Transcriptomic and Proteomic Analyses Reveal the Diversity of Venom Components from the Vaejovid Scorpion Serradigitus gertschi
Abstract
:1. Introduction
2. Results and Discussion
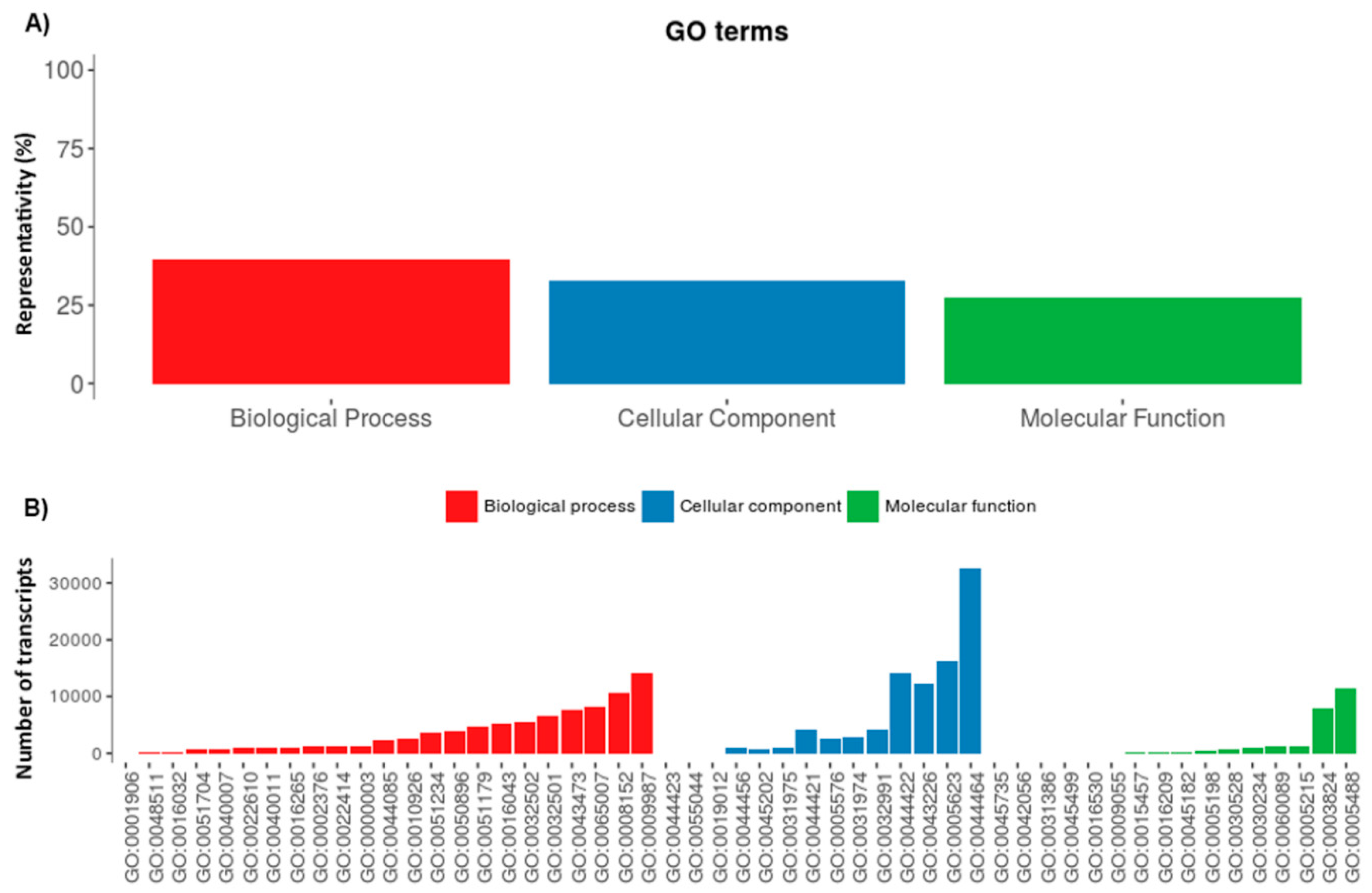
2.1. Serradigitus Gertschi Venom Gland Global Transcriptome Analysis
2.2. The Repertoire of Venom-Specific Transcripts in S. gertschi
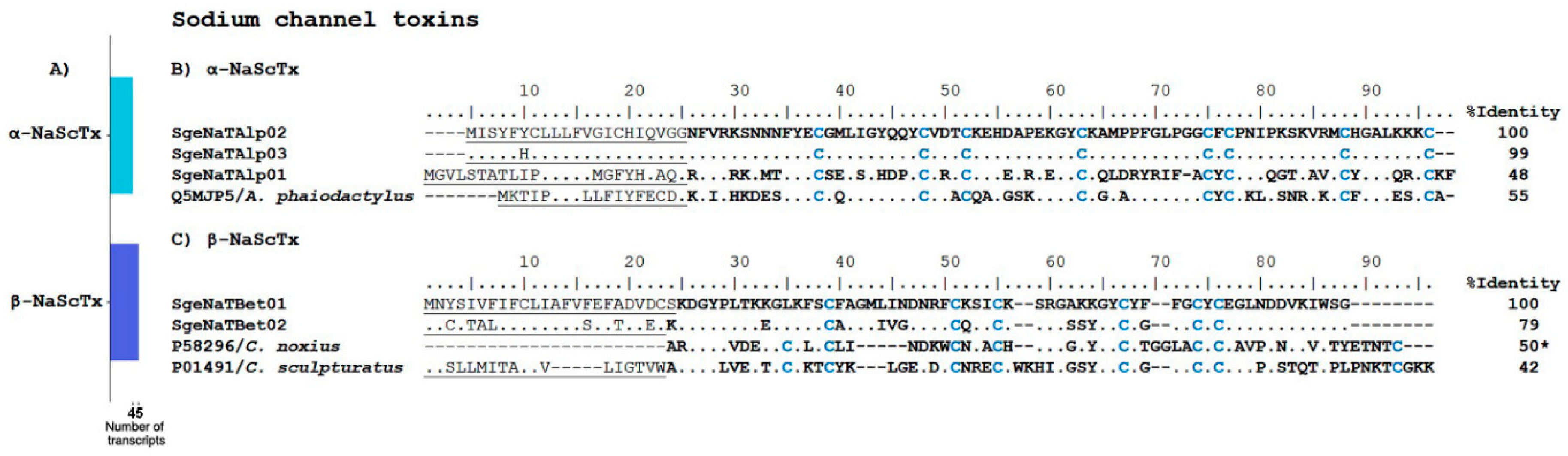
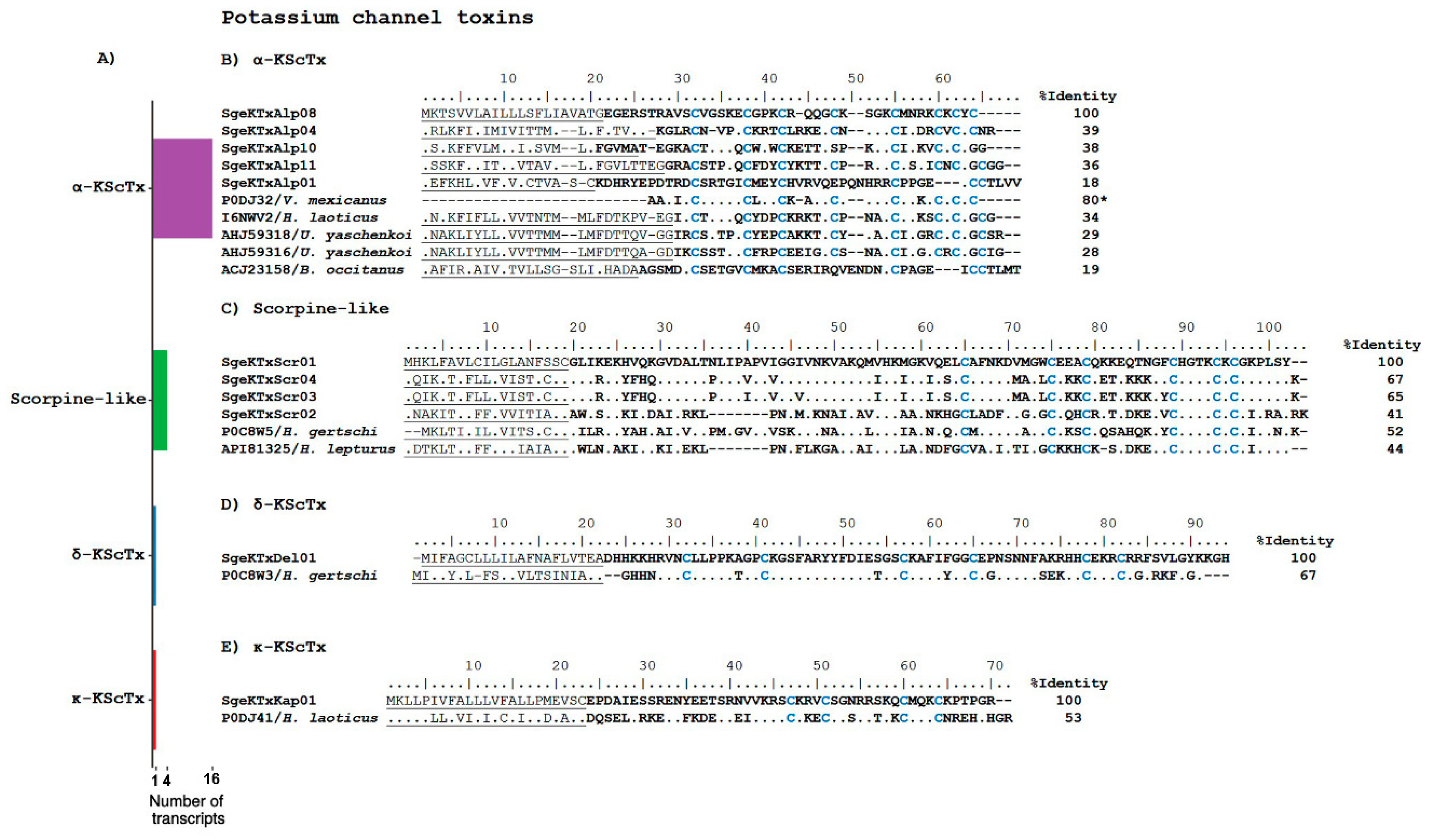
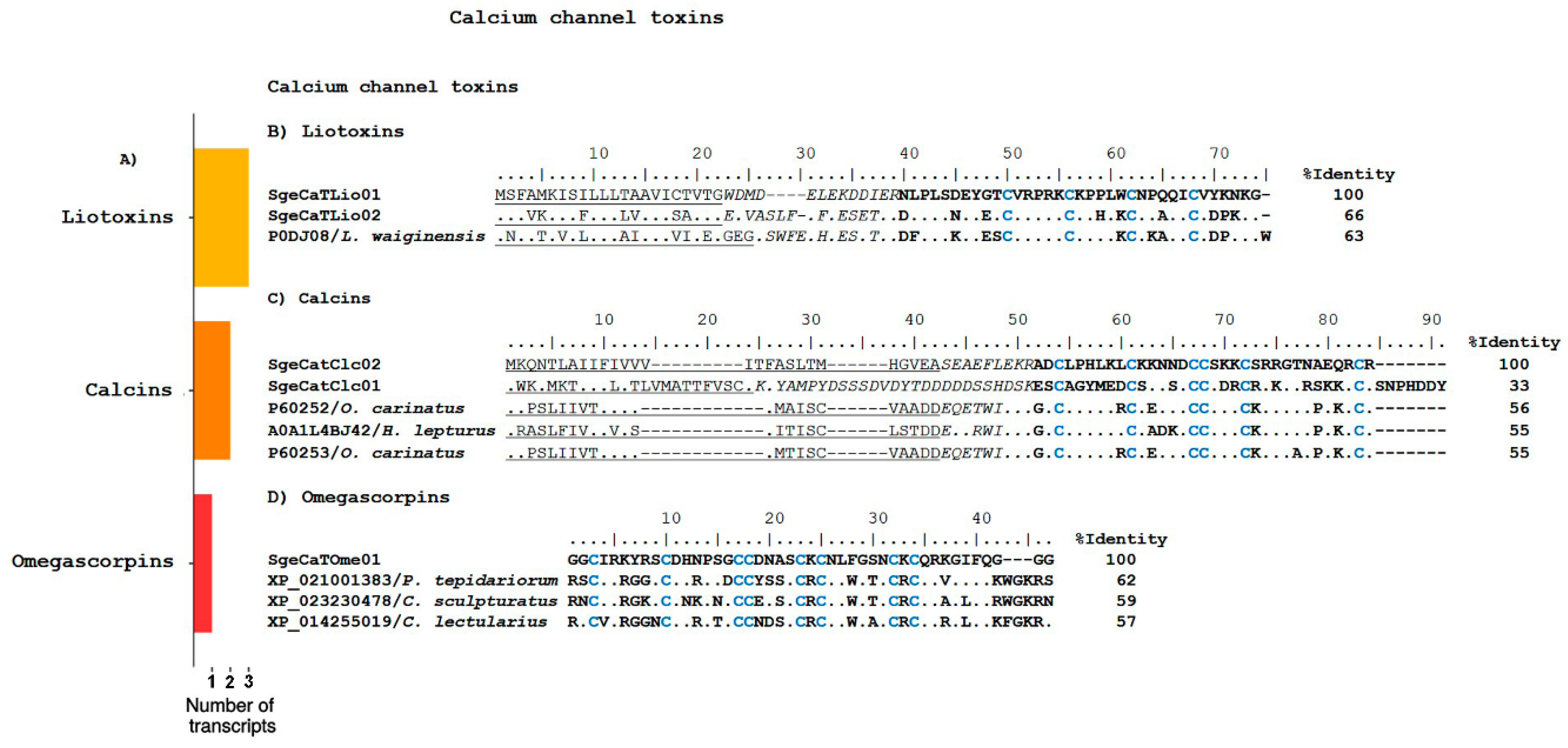
2.2.1. Ion Channel Toxins
Sodium Channel Toxins
Potassium Channel Toxins
Calcium Channel Toxins
Omegascorpins
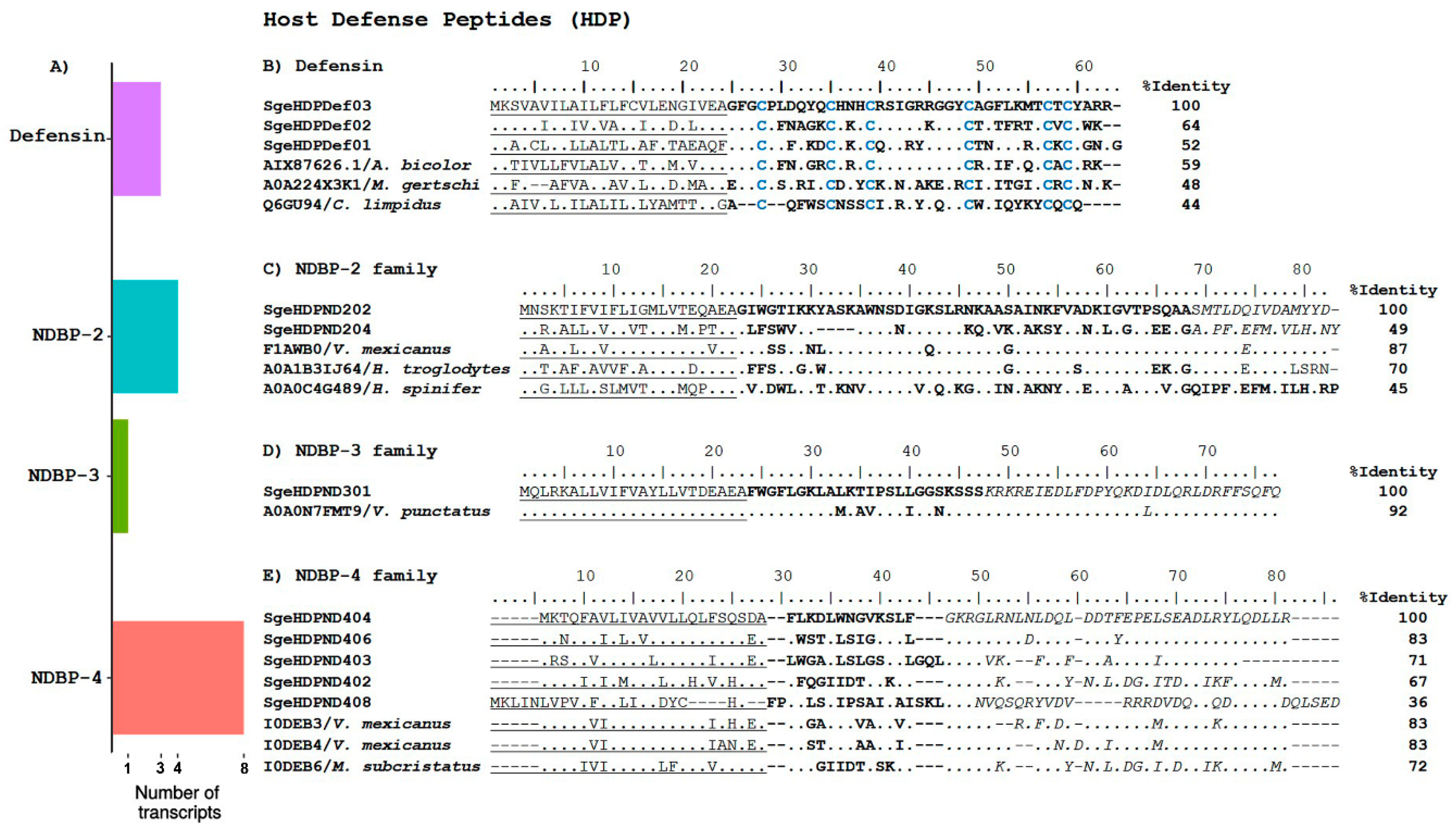
2.2.2. Host Defense Peptides
Non-Disulfide Bridged Peptides
2.2.3. Enzymes
2.2.4. Protease Inhibitors
2.2.5. Other Venom Components
La1-Like Peptides
CAP Superfamily
IGFBP Family
2.3. Proteomic Analyses
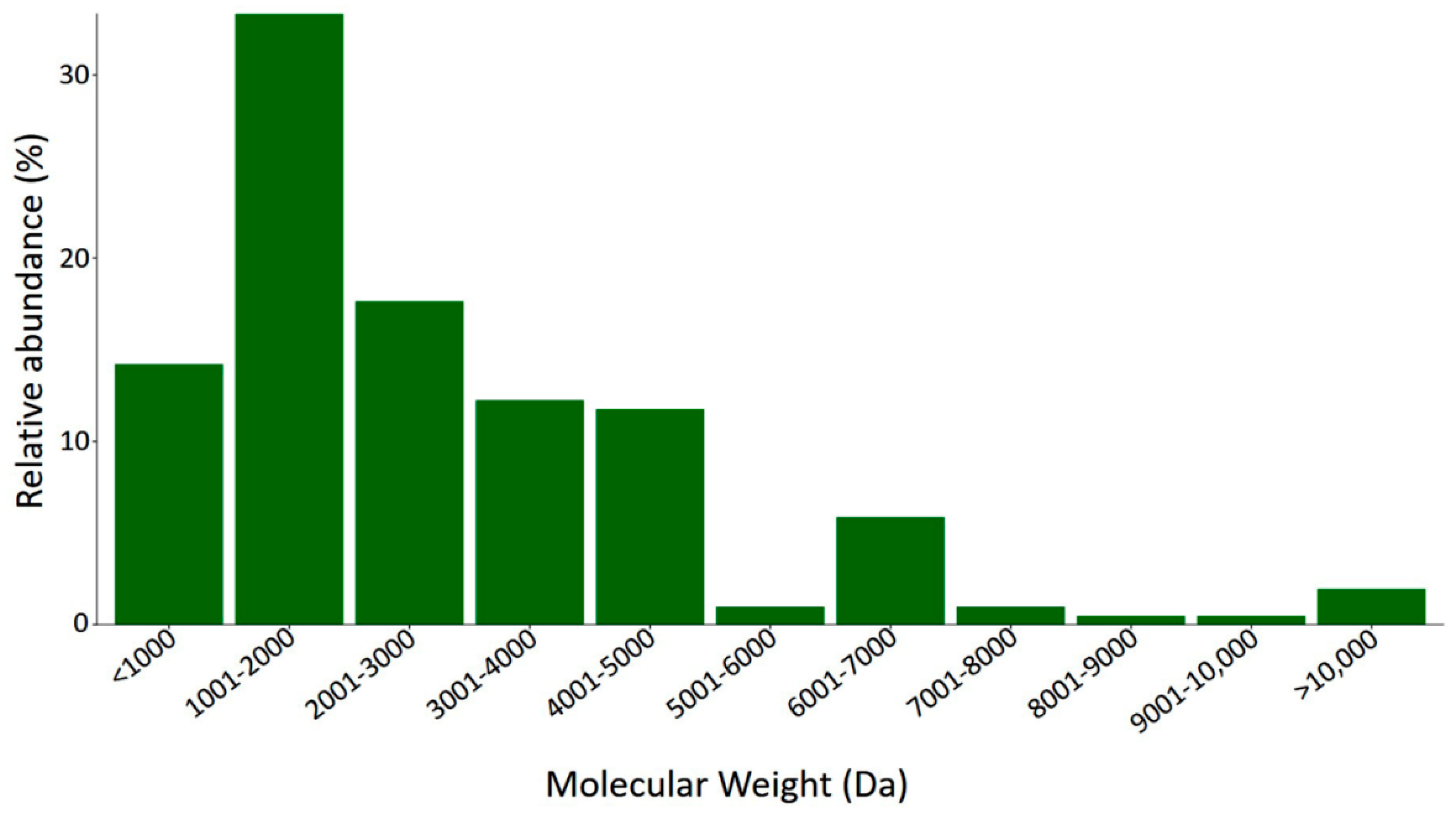
2.3.1. Mass Fingerprint Results and MS Data Analysis
2.3.2. LC-MS/MS Analysis of the Digested Venom of S. gertschi
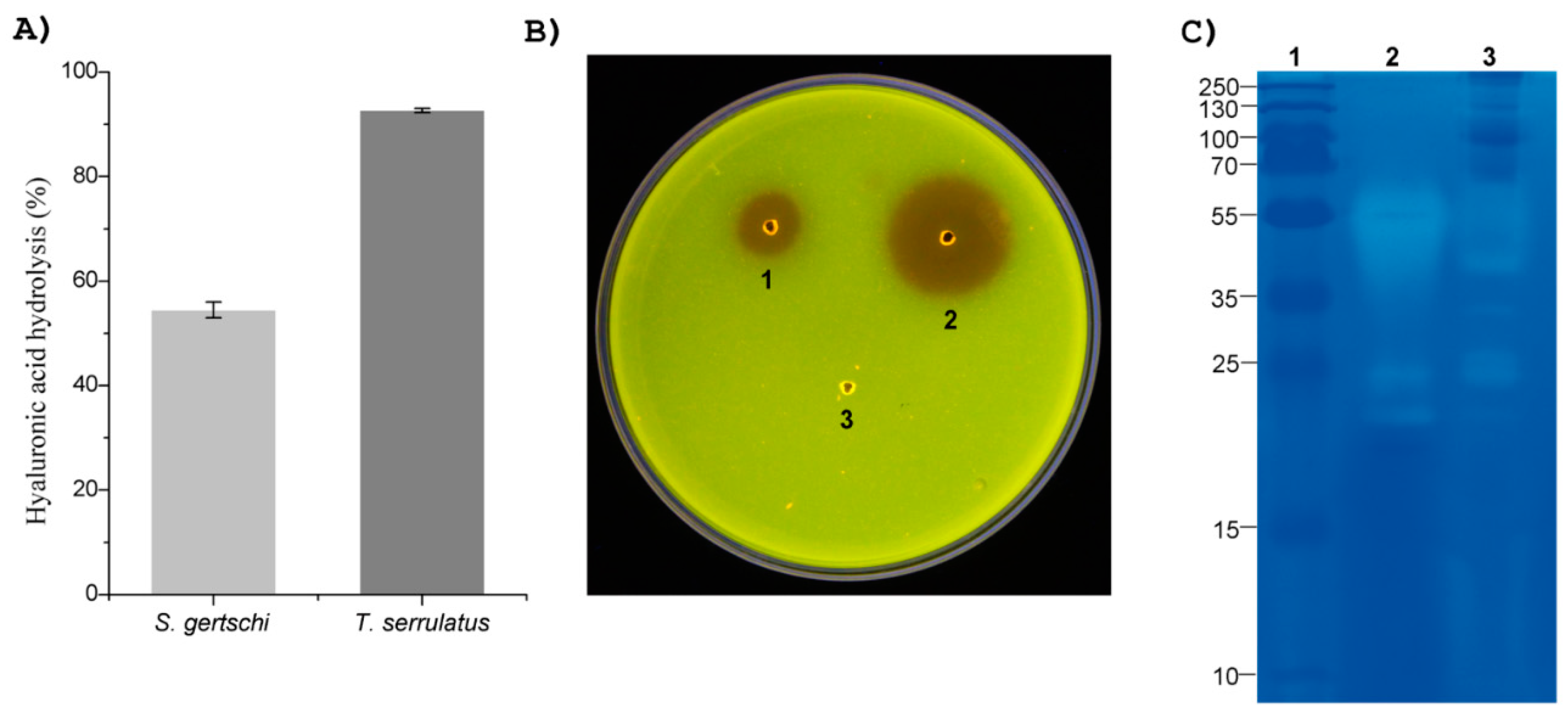
2.4. Venom Enzymatic Activities
3. Conclusions
4. Materials and Methods
4.1. Biological Material
4.2. RNA Extraction, RNA-Seq and Venom-Gland Transcriptome Assembly
4.3. Transcript Nomenclature
4.4. Bioinformatics
4.5. Molecular Mass Determination of the Venom Components
4.6. Identification of Proteome by Tryptic Digestion and LC-MS/MS Analysis
4.7. Venom Enzymatic Activities
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- González-Santillán, E.; Prendini, L. Redefinition and generic revision of the north american Vaejovid scorpion subfamily Syntropinae Kraepelin 1905, with descriptions of six new genera. Bull. Am. Museum Nat. Hist. 2013, 382, 1–71. [Google Scholar] [CrossRef]
- Santibáñez-López, C.E.; Francke, O.F.; Ureta, C.; Possani, L. Scorpions from Mexico: From species diversity to venom complexity. Toxins 2015, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Prendini, L. Substratum specialization and speciation in southern African scorpions: The Effect Hypothesis revisited. Scorpions 2001, 2001, 113–138. [Google Scholar]
- Hernández-Aponte, C.A.; Silva-Sanchez, J.; Quintero-Hernández, V.; Rodríguez-Romero, A.; Balderas, C.; Possani, L.D.; Gurrola, G.B. Vejovine, a new antibiotic from the scorpion venom of Vaejovis mexicanus. Toxicon 2011, 57, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Gurrola, G.B.; Hernandez-Lopez, R.A.; Rodriguez de la Vega, R.C.; Varga, Z.; Batista, C.V.F.; Salas-Castillo, S.P.; Panyi, G.; del Rio-Portilla, F.; Possani, L.D. Structure, function, and chemical synthesis of Vaejovis mexicanus peptide 24: A novel potent blocker of Kv1.3 potassium channels of human T lymphocytes. Biochemistry 2012, 51, 4049–4061. [Google Scholar] [CrossRef] [PubMed]
- Varga, Z.; Gurrola-Briones, G.; Papp, F.; Rodriguez de la Vega, R.C.; Pedraza-Alva, G.; Tajhya, R.B.; Gaspar, R.; Cardenas, L.; Rosenstein, Y.; Beeton, C.; et al. Vm24, a natural immunosuppressive peptide, potently and selectively blocks Kv1.3 potassium channels of human T cells. Mol. Pharmacol. 2012, 82, 372–382. [Google Scholar] [CrossRef] [PubMed]
- Quintero-Hernández, V.; Ramírez-Carreto, S.; Romero-Gutiérrez, M.T.; Valdez-Velázquez, L.L.; Becerril, B.; Possani, L.D.; Ortiz, E. Transcriptome analysis of scorpion species belonging to the Vaejovis genus. PLoS ONE 2015, 10, e0117188. [Google Scholar] [CrossRef] [PubMed]
- Romero-Gutierrez, T.; Peguero-Sanchez, E.; Cevallos, M.A.; Batista, C.V.F.; Ortiz, E.; Possani, L.D. A deeper examination of Thorellius atrox scorpion venom components with omic techonologies. Toxins 2017, 9, 399. [Google Scholar] [CrossRef] [PubMed]
- Cid-Uribe, J.I.; Santibáñez-López, C.E.; Meneses, E.P.; Batista, C.V.F.; Jiménez-Vargas, J.M.; Ortiz, E.; Possani, L.D. The diversity of venom components of the scorpion species Paravaejovis schwenkmeyeri (Scorpiones: Vaejovidae) revealed by transcriptome and proteome analyses. Toxicon 2018, 151, 47–62. [Google Scholar] [CrossRef] [PubMed]
