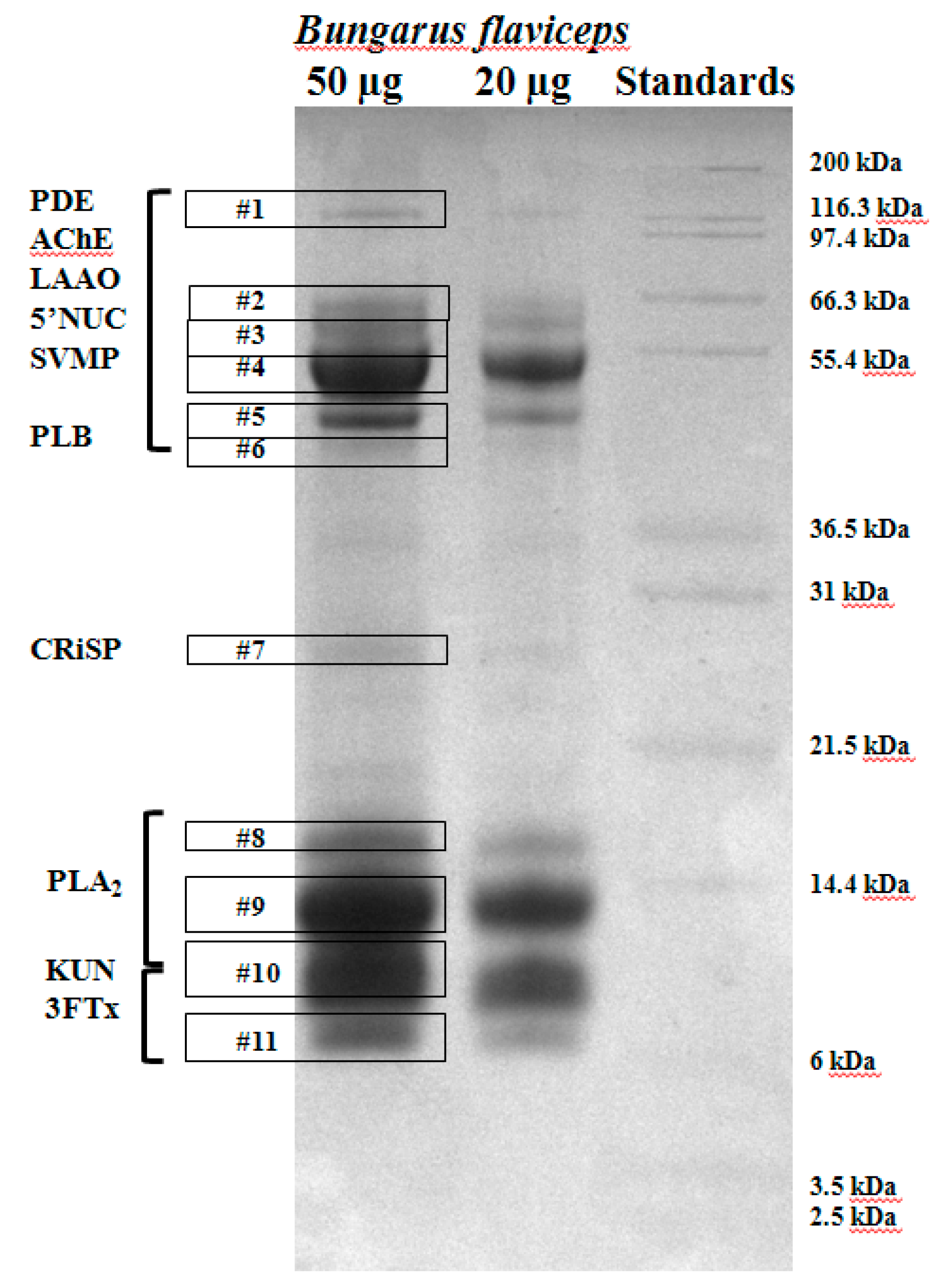
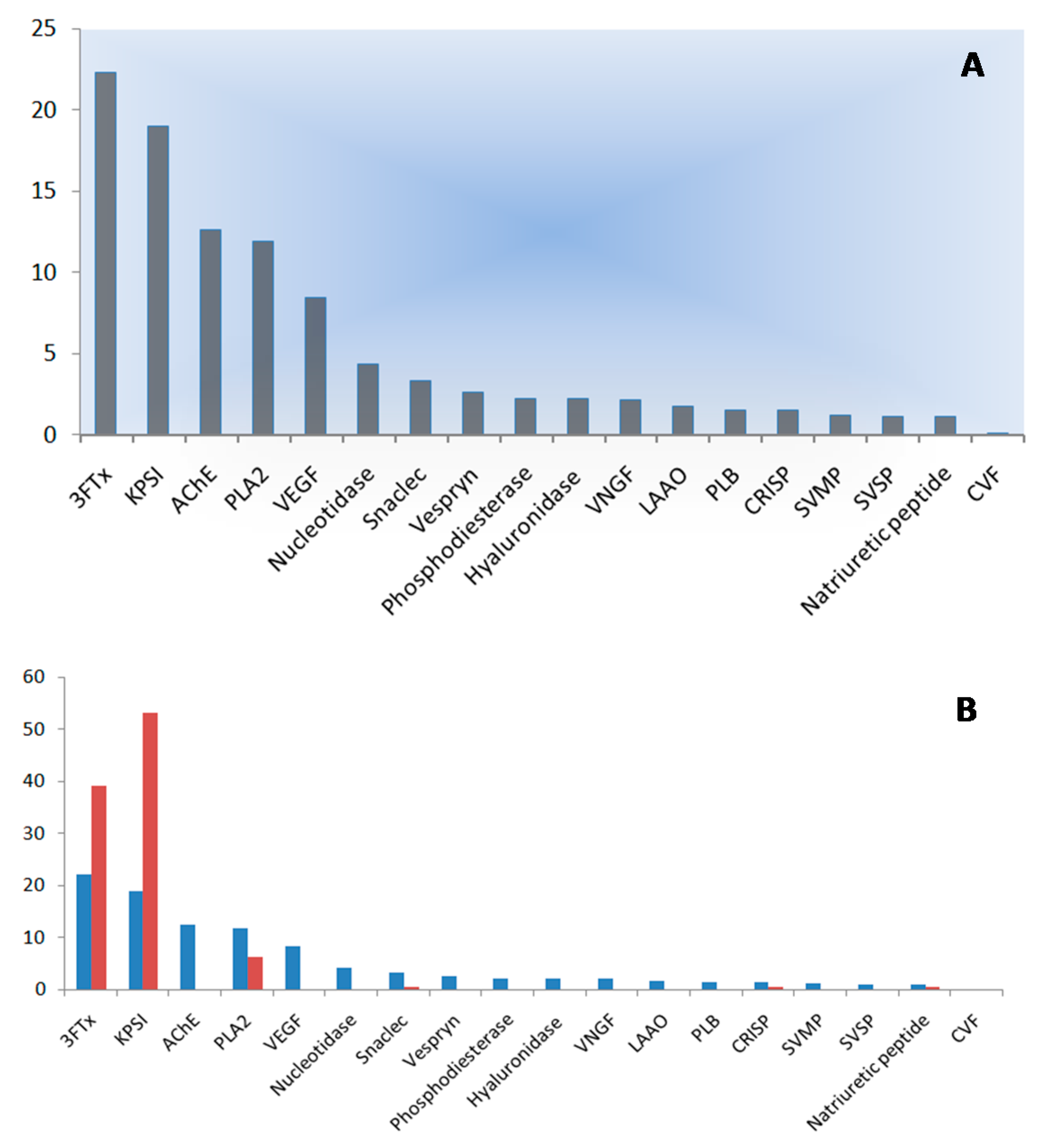
| Protein Family | Protein | Accession No. | Species | Number of Peptides Matched |
|---|---|---|---|---|
| 3FTx | Non-conventional three finger toxin isoform 1 | 294961050 | Bungarus flaviceps | 6 |
| 3FTx | Non-conventional three finger toxin isoform 6 | 294961060 | Bungarus flaviceps | 5 |
| 3FTx | Short-chain three finger toxin isoform 4 | 294961042 | Bungarus flaviceps | 4 |
| 3FTx | Short-chain three finger toxin isoform 7 | 294961048 | Bungarus flaviceps | 9 |
| 3FTx | Short-chain three finger toxin isoform 6 | 294961046 | Bungarus flaviceps | 7 |
| 3FTx | Short-chain three finger toxin isoform 1 | 294961036 | Bungarus flaviceps | 2 |
| 3FTx | κ-bungarotoxin | 809178 | Bungarus multicinctus | 1 |
| 3FTx | Short-chain three finger toxin isoform 3 | 294961040 | Bungarus flaviceps | 1 |
| 3FTx | κ-flavitoxin | 128938 | Bungarus flaviceps | 9 |
| 3FTx | Muscarinic toxin-like protein | 294961066 | Bungarus flaviceps | 9 |
| Serine protease inhibitor | β-bungarotoxin B chain precursor | 31745053 | Bungarus flaviceps | 7 |
| Serine protease inhibitor | Kunitz-type serine proteinase inhibitor isoform 5 | 294961076 | Bungarus flaviceps | 8 |
| Serine protease inhibitor | Kunitz-type serine proteinase inhibitor isoform 1 | 294961068 | Bungarus flaviceps | 4 |
| Acetylcholinesterase | Acetylcholinesterase | 1389604 | Bungarus flaviceps | 20 |
| Acetylcholinesterase | Acetylcholinesterase DEN | 476538388 | Denisoni adevisi | 4 |
| PLA2 | β-bungarotoxin A2 chain precursor | 31745049 | Bungarus flaviceps | 16 |
| PLA2 | β-bungarotoxin A1 chain precursor | 31745051 | Bungarus flaviceps | 3 |
| PLA2 | Phospholipase A2II precursor | 31745057 | Bungarus flaviceps | 21 |
