Comparative Proteomic Analysis of the Effect of Periplocoside P from Periploca sepium on Brush Border Membrane Vesicles in Midgut Epithelium of Mythimna separata Larvae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Compounds
2.2. Insect Rearing and Treatment
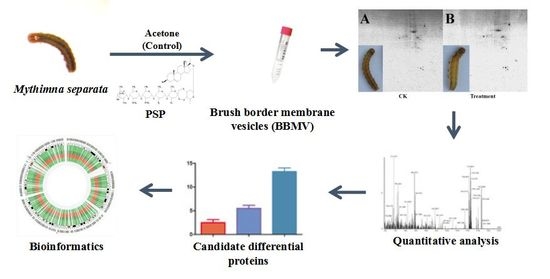
2.3. BBMVs Preparation
2.4. 2-DE
2.5. Image Acquisition and Data Analysis
2.6. Protein Identification and Bioinformatics Analysis
2.7. Quantitative Determination of Gene Expression Levels
2.8. Determination of Enzyme Activity
2.9. Data Statistics Analysis
3. Results
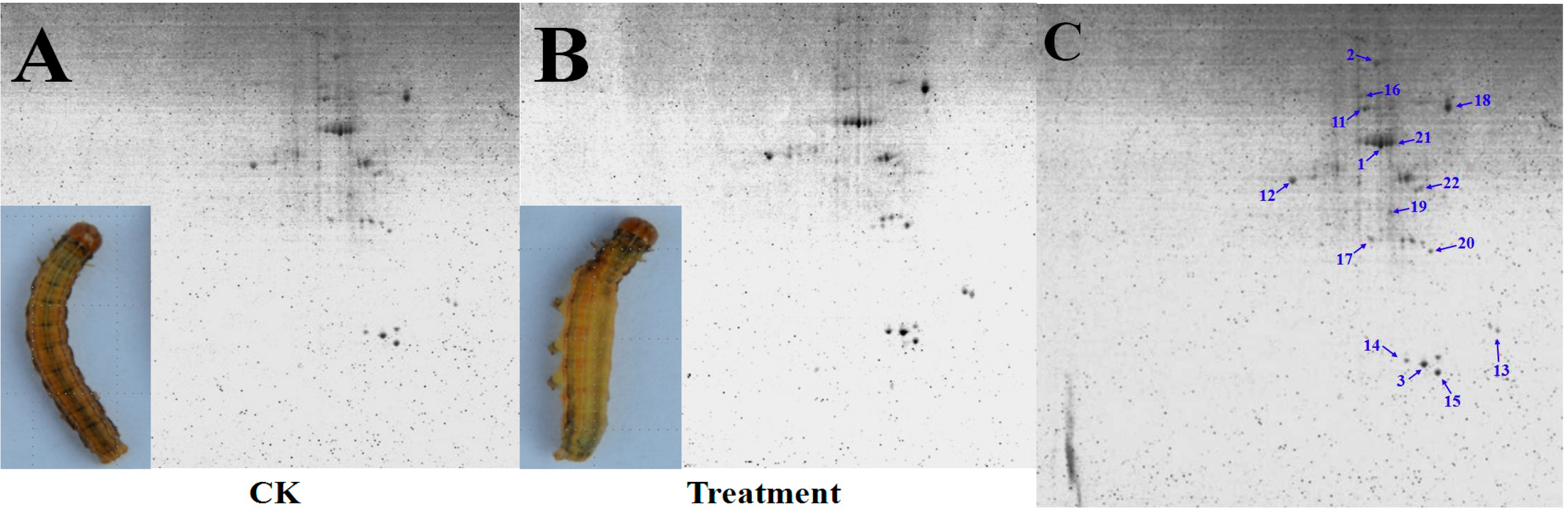
3.1. Quality Evaluation of BBMVs Protein
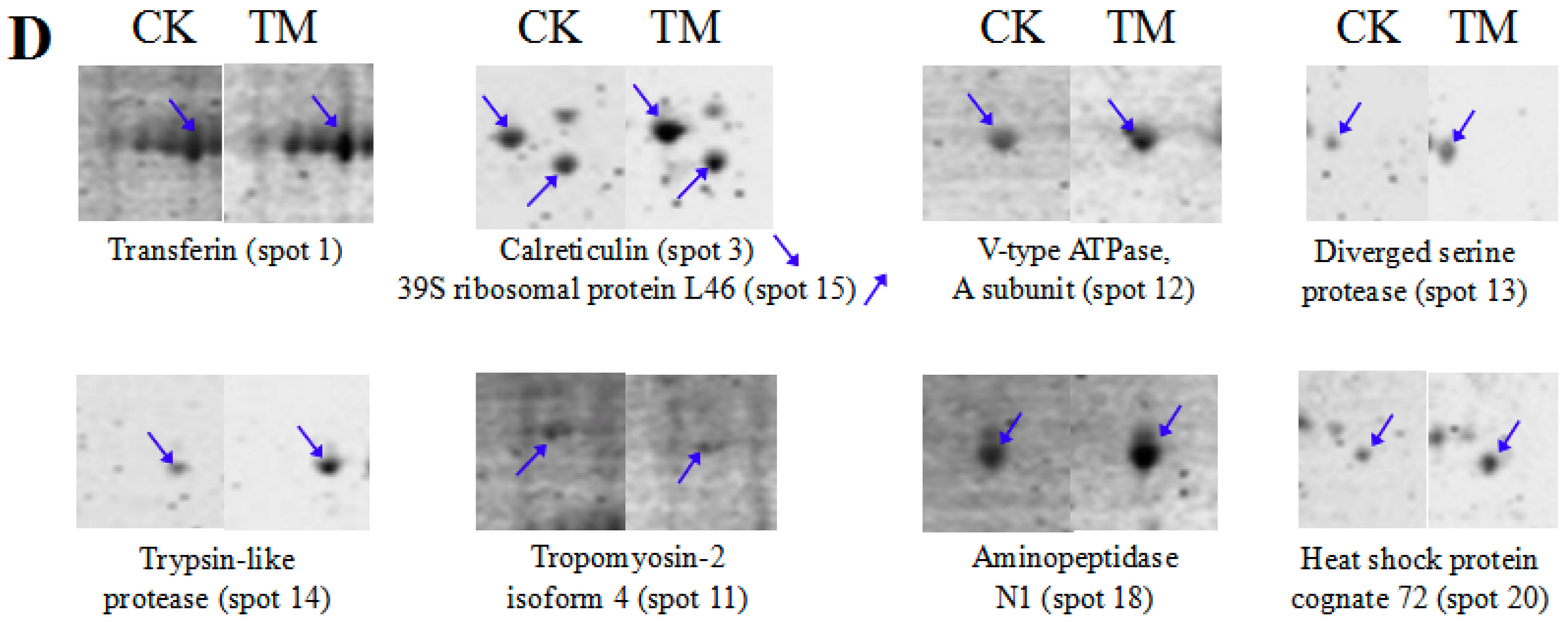
3.2. Detection and Comparative Analysis of Proteins Separated by 2-DE Gels
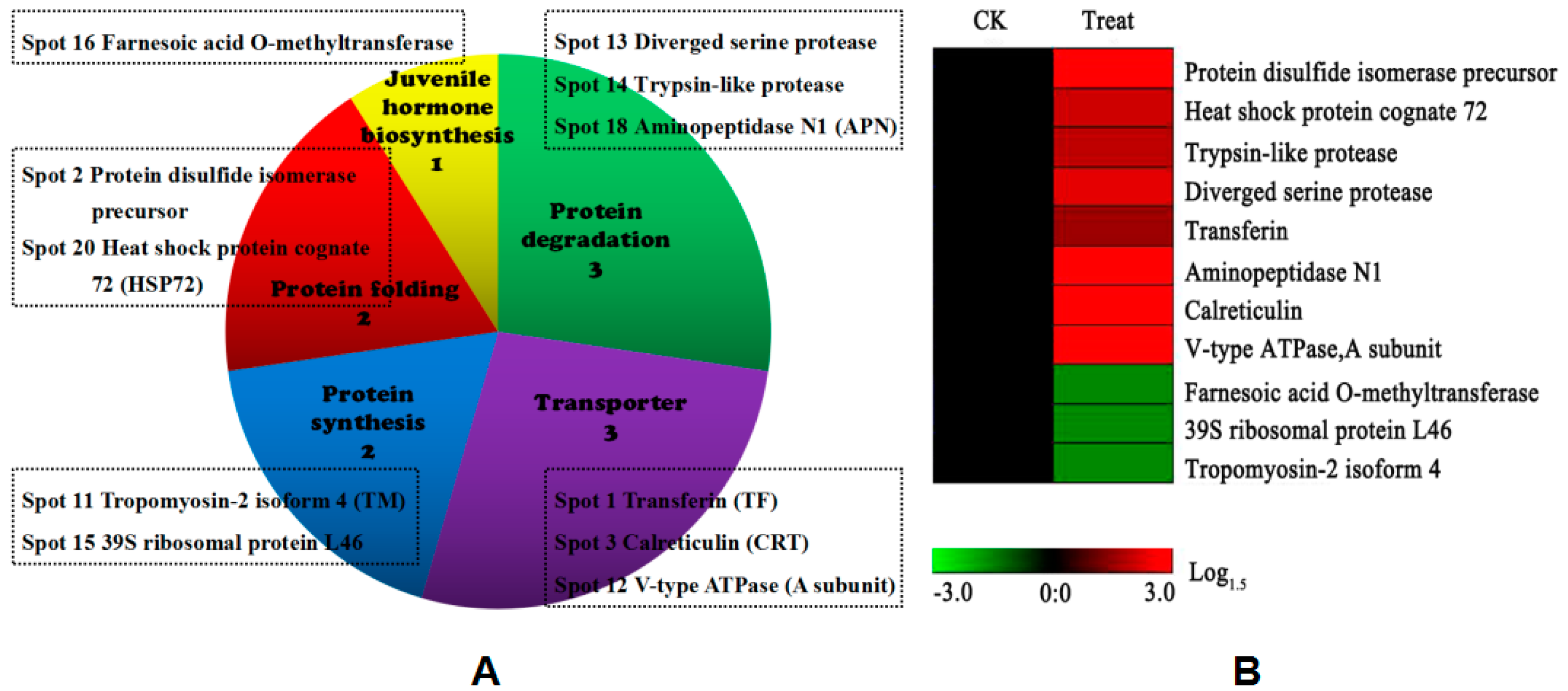
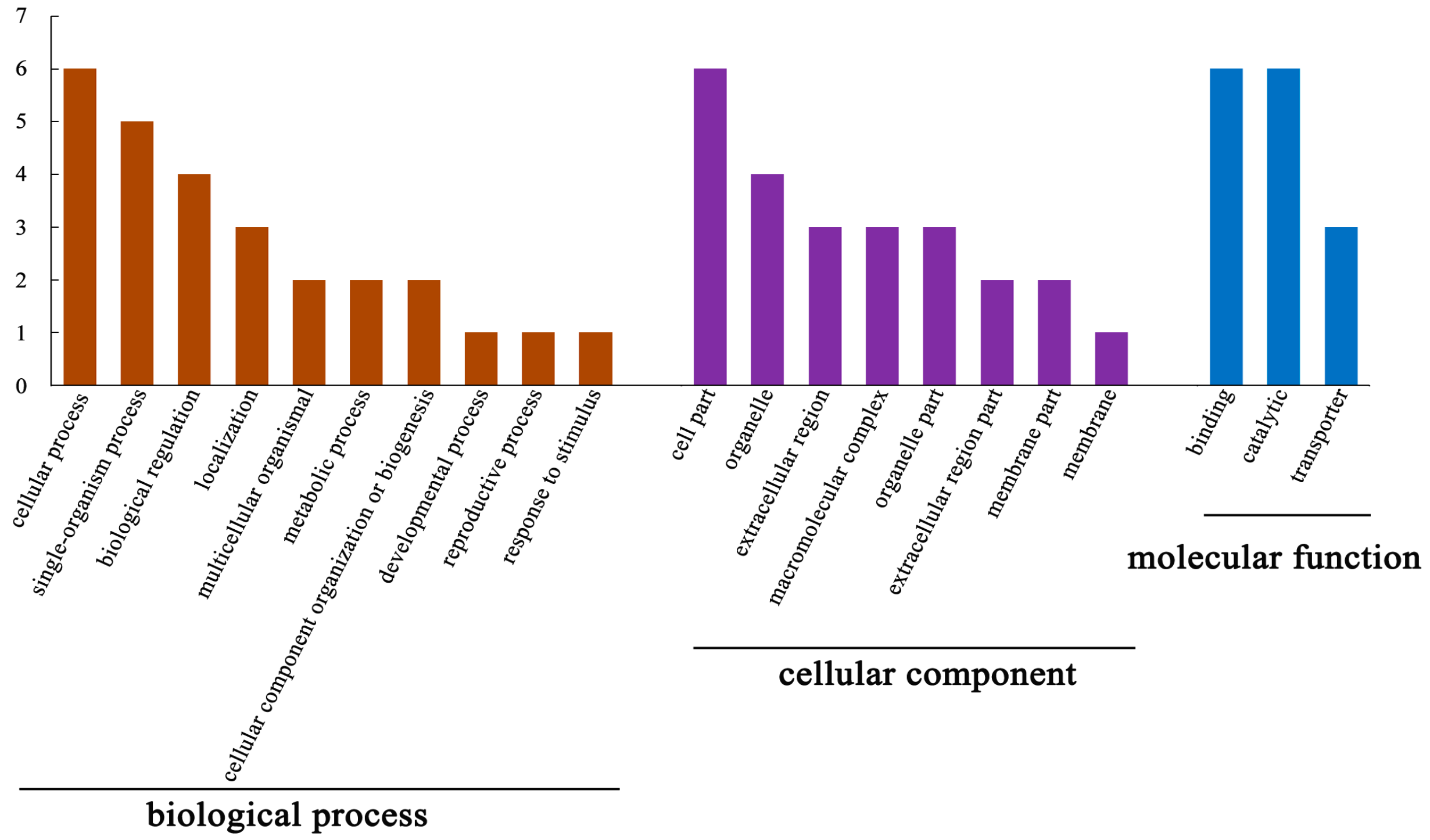
3.3. Functional Analysis of DEPs
3.4. Validation of Proteomic Data by RT-qPCR
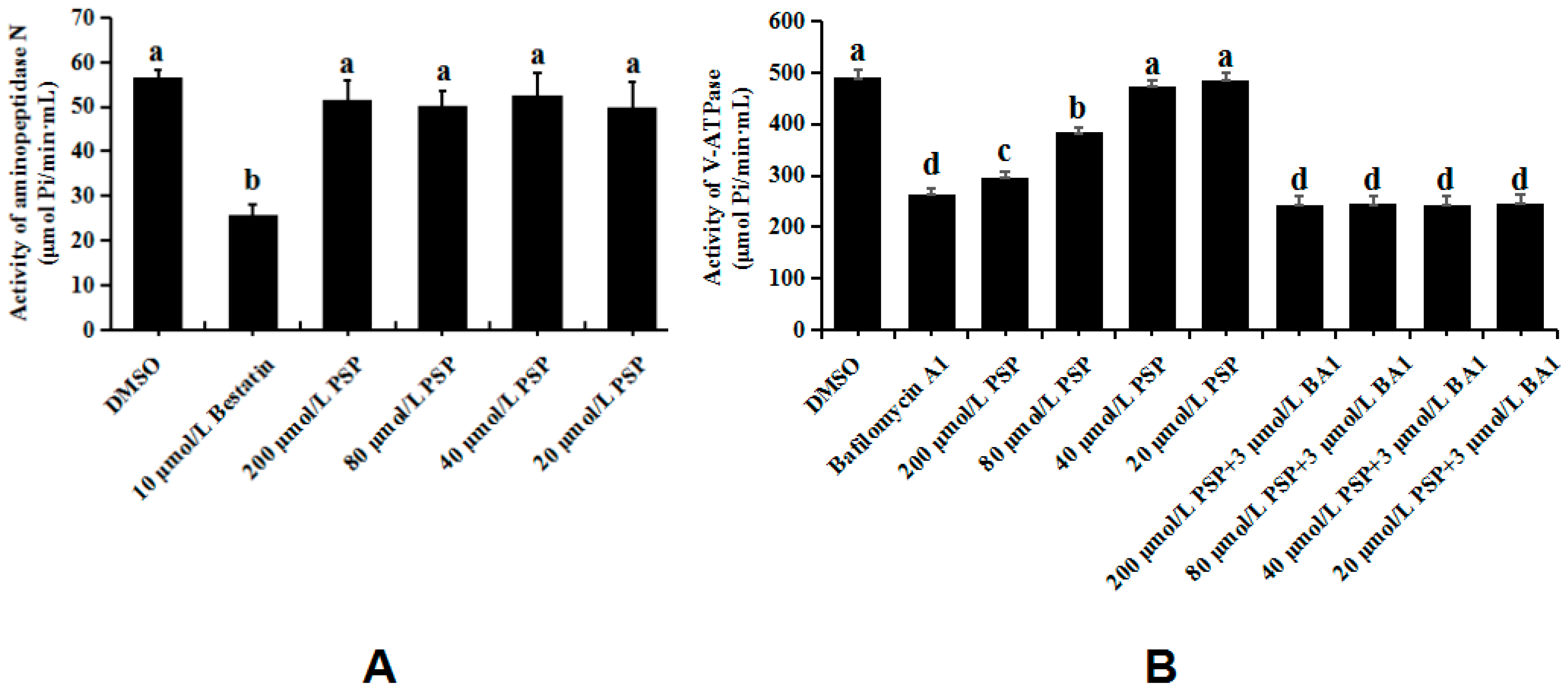
3.5. Enzymology Verification
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pant, M.; Dubey, S.; Patanjali, P.K. Recent Advancements in Bio-botanical Pesticide Formulation Technology Development. In Herbal Insecticides, Repellents and Biomedicines: Effectiveness and Commercialization; Springer: New Delhi, India, 2016. [Google Scholar]
- Scott, D.E.; Bayly, A.R.; Abell, C.; Skidmore, J. Small molecules, big targets: Drug discovery faces the protein-protein interaction challenge. Nat. Rev. Drug Discov. 2016, 15, 533–550. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.; Ursu, O.; Gaulton, A.; Bento, A.P.; Donadi, R.S.; Bologa, C.G.; Karlsson, A.; Lazikani, B.A.; Hersey, A.; Oprea, T.I.; et al. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 2017, 16, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Li, R.F.; Zhao, X.M.; Shi, B.J.; Wei, S.P.; Zhang, J.W.; Wu, W.J.; Hu, Z.N. Insecticidal pregnane glycosides from the root barks of periploca sepium. Nat. Prod. Commun. 2016, 11, 1425–1428. [Google Scholar]