- González-Santillán, E.; Prendini, L. Phylogeny of the north american vaejovid scorpion subfamily Syntropinae Kraepelin, 1905, based on morphology, mitochondrial and nuclear DNA. Cladistics 2015, 31, 341–405. [Google Scholar] [CrossRef]
- Williams, S.C. Scorpions of Baja California, Mexico, and adjacent islands. Occas. Pap. Calif. Acad. Sci. 1980, 135, 1–127. [Google Scholar]
- Williams, S. Scorpion bionomics. Ann. Rev. Entomol. 1987, 32, 275–295. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Shi, W.; Zeng, X.C.; Ge, F.; Yang, M.; Nie, Y.; Bao, A.; Wu, S.; Guoji, E. Unique diversity of the venom peptides from the scorpion Androctonus bicolor revealed by transcriptomic and proteomic analysis. J. Proteom. 2015, 128, 231–250. [Google Scholar] [CrossRef] [PubMed]
- Rokyta, D.; Ward, M. Venom-gland transcriptomics and venom proteomics of the black-back scorpion (Hadrurus spadix) reveal detectability challenges and an unexplored realm of animal toxin diversity. Toxicon 2017, 128, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Ward, M.; Ellsworth, S.; Rokyta, D. Venom-gland transcriptomics and venom proteomics of the Hentz striped scorpion (Centruroides hentzi; Buthidae) reveal high toxin diversity in a harmless member of a lethal family. Toxicon 2018, 142, 14–29. [Google Scholar] [CrossRef] [PubMed]
- Baradaran, M.; Jalali, A.; Soorki, M.; Jokar, M.; Galehdari, H. First transcriptome analysis of Iranian scorpion, Mesobuthus eupeus venom gland. Iran. J. Pharm. Res. 2018, in press. [Google Scholar]
- Zhong, J.; Zeng, X.C.; Zeng, X.; Nie, Y.; Zhang, L.; Wu, S.; Bao, A. Transcriptomic analysis of the venom glands from the scorpion Hadogenes troglodytes revealed unique and extremely high diversity of the venom peptides. J. Proteom. 2017, 150, 40–62. [Google Scholar] [CrossRef] [PubMed]
- Santibáñez-López, C.E.; Cid-Uribe, J.I.; Zamudio, F.Z.; Batista, C.V.F.; Ortiz, E.; Possani, L.D. Venom gland transcriptomic and venom proteomic analyses of the scorpion Megacormus gertschi Díaz-Najera, 1966 (Scorpiones: Euscorpiidae: Megacorminae). Toxicon 2017, 133, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Santibáñez-López, C.E.; Cid-Uribe, J.I.; Batista, C.V.F.; Ortiz, E.; Possani, L.D. Venom gland transcriptomic and proteomic analyses of the enigmatic scorpion Superstitionia donensis (Scorpiones: Superstitioniidae), with insights on the evolution of its venom components. Toxins 2016, 8, 367. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Gu, J.; Yan, Z.; Wang, M.; Ma, C.; Zhang, J.; Jiang, G.; Ge, M.; Xu, S.; Xu, Z.; et al. De novo transcriptomic analysis of the venomous glands from the scorpion Heterometrus spinifer revealed unique and extremely high diversity of the venom peptides. Toxicon 2018, 143, 1–19. [Google Scholar] [CrossRef] [PubMed]
- De la Vega, R.C.R.; Possani, L.D. Novel paradigms on scorpion toxins that affects the activating mechanism of sodium channels. Toxicon 2007, 49, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Gurevitz, M. Mapping of scorpion toxin receptor sites at voltage-gated sodium channels. Toxicon 2012, 60, 502–511. [Google Scholar] [CrossRef] [PubMed]
- Soleglad, M.; Fet, V. The systematics of the scorpion subfamily Uroctoninae (Scorpiones: Chactidae). Rev. Ibér. Aracnol. 2004, 10, 81–128. [Google Scholar]
- Valdez-Cruz, N.A.; Batista, C.V.F.; Zamudio, F.Z.; Bosmans, F.; Tytgat, J.; Possani, L.D. Phaiodotoxin, a novel structural class of insect-toxin isolated from the venom of the Mexican scorpion Anuroctonus phaiodactylus. Eur. J. Biochem. 2004, 271, 4753–4761. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Dominguez, M.E.; Olamendi-Portugal, T.; Garcia, U.; Garcia, C.; Arechiga, H.; Possani, L.D. Cn11, the first example of a scorpion toxin that is a true blocker of Na+ currents in crayfish neurons. J. Exp. Biol. 2002, 205, 869–876. [Google Scholar] [PubMed]