| PLA2 | Phospholipase A2 | 263083 | Bungarus fasciatus | 8 |
| PLA2 | Phospholipase A2 precursor | 31745055 | Bungarus flaviceps | 23 |
| PLA2 | Phospholipase A2 isoform 3 | 294961092 | Bungarus flaviceps | 5 |
| PLA2 | Phospholipase A2Kbf-III | 110559306 | Bungarus fasciatus | 4 |
| PLA2 | Phospholipase A2 isozyme 1 | 24638470 | Laticauda semifasciata | 1 |
| PLA2 | Phospholipase A2 | 29422777 | Bungarus candidus | 2 |
| PLA2 | Phospholipase A2 | 5924345 | Austrelaps superbus | 3 |
| PLA2 | Phospholipase A2 | 152032644 | Bungarus fasciatus | 3 |
| PLA2 | Phospholipase A2 precursor | 156257593 | Bungarus fasciatus | 1 |
| PLA2 | Phospholipase A2 | 48425218 | Bungarus caeruleus | 1 |
| PLA2 | Phospholipase A2 | 129428 | Laticauda colubrina | 1 |
| VEGF | Hypothetical protein L345_04144 | 565318860 | Ophiophagus hannah | 4 |
| Nucleotidase | Ecto-5′-nucleotidase 1 | 537444870 | Micrurus fulvius | 11 |
| Snaclec | Snaclec factor IX/factor X-binding protein B chain | 398488 | Bothrops jararaca | 1 |
| Snaclec | C-type lectin-like protein 1 | 13876735 | Bungarus fasciatus | 3 |
| Vespryn | Ohanin precursor | 70907886 | Ophiophagus hannah | 2 |
| Vespryn | Vespryn22 | 336042222 | Drysdalia coronoides | 1 |
| Phosphodiesterase | Phosphodiesterase 1 | 537444868 | Micrurus fulvius | 20 |
| Phosphodiesterase | Phosphodiesterase 1 | 338855302 | Crotalus adamanteus | 1 |
| Hyaluronidase | Hyaluronidase | 113203681 | Bitis arietans | 4 |
| VNGF | Venom nerve growth factor precursor | 266299 | Bungarus multicinctus | 4 |
| LAAO | L-amino-acid oxidase | 126035653 | Bungarus fasciatus | 5 |
| LAAO | L-amino acid oxidase | 126035649 | Bungarus multicinctus | 2 |
| LAAO | L-amino-acid oxidase | 426205815 | Crotalus durissus cumanensis | 2 |
| PLB | Phospholipase B | 537444729 | Micrurus fulvius | 3 |
