Automatic Forest Mapping at Individual Tree Levels from Terrestrial Laser Scanning Point Clouds with a Hierarchical Minimum Cut Method
Abstract
:1. Introduction
- precisely segment the crown points to the corresponding tree trunks in boreal coniferous forest plots with heterogeneous structures;
- implement a hierarchical strategy to isolate single trees from point clouds reliably; and
- estimate structure metrics at the individual tree level.
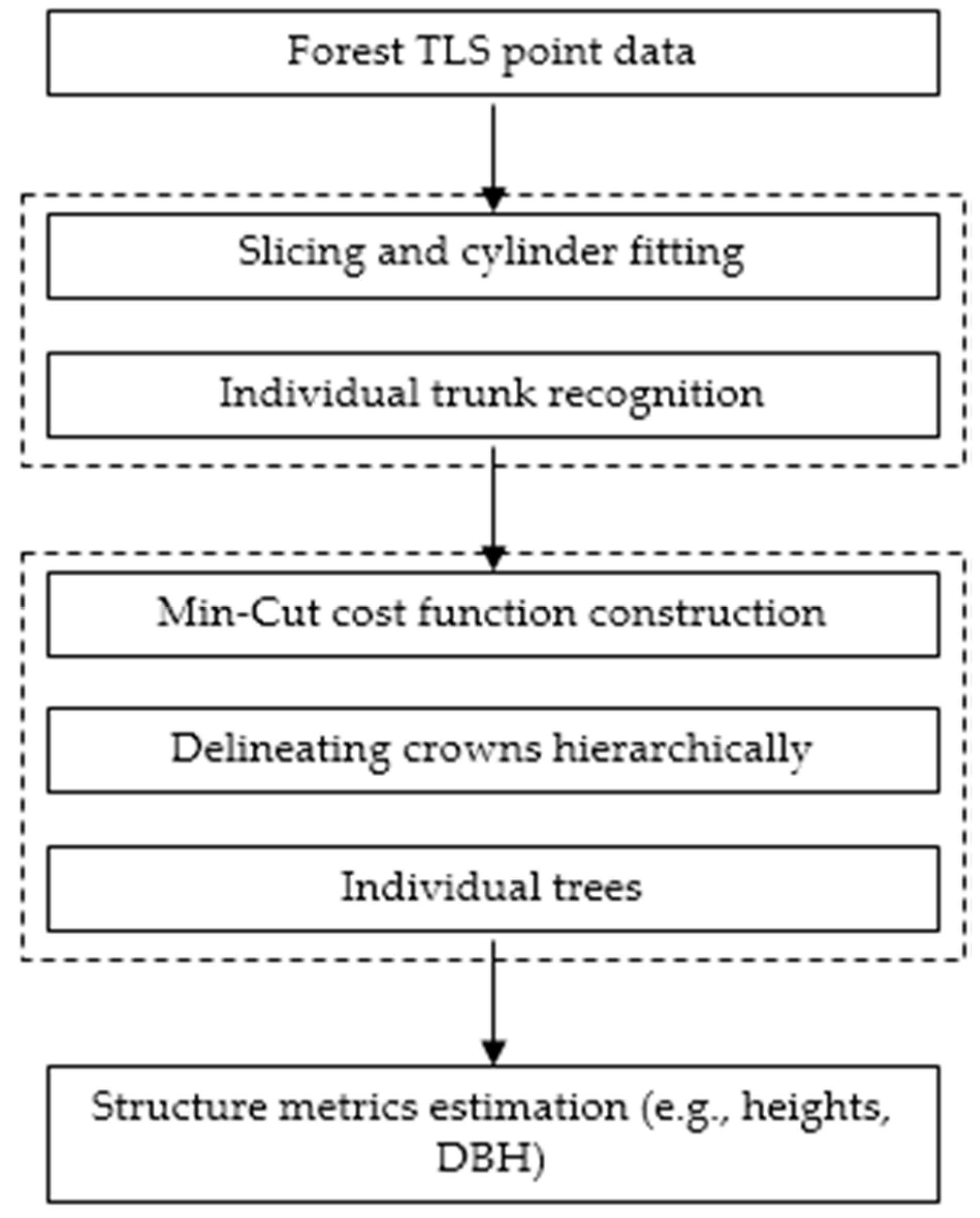
2. Methodology
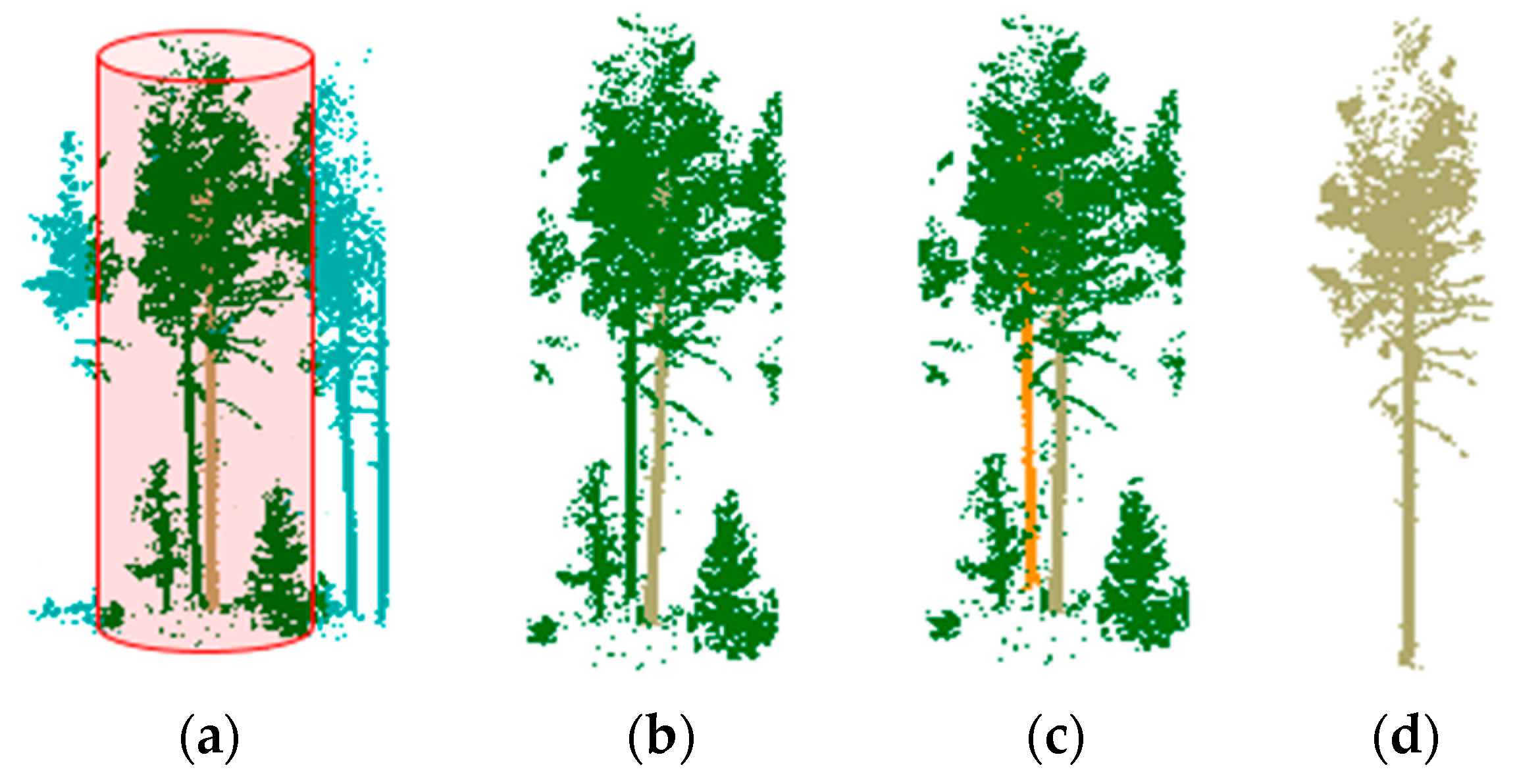
2.1. Localization of Candidate Trees
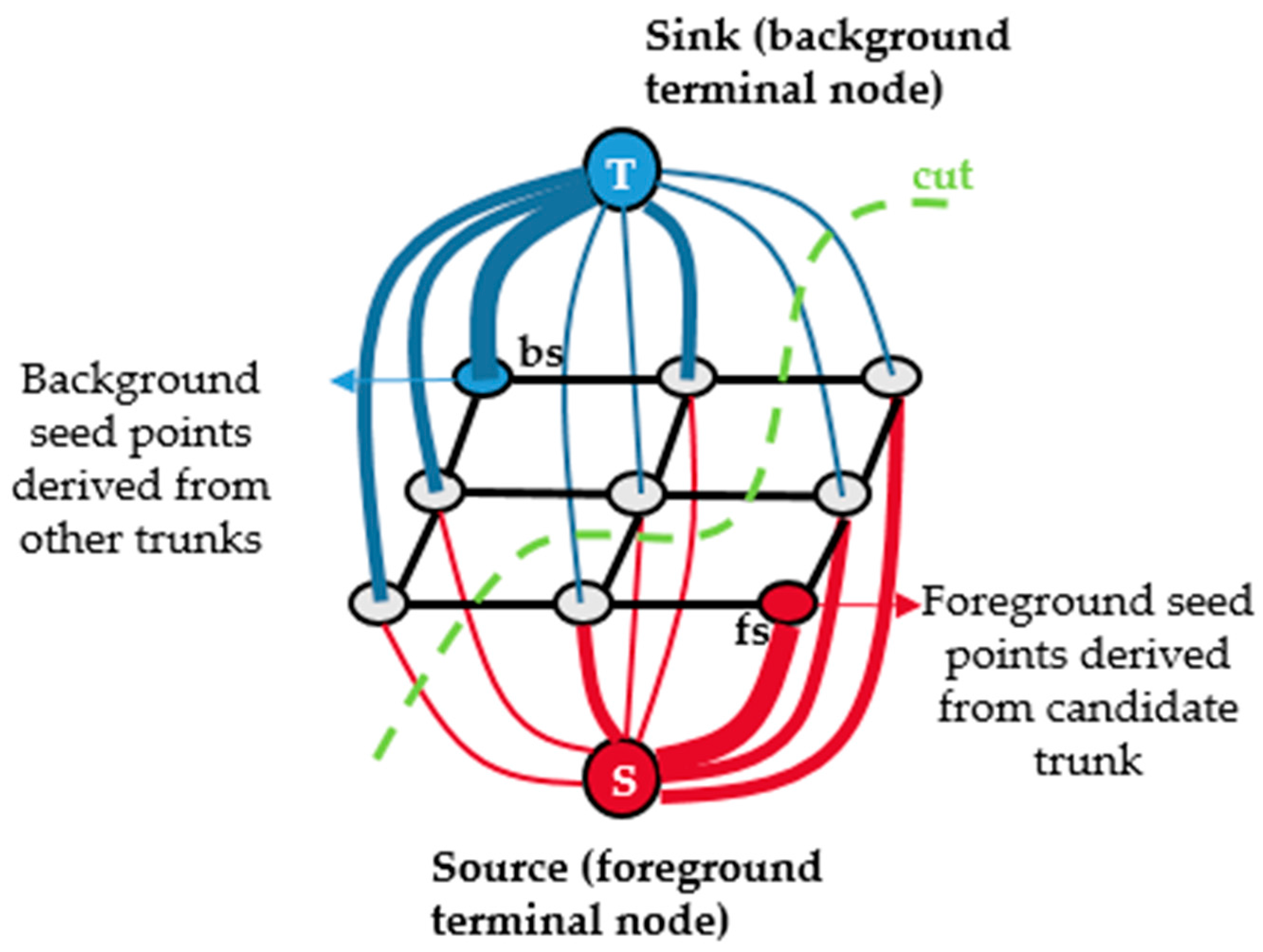