- Carcamo-Noriega, E.N.; Olamendi-Portugal, T.; Restano-Cassulini, R.; Rowe, A.; Uribe-Romero, S.J.; Becerril, B.; Possani, L.D. Intraspecific variation of Centruroides sculpturatus scorpion venom from two regions of Arizona. Arch. Biochem. Biophys. 2018, 638, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Kuzmenkov, A.I.; Krylov, N.A.; Chugunov, A.O.; Grishin, E.V.; Vassilevski, A.A. Kalium: A database of potassium channel toxins from scorpion venom. Database 2016, 2016, baw056. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Vargas, J.M.; Possani, L.D.; Luna-Ramírez, K. Arthropod toxins acting on neuronal potassium channels. Neuropharmacology 2017, 127, 139–160. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.J.; Hill, J.M.; Little, M.J.; Nicholson, G.M.; King, G.F.; Alewood, P.F. Unique scorpion toxin with a putative ancestral fold provides insight into evolution of the inhibitor cystine knot motif. Proc. Natl. Acad. Sci. USA 2011, 108, 10478–10483. [Google Scholar] [CrossRef] [PubMed]
- Torabi, E.; Asgari, S.; Khalaj, V.; Behdani, M.; Kazemi-Lomedasht, F.; Bagheri, K.P.; Shahbazzadeh, D. Corrigendum to “The first report on transcriptome analysis of the venom gland of Iranian scorpion, Hemiscorpius lepturus” [Toxicon 125 (2017) 123–130]. Toxicon 2017, 128, 60. [Google Scholar] [CrossRef] [PubMed]
- Luna-Ramirez, K.; Quintero-Hernandez, V.; Vargas-Jaimes, L.; Batista, C.V.F.; Winkel, K.D.; Possani, L.D. Characterization of the venom from the Australian scorpion Urodacus yaschenkoi: Molecular mass analysis of components, cDNA sequences and peptides with antimicrobial activity. Toxicon 2013, 63, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Possani-Postay, L.D.; Gurrola-Briones, G.; Salas-Castillo, S.P.; Batista, C.V.F.; Varga, Z.S.; Panyi, G.; Gáspár, R. Vm23 and Vm24, Two Scorpion Peptides that Block Human T-Lymphocyte Potassium Channels (Sub-Type Kv1.3) with High Selectivity and Decrease the In Vivo DTH-Response in Rats. European Patent EP2158213A1, 14 May 2007. [Google Scholar]
- Diego-Garcia, E.; Schwartz, E.F.; D’Suze, G.; Gonzalez, S.A.R.; Batista, C.V.F.; Garcia, B.I.; de la Vega, R.C.R.; Possani, L.D. Wide phylogenetic distribution of Scorpine and long-chain beta-KTx-like peptides in scorpion venoms: Identification of “orphan” components. Peptides 2007, 28, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Soorki, M.; Galehdari, H.; Baradaran, M.; Jalali, A. First venom gland transcriptomic analysis of Iranian yellow scorpion Odonthubuthus doriae. Toxicon 2016, 120, 69–77. [Google Scholar]
- Schwartz, E.F.; Diego-Garcia, E.; de la Vega, R.C.R.; Possani, L.D. Transcriptome analysis of the venom gland of the Mexican scorpion Hadrurus gertschi (Arachnida: Scorpiones). BMC Genom. 2007, 8, 119. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Luo, F.; Feng, J.; Yang, W.; Zeng, D.; Zhao, R.; Cao, Z.; Liu, M.; Li, W.; Jiang, L.; et al. Genomic and structural characterization of Kunitz-Type peptide LmKTT-1a highlights diversity and evolution of scorpion potassium channel toxins. PLoS ONE 2013, 8, e60201. [Google Scholar] [CrossRef] [PubMed]
- García-Fernández, R.; Peigneur, S.; Pons, T.; Alvarez, C.; González, L.; Chávez, M.A.; Tytgat, J. The kunitz-type protein ShPI-1 inhibits serine proteases and voltage-gated potassium channels. Toxins 2016, 8, 110. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Huys, I.; Dyason, K.; Verdonck, F.; Tytgat, J. Evolutionary trace analysis of scorpion toxins specific for K+-channels. Proteins Struct. Funct. Genet. 2004, 54, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Camargos, T.S.; Restano-Cassulini, R.; Possani, L.D.; Peigneur, S.; Tytgat, J.; Schwartz, C.A.; Alves, E.M.C.; de Freitas, S.M.; Schwartz, E.F. The new kappa-KTx 2.5 from the scorpion Opisthacanthus cayaporum. Peptides 2011, 32, 1509–1517. [Google Scholar] [CrossRef] [PubMed]
- Vandendriessche, T.; Kopljar, I.; Jenkins, D.P.; Diego-Garcia, E.; Abdel-Mottaleb, Y.; Vermassen, E.; Clynen, E.; Schoofs, L.; Wulff, H.; Snyders, D.; et al. Purification, molecular cloning and functional characterization of HelaTx1 (Heterometrus laoticus): The first member of a new k-KTX subfamily. Biochem. Pharmacol. 2012, 83, 1307–1317. [Google Scholar] [CrossRef] [PubMed]