| CRISP | Cysteine-rich seceretory protein | 190195343 | Bungarus candidus | 3 |
| CRISP | Opharin precursor | 225547744 | Ophiophagus hannah | 2 |
| SVMP | Scutatease-1 (PIII) | 145982766 | Notechis scutatus | 8 |
| SVMP | Metalloproteinase (PIII) | 126035640 | Bungarus multicinctus | 6 |
| SVMP | Metalloproteinase MTP9 (PIII) | 336042214 | Drysdalia coronoides | 4 |
| SVMP | Metalloproteinase (PIII) | 126035635 | Bungarus fasciatus | 4 |
| SVMP | P-III | 633276509 | Micropechis ikaheka | 4 |
| SVMP | MTP4 (PIII) | 537463069 | Micrurus fulvius | 3 |
| SVMP | Atragin precursor(PIII) | 224482347 | Naja atra | 3 |
| SVMP | Metalloproteinase isoform 3 (PIII) | 109254964 | Sistrurus catenatus edwardsi | 2 |
| SVMP | SVMP-Hop-14, partial (PIII) | 476539284 | Hoplocephalus bungaroides | 2 |
| SVMP | SVMP-Hop-46, partial(PIII) | 476539268 | Hoplocephalus bungaroides | 2 |
| SVMP | SVMP 1 | 537444726 | Micrurus fulvius | 2 |
| SVMP | Metalloproteinase (PII) | 82466485 | Bothrops asper | 1 |
| SVMP | Fur-1, partial (PI) | 476538467 | Furinaor nata | 1 |
| SVMP | jararhagin (PIII) | 62468 | Bothrops jararaca | 1 |
| SVMP | Metalloproteinase (PIII) | 241995585 | Philodrya solfersii | 1 |
| SVMP | Leucurolysin-B (PIII) | 223635807 | Bothrops leucurus | 1 |
| SVMP | Ech-32 (PIII) | 476538400 | Echiopsis curta | 1 |
| SVMP | Cobrin precursor(PIII) | 6006966 | Naja naja | 1 |
| SVMP | Metalloproteinase (PII) | 297594122 | Echis pyramidum leakeyi | 1 |
| SVMP | CohPH-3 (PII) | 522802426 | Crotalus oreganus helleri | 1 |
| SVSP | Serine proteinase isoform 2 | 109254940 | Sistrurus catenatus edwardsi | 2 |
| SVSP | SVSP 11 | 387014258 | Crotalus adamanteus | 1 |
| Natriuretic peptide | Natriuretic peptide | 294961100 | Bungarus flaviceps | 1 |