- Liu, M.; Liu, G.; Liu, Y.C.; Feng, J.; Jiang, R.B.; Zhou, T. Screening of active fractions with analgesic and anti-inflammatory effect from Miao Medicine Periploca forrestii. Drug Eval. Res. 2014, 4, 007. [Google Scholar]
- Zhang, M.S.; Bang, I.S.; Park, C.B. Lack of Mutagenicity Potential of Periploca sepium Bge. in Bacterial Reverse Mutation (Ames) Test, Chromosomal Aberration and Micronucleus Test in Mice. Environ. Health Toxicol. 2012, 27, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Chu, S.S.; Jiang, G.H.; Liu, W.L.; Liu, Z.L. Insecticidal activity of the root bark essential oil of Periploca sepium Bunge and its main component. Nat. Prod. Res. 2012, 26, 926–932. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.S.; Qiao, X.; Wang, J.; Qin, S. Study on antifeedant and insecticidal activities of extracts and fractions from Periploca spepium Bunge against Plutella xylostella (L.). Chin. J. Pestic. Sci. 2004, 6, 48–52. [Google Scholar]
- Li, Y. Chemical constituents from the roots of Periploca sepium with insecticidal activity. J. Asian Nat. Prod. Res. 2012, 14, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.C.; Shi, B.J.; Hu, Z.N. The insecticidal activity of Periplocoside NW. Chin. Bull. Entomol. 2008, 45, 950–952. [Google Scholar]
- He, L.; Zhao, J.; Shi, B.J.; Hu, Z.N.; Wu, W.J. Effects of insecticidal fraction F3-28 from Periploca sepium on the activities of digestive enzymes in the midgut of larvae of Mythimna separata and Agrotis ipsilon (Lepidoptera: Noctuidae). Acta Entomol. Sin. 2010, 53, 1248–1255. [Google Scholar]
- Shi, B.J.; Gao, L.T.; Ji, Z.Q.; Zhang, J.W.; Wu, W.J.; Hu, Z.N. Isolation of the insecticidal ingredients from Periploca sepium. Chin. J. Pestic. Sci. 2012, 1, 103–106. [Google Scholar]
- Feng, M.X.; Shi, B.J.; Zhao, Y.C.; Wu, W.J.; Hu, Z.N. Histopathological effects and immunolocalization of periplocoside NW from Periploca sepium Bunge on the midgut epithelium of Mythimna separata Walker larvae. Pestic. Biochem. Physiol. 2014, 115, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.X.; Zhao, J.; Zhang, J.W.; Wu, W.J.; Hu, Z.N. Fluorescence localization and comparative ultrastructural study of periplocoside NW from Periploca sepium Bunge in the midgut of the Oriental Amyworm, Mythimna separata Walker (Lepidoptera: Noctuidae). Toxins 2014, 6, 1575–1585. [Google Scholar] [CrossRef] [PubMed]