- Valdivia, H.H.; Kirbyf, M.S.; Lederer, W.J.; Coronado, R. Scorpion toxins targeted against the sarcoplasmic reticulum Ca(2+)-release channel of skeletal and cardiac muscle. Physiology 1992, 89, 12185–12189. [Google Scholar] [CrossRef]
- Xiao, L.; Gurrola, G.B.; Zhang, J.; Valdivia, C.R.; SanMartin, M.; Zamudio, F.Z.; Zhang, L.; Possani, L.D.; Valdivia, H.H. Structure–function relationships of peptides forming the calcin family of ryanodine receptor ligands. J. Gen. Physiol. 2016, 147, 375–394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, J.; Vetter, I.; Lewis, R.; Peigneur, S.; Tytgat, J.; Lam, A.; Gallant, E.; Beard, N.; Alewood, P.; Dulhunty, A. Multiple actions of phi-LITX-Lw1a on ryanodine receptors reveal a functional link between scorpion DDH and ICK toxins. Proc. Natl. Acad. Sci. USA 2013, 110, 8906–8911. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Darbon, H.; Dyason, K.; Verdonck, F.; Tytgat, J. Evolutionary origin of inhibitor cystine knot peptides. FASEB J. 2003, 17, 1765–1767. [Google Scholar] [CrossRef] [PubMed]
- Shahbazzadeh, D.; Srairi-Abid, N.; Feng, W.; Ram, N.; Borchani, L.; Ronjat, M.; Akbari, A.; Pessah, I.N.; De Waard, M.; El Ayeb, M. Hemicalcin, a new toxin from the Iranian scorpion Hemiscorpius lepturus which is active on ryanodine-sensitive Ca2+ channels. Biochem. J. 2007, 404, 89–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lundy, P.M.; Frew, R. Effect of omega-agatoxin-IVA on autonomic neurotransmission. Eur. J. Pharmacol. 1994, 261, 79–84. [Google Scholar] [CrossRef]
- Oppenheim, J.J.; Biragyn, A.; Kwak, L.W.; Yang, D. Roles of antimicrobial peptides such as defensins in innate and adaptive immunity. Ann. Rheum. Dis. 2003, 62, ii17–ii21. [Google Scholar] [CrossRef] [PubMed]
- Hazlett, L.; Wu, M. Defensins in innate immunity. Cell Tissue Res. 2011, 343, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Jenssen, H.; Hamill, P.; Hancock, R. Peptide antimicrobial agents. Clin. Microbiol. Rev. 2006, 19, 491–511. [Google Scholar] [CrossRef] [PubMed]
- De la Vega, R.C.R.; García, B.I.; D’Ambrosio, C.; Diego-García, E.; Scaloni, A.; Possani, L.D. Antimicrobial peptide induction in the haemolymph of the Mexican scorpion Centruroides limpidus limpidus in response to septic injury. Cell. Mol. Life Sci. 2004, 61, 1507–1519. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.; Tan, C.; Chanhome, L.; Tan, N. Comparative venom gland transcriptomics of Naja kaouthia (monocled cobra) from Malaysia and Thailand: Elucidating geographical venom variation and insights into sequence novelty. PeerJ 2017, 5, e3142. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Wang, Y.; Wei, L.; Ye, H.; Liu, H.; Wang, L.; Liu, R.; Li, D.; Lai, R. Snake venom-like waprin from the frog of Ceratophrys calcarata contains antimicrobial function. Gene 2013, 514, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.; Wong, H.; Desai, M.; Moochhala, S.; Kuchel, P.; Kini, R. Identification of a novel family of proteins in snake venoms. Purification and structural characterization of nawaprin from Naja nigricollis snake venom. J. Biol. Chem. 2003, 278, 40097–40104. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, H. Snake venom proteases inhibitors: Enhanced identification, expanding biological function, and promising future. In Snake Venoms, Toxinology; Springer: Dordrecht, The Netherland, 2017; pp. 161–186. [Google Scholar]
- Almaaytah, A.; Albalas, Q. Scorpion venom peptides with no disulfide bridges: A review. Peptides 2014, 51, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, E.; Gurrola, G.B.; Schwartz, E.F.; Possani, L.D. Scorpion venom components as potential candidates for drug development. Toxicon 2015, 93, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Carreto, S.; Jimenez-Vargas, J.M.; Rivas-Santiago, B.; Corzo, G.; Possani, L.D.; Becerril, B.; Ortiz, E. Peptides from the scorpion Vaejovis punctatus with broad antimicrobial activity. Peptides 2015, 73, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Carreto, S.; Quintero-Hernandez, V.; Jimenez-Vargas, J.M.; Corzo, G.; Possani, L.D.; Becerril, B.; Ortiz, E. Gene cloning and functional characterization of four novel antimicrobial-like peptides from scorpions of the family Vaejovidae. Peptides 2012, 34, 290–295. [Google Scholar] [CrossRef] [PubMed]