| Complement-depleting factor | Complement-depleting factor | 126035660 | Bungarus fasciatus | 1 |
| Protein Family | Protein | Accession No. | Species | Database Sequence | Alignment Score | De Novo Sequence | ALC (%) | m/z | ppm to De Novo Derived Sequence |
|---|---|---|---|---|---|---|---|---|---|
| 3FTx | Muscarinic toxin-like protein | 117606606 | Ophiophagus hannah | TFTCPELTPN | 64 | CGTYTCPELTPDRR | 52 | 575.9354 | 13.5 |
| PLA2 | Phospholipase A2 | 9453880 | Laticauda semifasciata | VHDDCYGEAEK | 87 | VHDDCYGEAEK | 58 | 563.5938 | −1.8 |
| LVQFTYLIQCANKGSRASYHYADYGCY | 84 | RLAQFACLLQCADEEVGVDLHYADYGCYNCK | 46 | 906.911 | 5.8 | ||||
| Nucleotidase | Snake venom 5′-nucleotidase | 752779244 | Boiga irregularis | VVQFMNSLR | 69 | EAAAVVQFMNSLR | 51 | 479.2537 | 7.1 |
| HGQGTGELLQVSGIKVVYDLS | 99 | HPQGTGELLQVSLGKVVYDLSKGAPVNK | 52 | 587.7292 | 4.2 | ||||
| Snaclec | C-type lectin 2 | 537444936 | Micrurus fulvius | DHHCPSDWYSFDKFCYKFI | 105 | DNHCNADWSSFDKFCYQFLR | 52 | 653.2799 | 2 |
| Vespryn | Vespryn 23 | 336042224 | Drysdalia coronoides | HFFEVK | 49 | HFFEVK | 60 | 403.7129 | −1.1 |
| IVVFLDYSEGK | 68 | LVVFLDYKEGK | 60 | 437.5828 | −1.2 | ||||
| IVVFLDY | 51 | LVVFLDYK | 71 | 498.791 | −1.7 | ||||
| Phosphodiesterase | Phosphodiesterase | 675402318 | Protobothrops elegans | FYTLYIEEPDTTGHK | 106 | QFLPVFFTLYLEEPDTTGHK | 57 | 592.0533 | 7 |
| RPDFYTLYIEEPDTTGHK | 130 | RPDFFTLYLEEPDTTGHK | 59 | 542.2665 | −2.6 | ||||
| VNGF | Venom nerve growth factor | 335892638 | Bungarus fasciatus | YFFETK | 51 | AGYFFETK | 51 | 481.7344 | −0.2 |
| LAAO | L-amino acid oxidase | 126035653 | Bungarus fasciatus | SYVTADYVIVCAT | 76 | LTSYVTADYLLVSATMGR | 51 | 981.0001 | −6.1 |
| TADYVIVCATSR | 80 | TLSAAHTADYLLVCATSR | 52 | 650.6723 | 14.1 | ||||