- Candas, M.; Loseva, O.; Oppert, B.; Kosaraju, P.; Bulla, L.A. Insect resistance to Bacillus thuringiensis: Alterations in the indianmeal moth larval gut proteome. Mol. Cell. Proteom. 2003, 2, 19–28. [Google Scholar] [CrossRef]
- Mcnall, R.J.; Adang, M.J. Identification of novel Bacillus thuringiensis Cry1Ac binding proteins in Manduca sexta midgut through proteomic analysis. Insect Biochem. Mol. Biol. 2003, 33, 999–1010. [Google Scholar] [CrossRef]
- Xia, J.X.; Guo, Z.J.; Yang, Z.Z.; Zhu, X.; Kang, S.; Yang, X.; Yang, F.S.; Wu, Q.J.; Wang, S.L.; Xie, W.; et al. Proteomics-based identification of midgut proteins correlated with Cry1Ac resistance in Plutella xylostella (L.). Pestic. Biochem. Physiol. 2016, 132, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Ding, X.; Xia, L.; Huang, S.; Huang, F. Proteomic analysis of BBMV in Helicoverpa armigera midgut with and without Cry1Ac toxin treatment. Biocontrol Sci. Technol. 2011, 21, 139–151. [Google Scholar] [CrossRef]
- Shi, B.J.; Zhang, J.W.; Gao, L.T.; Chen, C.C.; Ji, Z.Q.; Hu, Z.N.; Wu, W.J. A new pregnane glycoside from the root barks of Periploca sepium. Chem. Nat. Compd. 2014, 49, 1043–1047. [Google Scholar] [CrossRef]
- Lu, L.N.; Qi, Z.J.; Zhang, J.W.; Wu, W.J. Separation of Binding Protein of Celangulin V from the Midgut of Mythimna separata Walker by Affinity Chromatography. Toxins 2015, 7, 1738–1748. [Google Scholar] [CrossRef] [PubMed]
- Wolfersberger, M.G.; Luethy, P.; Maurer, A.; Parenti, P.; Sacchi, F.V.; Giordana, B.; Hanozet, G.M. Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the cabbage butterfly (Pieris brassicae). Comp. Biochem. Physiol. 1987, 86, 301–308. [Google Scholar] [CrossRef]