- De la Vega, R.C.R.; Possani, L.D. Current views on scorpion toxins specific for K+-channels. Toxicon 2004, 43, 865–875. [Google Scholar] [CrossRef] [PubMed]
- Vanuopadath, M.; Sajeev, N.; Murali, A.R.; Sudish, N.; Kangosseri, N.; Sebastian, I.R.; Jain, N.D.; Pal, A.; Raveendran, D.; Nair, B.G.; et al. Mass spectrometry-assisted venom profiling of Hypnale hypnale found in the Western Ghats of India incorporating de novo sequencing approaches. Int. J. Biol. Macromol. 2018, 118, 1736–1746. [Google Scholar] [CrossRef] [PubMed]
- Walter, A.; Bechsgaard, J.; Scavenius, C.; Dyrlund, T.S.; Sanggaard, K.W.; Enghild, J.J.; Bilde, T. Characterisation of protein families in spider digestive fluids and their role in extra-oral digestion. BMC Genom. 2017, 18, 600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, T.; Bjourson, A.J.; Orr, D.F.; Kwok, H.; Rao, P.; Ivanyi, C.; Shaw, C. Unmasking venom gland transcriptomes in reptile venoms. Anal. Biochem. 2002, 311, 152–156. [Google Scholar] [CrossRef]
- De Oliveira, U.C.; Candido, D.M.; Coronado Dorce, V.A.; Junqueira-De-Azevedo, I.D.L.M. The transcriptome recipe for the venom cocktail of Tityus bahiensis scorpion. Toxicon 2015, 95, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, J.; Rucavado, A.; Escalante, T.; Díaz, C. Hemorrhage induced by snake venom metalloproteinases: Biochemical and biophysical mechanisms involved in microvessel damage. Toxicon 2005, 45, 997–1011. [Google Scholar] [CrossRef] [PubMed]
- Hass, E.; Stanley, D. Phospholipases. In xPharm: The Comprehensive Pharmacology Reference; Elsevier: New York, NY, USA, 2007; pp. 1–3. [Google Scholar]
- Hmed, B.; Serria, H.; Mounir, Z. Scorpion peptides: Potential use for new drug development. J. Toxicol. 2013, 2013, 958797. [Google Scholar] [CrossRef] [PubMed]
- Borchani, L.; Sassi, A.; Shahbazzadeh, D.; Strub, J.; Tounsi-Guetteti, H.; Boubaker, M.; Akbari, A.; Dorsselaer, A.; El Ayeb, M. Heminecrolysin, the first hemolytic dermonecrotic toxin purified from scorpion venom. Toxicon 2011, 58, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Vines, C.M.; Bill, C.A. Phospholipases. In eLS; Wiley Online Library: Hoboken, NJ, USA, 2015; pp. 1–9. [Google Scholar]
- Bordon, K.C.F.; Wiezel, G.A.; Amorim, F.G.; Arantes, E.C. Arthropod venom Hyaluronidases: Biochemical properties and potential applications in medicine and biotechnology. J. Venom. Anim. Toxins Incl. Trop. Dis. 2015, 21, 43. [Google Scholar] [CrossRef] [PubMed]
- King, T.P.; Wittkowski, K.M. Hyaluronidase and hyaluronan in insect venom allergy. Int. Arch. Allergy Immunol. 2011, 156, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Sutti, R.; Tamascia, M.L.; Hyslop, S.; Rocha-E-Silva, T.A.A. Purification and characterization of a hyaluronidase from venom of the spider Vitalius dubius (Araneae, Theraphosidae). J. Venom. Anim. Toxins Incl. Trop. Dis. 2014, 20, 2. [Google Scholar] [CrossRef] [PubMed]
- Luna-Ramírez, K.; Quintero-Hernández, V.; Juárez-González, V.R.; Possani, L.D. Whole transcriptome of the venom gland from Urodacus yaschenkoi scorpion. PLoS ONE 2015, 10, e0127883. [Google Scholar] [CrossRef] [PubMed]
- Amorim, F.G.; Costa, T.R.; Baiwir, D.; De Pauw, E.; Quinton, L.; Sampaio, S.V. Proteopeptidomic, functional and immunoreactivity characterization of Bothrops moojeni snake venom: Influence of snake gender on venom composition. Toxins 2018, 10, 177. [Google Scholar] [CrossRef] [PubMed]