| LAAO | L-amino acid oxidase | 401021343 | Lachesis muta | LSAAYVLAEAGHQVTVLEASER | 105 | LSAAYVLSEKHQVTVLQASER | 52 | 583.3127 | −2.5 |
| LAAO | L-amino acid oxidase | 537444909 | Micrurus fulvius | SAAYVLAEAGHKVTLLE | 103 | QLLVVQGMQDSAAYVLSEAGHKVTVLESNNK | 50 | 669.7629 | 20.9 |
| AGMAGLSAAYVLAEAGHK | 116 | VKHHSVPPRVAGMAGLSAAYVLSEAGHK | 52 | 574.7169 | 10.5 | ||||
| LAAO | L-amino acid oxidase | 3426324 | Crotalus adamanteus | RVVIVGAGMAGLSAAYVL | 104 | RLALVGAGMAGLSAAYVLNPSSPGVLSQLLWSR | 53 | 671.7708 | −3.1 |
| LAAO | L-amino acid oxidase 1a | 537444909 | Micrurus fulvius | GMAGLSAAYVLAEAGHK | 110 | RTPVVKGMAGLSAAYVLSEAGHK | 48 | 590.3197 | −1.2 |
| Phospholipase B | Phospholipase B | 537444729 | Micrurus fulvius | KGYWPSYNIPFHK | 94 | KPYWPSYNLPFHK | 56 | 559.6232 | −1.9 |
| CRISP | CRISP precursor | 158262802 | Austrelaps superbus | VDKHNALR | 58 | VAAVDKHNALR | 45 | 398.5624 | −2 |
| YLYVCQYCPAGNIRGSIATPYK | 84 | YLYVCQYCFRAFGAPYNNKHGSLATPYK | 43 | 847.8989 | −8.6 | ||||
| CRISP | Cysteine-rich secretory protein A | 698375481 | Opheodrys aestivus | WNSNAAQNAK | 76 | EMWNSNAAQNAK | 44 | 682.3055 | −1.5 |
| PAGNIVGSIATPYK | 95 | MENTQAAVCSDPAGNLVGSLATPYK | 44 | 846.4114 | 7.4 | ||||
| PAGNIVGSIATPYK | 95 | LYYCMNEGQMAPAGNLVGSLATPYK | 44 | 897.7457 | −14.4 | ||||
| ASCFCH | 50 | HCASCFCHGGK | 48 | 632.2381 | −10.1 | ||||
| SVSP | Venom thrombin-like enzyme | 118500915 | Deinagkistrodon acutus | DECDINEHR | 72 | VVNDECDLNEHR | 51 | 500.5556 | −1.3 |
| SVMP | Metalloproteinase 13b | 699236656 | Hypsiglena sp. JMG-2014 | DPDNGMVEPGTK | 82 | LNMMSSSPNACADPDEGMVEPGTK | 50 | 846.6896 | −2.9 |
| SVMP | Metalloproteinase 4a | 752782853 | Boiga irregularis | YLVVDNR | 53 | LYLVVDNR | 72 | 496.282 | −0.4 |
| AYEMVNILNVIFR | 81 | AYEMVNLLNTMFR | 61 | 801.3932 | −1.5 | ||||