- Viswanathan, S.; Ünlü, M.; Minden, J.S. Two-dimensional difference gel electrophoresis. Nat. Protoc. 2006, 1, 1351–1358. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Tomas, H.; Havlis, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef] [PubMed]
- Hafkenscheid, J.C.M. Aminopeptidases and amino acid arylamidases. Method Enzym. Anal. 1984, 5, 32–34. [Google Scholar]
- Lowry, O.H.; Roberts, N.R.; Wu, M.; Hixon, W.S.; Crawford, E.J. The quantitative histochemistry of brain; II enzyme measurements. J. Biol. Chem. 1954, 207, 19–37. [Google Scholar] [PubMed]
- Tiburcy, F.; Beyenbach, K.W.; Wieczorek, H. Protein kinase A-dependent and-independent activation of the V-ATPase in malpighian tubules of Aedes aegypti. J. Exp. Biol. 2013, 216, 881–891. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, H.; Cioffi, M.; Klein, U.; Harvey, W.R.; Schweikl, H.; Wolfersberger, M.G. Isolation of goblet cell apical membrane from tobacco hornworm midgut and purification of its vacuolar-type ATPase. Method Enzymol. 1990, 192, 608–616. [Google Scholar]
- Wolfersberger, M.G. Enzymology of plasma membrane of insect instestinal cells. Am. Zool. 1984, 24, 187–197. [Google Scholar] [CrossRef]
- Bowman, E.J.; Siebers, A.; Altendorf, K. Bafilomycins: A class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc. Nat. Acad. Sci. USA 1988, 85, 7972–7976. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Feng, Y.; Forgac, M. Proton conduction and bafilomycin binding by the V0 domain of the coated vesicle V-ATPase. J. Biol. Chem. 1994, 269, 23518–23523. [Google Scholar] [PubMed]
- Scapin, G.; Patel, D.; Arnold, E. Multifaceted Roles of Crystallography in Modern Drug Discovery; NATO Science for Peace and Security; Springer: Dordrecht, The Netherlands, 2015; pp. 107–114. [Google Scholar]
- Johnson, A.T. A Prospective Method to Guide Small Molecule Drug Design. J. Chem. Educ. 2015, 92, 836–842. [Google Scholar] [CrossRef]
- Colangelo, T.; Polcaro, G.; Ziccardi, P.; Pucci, B.; Muccillo, L.; Galgani, M. Proteomic screening identifies calreticulin as a miR-27a direct target repressing MHC class I cell surface exposure in colorectal cancer. Cell Death Dis. 2016, 7, e2120. [Google Scholar] [CrossRef] [PubMed]