- Kazuma, K.; Masuko, K.; Konno, K.; Inagaki, H. Combined venom gland transcriptomic and venom peptidomic analysis of the predatory ant Odontomachus monticola. Toxins 2017, 9, 323. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, K.; Marciniak, P.; Rosiñski, G.; Rychlik, L. Toxic activity and protein identification from the parotoid gland secretion of the common toad Bufo bufo. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2018, 205, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Silva-Libério, M.; Bastos, I.; Júnior, O.; Fontes, W.; Santana, J.; Castro, M. The crude skin secretion of the pepper frog Leptodactylus labyrinthicus is rich in metallo and serine peptidases. PLoS ONE 2014, 9, e96893. [Google Scholar]
- León, D.; Fellay, I.; Mantel, P.; Walch, M. Killing bacteria with cytotoxic effector proteins of human killer immune cells: Granzymes, granulysin, and perforin. Methods Mol. Biol. 2017, 1535, 275–284. [Google Scholar] [PubMed]
- Vema, M.; Xavier, F.; Verma, Y.; Sobha, K. Evaluation of cytotoxic and anti-tumor activity of partially purified serine protease isolate from the Indian earthworm Pheretima posthuma. Asian Pac. J. Trop. Biomed. 2013, 3, 896–901. [Google Scholar]
- Menaldo, D.; Bernardes, C.; Pereira, J.; Silveira, D.; Mamede, C.; Stanziola, L.; de Oliveira, F.; Pereira-Crott, L.; Faccioli, L.; Sampaio, S. Effects of two serine proteases from Bothrops pirajai snake venom on the complement system and the inflammatory response. Int. Immunopharmacol. 2013, 15, 764–771. [Google Scholar] [CrossRef] [PubMed]
- Pérez, J.; Sánchez, E. Natural protease inhibitors to hemorrhagins in snake venoms and their potential use in medicine. Toxicon 1999, 37, 703–728. [Google Scholar] [CrossRef]
- Ma, H.; Xiao-Peng, T.; Yang, S.; Lu, Q.; Lai, R. Protease inhibitor in scorpion (Mesobuthus eupeus) venom prolongs the biological activities of the crude venom. Chin. J. Nat. Med. 2016, 14, 607–614. [Google Scholar] [CrossRef]
- Proaño-Bolaños, C.; Li, R.; Zhou, M.; Wang, L.; Xi, X.; Tapia, E.; Coloma, L.; Chen, T.; Shaw, C. Novel Kazal-tyoe proteinase inhibitors from the skin secretion of the Splendid leaf frog, Cruziohyla calcarifer. EuPA Open Proteom. 2017, 15, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, Y.; Lu, Z.; Zhain, L.; Jiang, J.; Liu, J.; Yu, H. A novel serine protease inhibitor from the venom of Vespa bicolor Fabricius. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2009, 153, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Wang, X.; Liu, H.; San, M.; Xu, Y.; Li, J.; Li, S.; Cao, Z.; Li, W.; Wu, Y.; et al. A new Kunitz-type plasmin inhibitor from scorpion venom. Toxicon 2015, 106, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Nagao, J.; Miyashita, M.; Nakagawa, Y.; Miyagawa, H. Chemical synthesis of La1 isolated from the venom of the scorpion Liocheles australasiae and determination of its disulfide bonding pattern. J. Pept. Sci. 2015, 21, 636–643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyashita, M.; Otsuki, J.; Hanai, Y.; Nakagawa, Y.; Miyagawa, H. Characterization of peptide components in the venom of the scorpion Liocheles australasiae (Hemiscorpiidae). Toxicon 2007, 50, 428–437. [Google Scholar] [CrossRef] [PubMed]
- Martinez, G.; Hograindleur, J.; Voisin, S.; Abi Nahed, R.; Abd El Aziz, T.; Escoffier, J.; Bessonnat, J.; Fovet, C.; De Waard, M.; Hennebicg, S.; et al. Spermaurin, an La1-like peptide from the venom of the scorpion Scorpio maurus plamatus, improves sperm motility and fertilization in different mammalian species. Mol. Hum. Reprod. 2017, 23, 116–131. [Google Scholar] [PubMed]
- Gibbs, G.M.; Roelants, K.; O’Bryan, M.K. The CAP superfamily: Cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins—Roles in reproduction, cancer, and immune defense. Endocr. Rev. 2008, 29, 865–897. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.; Vidal, N.; Norman, J.; Vonk, F.; Scheib, H.; Ramjan, S.; Kuruppu, S.; Fung, K.; Hedges, S.; Richardson, M.; et al. Early evolution of the venom system in lizards and snakes. Nature 2006, 439, 584–588. [Google Scholar] [CrossRef] [PubMed]