| YLVVDNR | 53 | TNTARCLTLYLKLKYQPPAYLVVDNR | 51 | 622.9447 | 4 | ||||
| SPDYGMVEPGTK | 64 | DYPGHHACALEDDQLLMCSEDYGMVEPASK | 51 | 850.1182 | 11.6 | ||||
| SVMP | Metalloproteinase | 172046653 | Naja mossambica | TSMVAITMAHQMGHNLGMNDDR | 122 | NTDMVAGTMAHQMGHNLGLNDHR | 52 | 841.0485 | 7.2 |
| AHQMGHNLGMNDDR | 81 | EFVRQAFANALYPPWWMPKYMVPQKCKAHEMGHNLGLNDHR | 50 | 986.0939 | 12.5 | ||||
| SVMP | Metalloproteinase (type III) 2a | 752782867 | Boiga irregularis | TEGMVEPGTK | 75 | QTEGMVEPGTK | 60 | 580.2688 | 0.3 |
| TESFVASTMAHELGHNLGINHDR | 136 | TNPFVGSTMAHEMGHNLGLNHDR | 60 | 634.5504 | 7.2 | ||||
| SVMP | Atrase B, partial | 289655973 | Naja atra | FGEWRETVLLPR | 92 | AFGEWRETVLLPR | 63 | 525.2878 | 0.2 |
| SVMP | Metalloproteinase 11a | 699236668 | Hypsiglena sp. JMG-2014 | QPCQNNQGYCYNGK | 94 | GPGLQPCGNNQGYCYDGK | 44 | 964.4064 | −0.3 |
| KYIEFYIVVDH | 78 | SMTKPCLMRVCDLVKYLQFYLVVDHVKSYVLCYR | 48 | 823.6322 | 11.2 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chapeaurouge, A.; Silva, A.; Carvalho, P.; McCleary, R.J.R.; Modahl, C.M.; Perales, J.; Kini, R.M.; Mackessy, S.P. Proteomic Deep Mining the Venom of the Red-Headed Krait, Bungarus flaviceps. Toxins 2018, 10, 373. https://doi.org/10.3390/toxins10090373
Chapeaurouge A, Silva A, Carvalho P, McCleary RJR, Modahl CM, Perales J, Kini RM, Mackessy SP. Proteomic Deep Mining the Venom of the Red-Headed Krait, Bungarus flaviceps. Toxins. 2018; 10(9):373. https://doi.org/10.3390/toxins10090373
Chicago/Turabian StyleChapeaurouge, Alex, Andreza Silva, Paulo Carvalho, Ryan J. R. McCleary, Cassandra Marie Modahl, Jonas Perales, R. Manjunatha Kini, and Stephen P. Mackessy. 2018. "Proteomic Deep Mining the Venom of the Red-Headed Krait, Bungarus flaviceps" Toxins 10, no. 9: 373. https://doi.org/10.3390/toxins10090373