- Schneider, S.M.; Zuhl, M.N. HSP72 Up-regulation with heat acclimation. Temp. Multidiscip. Biomed. J. 2016, 3, 28–30. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Binder, R.; Ramalingam, T. CD91 is a common receptor for heat shock proteins gp96, hsp90, hsp70, and calreticulin. Immunity 2001, 14, 303–313. [Google Scholar] [CrossRef]
- Grover, S.; Kaur, S.; Gupta, A.K.; Taggar, G.K.; Kaur, J. Characterization of Trypsin Like Protease from Helicoverpa armigera, (Hubner) and Its Potential Inhibitors. Proc. Natl. Acad. Sci. India 2016, 1–8. [Google Scholar] [CrossRef]
- Zuo, J.N. Cloning of the cDNA of Trypsin and Prokaryotic Expression from Midgut of Mythimna Separata and Activities Effects of Perilocosides Compounds on Trypsin; Northwest A&F University: Xianyang, China, 2014. [Google Scholar]
- Salahuddin, P. Protein disulfide isomerase: Structure, mechanism of oxidative protein folding and multiple functional roles. J. Biochem. Mol. Biol. Res. 2016, 2, 173–179. [Google Scholar]
- Feng, M.X.; He, Z.Y.; Wang, Y.Y.; Yan, X.F.; Wu, W.J.; Hu, Z.N. Isolation of the binding protein of Periplocoside E from BBMVs in midgut of the Oriental amyworm Mythimna separata Walker (Lepidoptera: Noctuidae) through affinity chromatography. Toxins 2016, 8, 139. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, H.; Huss, M.; Merzendorfer, H.; Reineke, S.; Vitavska, O.; Zeiske, W. The insect plasma membrane H+ V-ATPase: Intra-, inter-, and supramolecular aspects. J. Bioenerg. Biomembr. 2003, 35, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, H.; Grber, G.; Harvey, W.R.; Huss, M.; Merzendorfer, H.; Zeiske, W. Structure and regulation of insect plasma membrane H(+)V-ATPase. J. Exp. Biol. 2000, 203, 127–135. [Google Scholar] [PubMed]