- Reddy, T.; Gibbs, G.; Merriner, D.; Kerr, J.; O’Bryan, M. Cysteine-rich secretory proteins are not exclusively expressed in the male reproductive tract. Dev. Dyn. 2008, 237, 3313–3323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almeida, D.; Scortecci, K.; Kobashi, L.; Agnez-Lima, L.; Medeiros, S.; Silva-Junior, A.; Junqueira-de-Azevedo, I.; Fernandes-Pedrosa, M. Profiling the resting venom gland of the scorpion Tityus stigmurus through a transcriptomic survey. BMC Genom. 2012, 13, 362. [Google Scholar] [CrossRef] [PubMed]
- Hwa, V.; Oh, Y.; Rosenfeld, R. The insulin-like growth factor-binding protein (IGFBP) superfamily. Endocr. Rev. 1999, 20, 761–787. [Google Scholar] [CrossRef] [PubMed]
- Bryant, D.M.; Johnson, K.; DiTommaso, T.; Tickle, T.; Couger, M.B.; Payzin-Dogru, D.; Lee, T.J.; Leigh, N.D.; Kuo, T.H.; Davis, F.G.; et al. A tissue-mapped axolotl de novo transcriptome enables identification of limb regeneration factors. Cell Rep. 2017, 18, 762–776. [Google Scholar] [CrossRef] [PubMed]
- Tolksdorf, S.; McCready, M.H. The turbidimetric assay of hyaluronidase. J. Lab. Clin. Med. 1949, 34, 74–89. [Google Scholar] [PubMed]
- Horta, C.C.R.; de Magalhães, B.F.; Oliveira-Mendes, B.B.R.; Carmo, A.O.D.; Duarte, C.G.; Felicori, L.F.; Machado-de-Ávila, R.A.; Chávez-Olórtegui, C.; Kalapothakis, E. Molecular, immunological, and biological characterization of Tityus serrulatus venom hyaluronidase: New insights into its role in envenomation. PLoS Negl. Trop. Dis. 2014, 8, e2693. [Google Scholar] [CrossRef] [PubMed]
- Habermann, E.; Hardt, K.L. A sensitive and specific plate test for the quantitation of phospholipases. Anal. Biochem. 1972, 50, 163–173. [Google Scholar] [CrossRef]
- Ramírez-Avila, J.; Quevedo, B.E.; López, E.; Renjifo, J.M. Purification and partial characterization of phospholipases A2 from Bothrops asper (barba amarilla) snake venom from Chiriguaná (Cesar, Colombia). J. Venom. Anim. Toxins Incl. Trop. Dis. 2004, 10, 242–259. [Google Scholar] [CrossRef]


| Retention Time (Min) | Molecular Mass (Da) |
|---|---|
| 0−20 | 444.26; 492.30; 673.37; 748.24; 775.41; 815.48; 888.44; 1018.52; 1105.66; 1158.64; 1234.61; 1245.64; 1374.74; 1434.69; 1460.76; 1484.79; 1573.83; 1633.82; 1694.86; 1731.90; 1832.95; 2290.88; 2350.31; 2452.07; 3464.44; 3565.49; 3616.63; 3648.56; 3701.77; 3773.73; 3928.89; 4052.58; 4361.81; 4544.96; 4744.10; 4815.13; 4879.28; 4882.28 |
| 20−40 | 2709.25; 3951.91; 4001.95; 4138.94; 4180.02; 4694.16 |
| 40−60 | 645.34; 683.30; 963.55; 1286.78; 1305.70; 1343.65; 1413.68; 1902.00; 2124.17; 3937.86; 4393.03; 5053.20; 5318.35 |
| 60−80 | 539.15; 722.39; 765.40; 836.46; 1004.54; 1193.63; 1264.68; 1408.59; 1643.90; 1750.81; 2036.03; 2103.22; 2229.20; 2411.28; 2467.34; 2580.42; 2629.31; 2757.39; 2855.56; 2837.54; 2984.55; 3338.73; 3350.48; 4068.67; 4348.99; 4444.81; 8029.68 |
| 80−100 | 707.35; 890.52; 978.51; 1518.92; 1647.01; 1744.06; 1859.09; 1972.17; 2561.42; 2950.62; 3274.43; 3421.02; 6230.62; 6418.87; 6568.88; 7088.10; 9023.88 |
| 100−120 | 1162.70; 1391.84; 1616.99; 1677.98; 1845.10; 1883.05; 2057.18; 2083.02; 2575.43; 3219.89; 3914.08; 6741.05; 6813.82; 7070.08 |
| 120−140 | 1380.75; 1495.78; 1582.81; 1683.86; 1721.82; 2248.22; 2361.30; 3001.49; 6457.78 |
| 140−160 | 804.39; 871.42; 1081.50; 1925.03; 2066.00; 3327.88; 3529.02; 6142.59; 6614.95; 6725.05 |
| 160−180 | 727.49; 876.43; 932.56; 1138.62; 1251.71; 1348.67; 1626.82; 1717.00; 1786.11; 1823.99; 2014.18; 2151.16; 2315.34; 2917.48; 3290.92; 6126.59; 6441.77; 6586.89 |
| 180−200 | 557.37; 989.52; 1046.54; 1119.63; 1567.83; 1603.92; 1807.94; 1888.12; 1956.00; 2146.18; 2485.44; 2674.46; 3031.66; 3310.88; 4256.28 |
| 200−220 | 599.40; 712.48; 1360.80; 1475.83; 1588.91; 1759.02; 2275.35; 2493.38; 2557.42; 2820.65; 3044.73; 3263.67; 3394.80; 3689.03; 4330.32; 4592.48; 4720.54; 4791.57; 4862.60; 4949.65; 12,869.20 |
| 220−240 | 657.36; 770.45; 883.51; 1027.60; 1096.62; 1185.67; 1298.75; 1438.81; 1525.84; 1838.02; 2371.36; 4428.40; 4652.41; 11,833.52; 12,386.79; 12,432.80 |
| 240−290 | N/D |
| Transcriptome ID | Score | Coverage | Protein Fragments | Xcorr of the Protein Fragments | Protein Type | Accession Number of the Reference Protein |