- Marshansky, V.; Futai, M. The V-type H+-ATPase in vesicular trafficking: Targeting, regulation and function. Curr. Opin. Cell Biol. 2008, 20, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Beyenbach, K.W.; Wieczorek, H. The V-type H+ ATPase: Molecular structure and function, physiological roles and regulation. J. Exp. Biol. 2006, 209, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Imamura, H.; Funamoto, S.; Yoshida, M.; Yokoyama, K. Reconstitution in vitro of V1 complex of Thermus thermophilus V-ATPase revealed that ATP binding to the A subunit is crucial for V1 formation. J. Biol. Chem. 2006, 281, 38582–38591. [Google Scholar] [CrossRef] [PubMed]
- Hunke, C.; Chen W, J.; Schäfer, H.J.; Grüber, G. Cloning, purification, and nucleotide-binding traits of the catalytic subunit A of the V-ATPase from Aedes albopictus. Protein Expr. Purif. 2007, 53, 378–383. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Qi, Z.J.; Qi, M.; Wu, W.J.; Hu, Z.N. Effects of Periplocoside P from Periploca sepium on the Midgut Transmembrane Potential of Mythimna separata Larvae. Sci. Rep. 2016, 6, 36982. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.J.; Hine, R.; Beyenbach, K.W.; Perplocoside, P. Inhibits electrolyte and fluid secretion by inhibiting the V-type H+ ATPase in malpighian tubules of the yellow fever mosquito Aedes aegypti. In Proceedings of the 1st International Symposium on Insecticide Toxicology, Guangzhou, China, 5–7 August 2014; p. 300. [Google Scholar]
- Khaitlina, S.Y. Chapter Seven—Tropomyosin as a Regulator of Actin Dynamics. Int. Rev. Cell Mol. Biol. 2015, 318, 255–291. [Google Scholar] [PubMed]
- Johnston, A.M.; Fallon, A.M. Characterization of the ribosomal proteins from mosquito (Aedes albopictus) cells. Eur. J. Biochem. 1985, 150, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Burtenshaw, S.M.; Su, P.P.; Zhang, J.R.; Tobe, S.S.; Dayton, L.; Bendena, W.G. A putative farnesoic acid O-methyltransferase (FAMeT) orthologue in Drosophila melanogaster (CG10527): Relationship to juvenile hormone biosynthesis? Peptides 2008, 29, 242–251. [Google Scholar] [CrossRef] [PubMed]


| Gene | Primer Sequence (5′-3′) |
|---|---|
| β-actin | GGTGTGATGGTTGGTATGGGT |
| TCGTTGTAGAAGGTGTGGTGC | |
| vma1 | TATCCTGGGCTCCATCTTTG |
| TTGATGCTCAATGGGTTGAA |
| Protein Sample | Concentration (mg/mL) | APN Activity (μmol/min·mL) | ALP Activity (μmol/min·mL) |
|---|---|---|---|
| BBMV | 0.309 | 28.169 | 11.747 |
| Crude enzyme fluid | 1.327 | 3.947 | 1.829 |
| Spot No. | Protein Name | Accession No. | Matched Peptide Sequences (m/z) | Score | Relative Molecular Mass (Mr) | Protein Isoelectric Point (pI) | Sequence Coverage (%) | Fold Change (t-Test p < 0.05) |
|---|---|---|---|---|---|---|---|---|
| 1 | Transferin | gi|556559879 | HIQALECLR (1139.599) | 79 | 84,436.6 | 5.79 | 4 | +1.9 |
| ETAAAQENITR (1203.5964) | ||||||||
| ASAYTLGIQPAISCQQR (1863.9382) | ||||||||
| 2 | Protein disulfide isomerase precursor | gi|112984454 | NFEEKR (822.4104) | 109 | 55782.2 | 4.6 | 11 | NA |
| QLVPIYDK (975.551) | ||||||||
| LAEEESPIK (1015.5306) | ||||||||
| GYPTLKFFR (1128.6201) | ||||||||
| LIALEQDMAK (1147.6028) | ||||||||
| LAEEESPIKLAK (1327.7467) | ||||||||
| 3 | Calreticulin | gi|389608333 | AVGEEVKK (859.4883) | 50 | 46,299.2 | 4.48 | 2.7 | +3.42 |
| VHVIFSYK (992.5564) | ||||||||
| KVHVIFSYK (1120.6514) | ||||||||
| DAGAIAGLNVMR (1491.6533) | ||||||||
| VESGELEADWDFLPPKK (1959.9698) | ||||||||
| 11 | Tropomyosin-2 isoform 4 | gi|153792609 | LIAEESDK (904.4622) | 205 | 29,623 | 4.77 | 4.257 | –1.93 |
| EAEARAEFAER (1278.6073) | ||||||||
| LSEASQAADESER (1392.6238) | ||||||||
| EEAESEVAALNRR (1473.7292) | ||||||||
| LSEASQAADESERIR (1661.809) | ||||||||
| IQLLEEDLERSEER (1758.8868) | ||||||||
| LLQEEMEATLHDIQNM (1946.8834) | ||||||||
| TNMEDDRVAILEAQLSQAK (2148.0601) | ||||||||
| 12 | V-type ATPase, A subunit | gi|71410785 | VIVVIPFILLGIP (1392.923) | 86 | 67,985.8 | 5.6 | 1.213 | +3.56 |
| MLFVPLGLGQWQLLR (1771.0088) | ||||||||
| ESGNHPLLTGQR (2002.0215) | ||||||||
| 13 | Diverged serine protease | gi|2463066 | EDMQPLMQDNSD (1438.5461) | 98 | 27,320.9 | 5.70 | 0.42 | +2.56 |
| VWLQTCVGSVLTSR (1728.9491) | ||||||||
| 14 | Trypsin-like protease | gi|2463084 | AAVTISSR (804.4574) | 107 | 26,948.1 | 9.35 | 5.776 | +2.18 |
| EVPKSEVK (915.5145) | ||||||||
| ALVSFKIDDK (1135.6357) | ||||||||
| ELNSLQEKGSK (1232.6481) | ||||||||