|---|---|---|---|---|---|---|
| SgeKTxScr02 | 1248.72 | 56.41 | HGCLADFDVGGGCEQHCR | 7.26 | Potassium channel toxin. Scorpine-like peptide. | API81325.1 |
| KIQDAIDR | 2.54 | |||||
| AWISEK | 1.91 | |||||
| SgeHDPND301 | 163.69 | 87.50 | FWGFLGK | 3.18 | HDP. NDBP-3 family. | ALG64975.1 |
| TIPSLLGGSK | 2.09 | |||||
| SgeHDPND204 | 153.72 | 36.67 | KAWNSNIGK | 3.61 | HDP. NDBP-2 family. | AGK88593.1 |
| AWNSNIGK | 2.31 | |||||
| SgeHDPND202 | 138.57 | 63.83 | AWNSDIGK | 3.59 | HDP. NDBP-2 family. | ALG64975.1 |
| GIWGTIK | 2.76 | |||||
| IGVTPSQAAS | 2.70 | |||||
| SgeEnzPA207 | 109.20 | 50.60 | YGLSnTGSYTLLNcDcEK | 6.04 | Enzyme. Phospholipase A2. | API81335.1 |
| WIYFTAYSPK | 2.85 | |||||
| cANPVGKWKADYK | 1.38 | |||||
| EGWIKK | 1.02 | |||||
| SgeCaTLio02 | 65.14 | 42.86 | DLPLSNEYETcVRPR | 3.74 | Calcium channel toxin. Liotoxin. | P0DJ08.1 |
| SgeEnzPA204 | 64.99 | 36.4 | TLLNcDcEEAFDHcLQTTADK | 4.38 | Enzyme. Phospholipase A2. | API81335.1 |
| TLLNcDcEEAFDHcLQTTADKLEGADKEDTK | 3.99 | |||||
| IIQNYYFNIFK | 3.18 | |||||
| cRmLnSTKEVAR | 1.17 | |||||
| SgeHDPND404 | 64.77 | 53.85 | DLWNGVK | 2.37 | HDP. NDBP-4 family. | I0DEB3.1 |
| SgeNaTBet02 | 49.29 | 47.54 | GSSYGYcYGFGcYcEGLNDDVK | 4.87 | Sodium channel toxin. Beta. | P01491.3 |
| FcQSIcK | 2.46 | |||||
| SgeOthLa104 | 14.30 | 65.82 | NVPGPVDAPFPDccPTSLcR | 4.24 | La1-like peptide | AOF40216.1 |
| SgeEnzPA206 | 13.61 | 40.08 | AFYFHLYGnGcYHVK | 3.79 | Enzyme. Phospholipase A2. | API81339.1 |
| cLDQVVDGTSWYDYHATLGLIK | 1.31 | |||||
| SENGRGLR | 1.09 | |||||
| SgeOthLa106 | 13.36 | 33.33 | TGQYLNEGEEWRDPNHcSIYQcR | 3.82 | La1-like peptide | API81334.1 |
| SgeNaTAlp02 | 12.80 | 64.79 | AmPPFGLPGGcFcPNIPK | 3.14 | Sodium channel toxin. Alpha. | Q5MJP5.1 |
| SgeCaTLio01 | 12.17 | 88.57 | NLPLSDEYGTcVRPR | 2.96 | Calcium channel toxin. Liotoxin. | P0DJ08.1 |
| cKPPLWcNPQQIcVYK | 2.86 | |||||
| SgeKTxScr01 | 10.51 | 71.08 | GVDALTNLIPAPVIGGIVNK | 4.70 | Potassium channel toxin. Scorpine-like peptide. | P0C8W5.1 |
| VQELcAFNK | 3.31 | |||||
| SgeOthCAP02 | 8.09 | 48.96 | STNDFIFYcDFSK | 4.15 | CAP superfamily | API81352.1 |
| VEHSGTAFWTIGmHMqFDQEMESTIKLEAYR | 1.33 | |||||
| VLYTcNYGPAGNmQGGTmYIKGEPcSQcPK | 1.21 | |||||
| SgeEnzHya01 | 4.11 | 31.30 | YFTDTTSVFDSK | 2.43 | Enzyme. Hyaluronidase. | API81375.1 |
| ILVNNK | 1.68 | |||||
| ILVNNKESFVGDK | 1.07 | |||||
| SgeEnzMtP02 | 3.85 | 27.14 | KFNYPHGTIK | 2.15 | Enzyme. Metalloproteinase. | AMO02513.1 |
| KEPKKPPVPK | 1.19 | |||||
| SgeCatClc02 | 2.27 | 24.24 | ADcLPHLK | 2.27 | Calcium channel toxin. Calcin. | API81327.1 |
| c15440_g1_i1 | 6.83 | 9.30 | ELTLEDVGTILR | 3.67 | N/D | N/D |
| c23802_g1_i1 | 7.08 | 10.48 | WYNDcIDYcDR | 3.59 | N/D | N/D |
| c27313_g1_i1 | 13.32 | 15.28 | FGDITLLGPEVTNR | 4.12 | N/D | N/D |
| IISSPFYK | 1.21 | N/D | N/D | |||
| c26154_g1_i1 | 7.74 | 10.53 | LAELETIVALAK | 3.85 | N/D | N/D |
| c26889_g1_i1 | 94.33 | 20.29 | EFFLGLLGLK | 3.63 | N/D | N/D |
| IDPKEFFLGLLGLK | 1.54 | N/D | N/D |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romero-Gutiérrez, M.T.; Santibáñez-López, C.E.; Jiménez-Vargas, J.M.; Batista, C.V.F.; Ortiz, E.; Possani, L.D. Transcriptomic and Proteomic Analyses Reveal the Diversity of Venom Components from the Vaejovid Scorpion Serradigitus gertschi. Toxins 2018, 10, 359. https://doi.org/10.3390/toxins10090359
Romero-Gutiérrez MT, Santibáñez-López CE, Jiménez-Vargas JM, Batista CVF, Ortiz E, Possani LD. Transcriptomic and Proteomic Analyses Reveal the Diversity of Venom Components from the Vaejovid Scorpion Serradigitus gertschi. Toxins. 2018; 10(9):359. https://doi.org/10.3390/toxins10090359
Chicago/Turabian StyleRomero-Gutiérrez, Maria Teresa, Carlos Eduardo Santibáñez-López, Juana María Jiménez-Vargas, Cesar Vicente Ferreira Batista, Ernesto Ortiz, and Lourival Domingos Possani. 2018. "Transcriptomic and Proteomic Analyses Reveal the Diversity of Venom Components from the Vaejovid Scorpion Serradigitus gertschi" Toxins 10, no. 9: 359. https://doi.org/10.3390/toxins10090359