| EVVVKEWYIK (1292.725) | ||||||||
| 15 | 39S ribosomal protein L46, Mitochondrial-like | gi|512919884 | SGNPTKSK (818.4366) | 135 | 30,310.5 | 8.51 | 2.435 | −1.91 |
| QTAERIVK (944.5523) | ||||||||
| YKYPSEMNGK (1232.5616) | ||||||||
| SNHEIQHENDK (1350.6033) | ||||||||
| IFFYYANYKSGNPTK (1812.8956) | ||||||||
| LGNDSKTLLPQGHWQEGETLR (2379.2051) | ||||||||
| 16 | Farnesoic acid O-methyltransferase | gi|528079470 | SIPPGALR (810.4832) | 30 | 12,599.4 | 9.05 | 1.382 | −1.54 |
| VNHDGCTTPGK (1185.5317) | ||||||||
| SSAEYECLVLM (1317.5702) | ||||||||
| KLLSIGDEVNEAVSSMC (1867.8776) | ||||||||
| 18 | Aminopeptidase N1 | gi|34100664 | ELAEQEK (846.4203) | 154 | 112,775.9 | 5.60 | 6.142 | +3.37 |
| IVEDKNR (873.4788) | ||||||||
| MNIGNIEAR (1033.5095) | ||||||||
| FSKCLMDCR (1232.5221) | ||||||||
| DMLSSLNQEESLK (1493.7152) | ||||||||
| 20 | Heat shock protein cognate 72 | gi|157658 | RGGECAR (805.3733) | 91 | 72,234.9 | 5.22 | 1.073 | +2.33 |
| YCHRNYGGSK (1241.5481) | ||||||||
| CPAGLGGNHCEVGRR (1639.754) | ||||||||
| DIRAEDQTQECNMGDA (1868.7385) | ||||||||
| FSGISMQVFAIVNGNISPYVLDPNFSHK (3097.5452) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, M.; Li, Y.; Chen, X.; Wei, Q.; Wu, W.; Hu, Z. Comparative Proteomic Analysis of the Effect of Periplocoside P from Periploca sepium on Brush Border Membrane Vesicles in Midgut Epithelium of Mythimna separata Larvae. Toxins 2018, 10, 7. https://doi.org/10.3390/toxins10010007
Feng M, Li Y, Chen X, Wei Q, Wu W, Hu Z. Comparative Proteomic Analysis of the Effect of Periplocoside P from Periploca sepium on Brush Border Membrane Vesicles in Midgut Epithelium of Mythimna separata Larvae. Toxins. 2018; 10(1):7. https://doi.org/10.3390/toxins10010007
Chicago/Turabian StyleFeng, Mingxing, Yankai Li, Xueting Chen, Quansheng Wei, Wenjun Wu, and Zhaonong Hu. 2018. "Comparative Proteomic Analysis of the Effect of Periplocoside P from Periploca sepium on Brush Border Membrane Vesicles in Midgut Epithelium of Mythimna separata Larvae" Toxins 10, no. 1: 7. https://doi.org/10.3390/toxins10010007