2.2. Isolating Tree Crown Points with the Hierarchical Minimum Cut Operator
2.3. Estimating Structure Metrics at the Tree Level
3. Results and Analysis
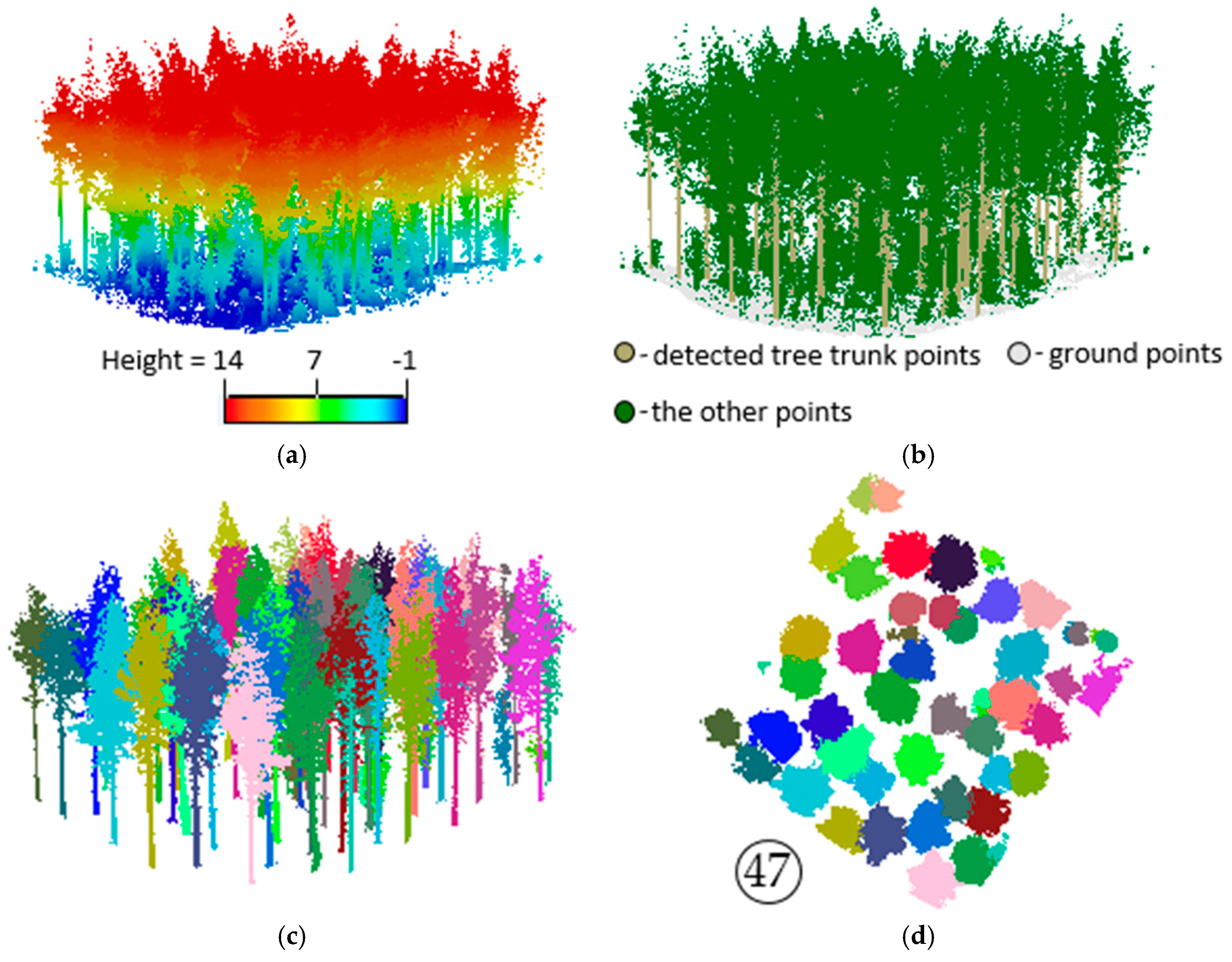
3.1. Data Description
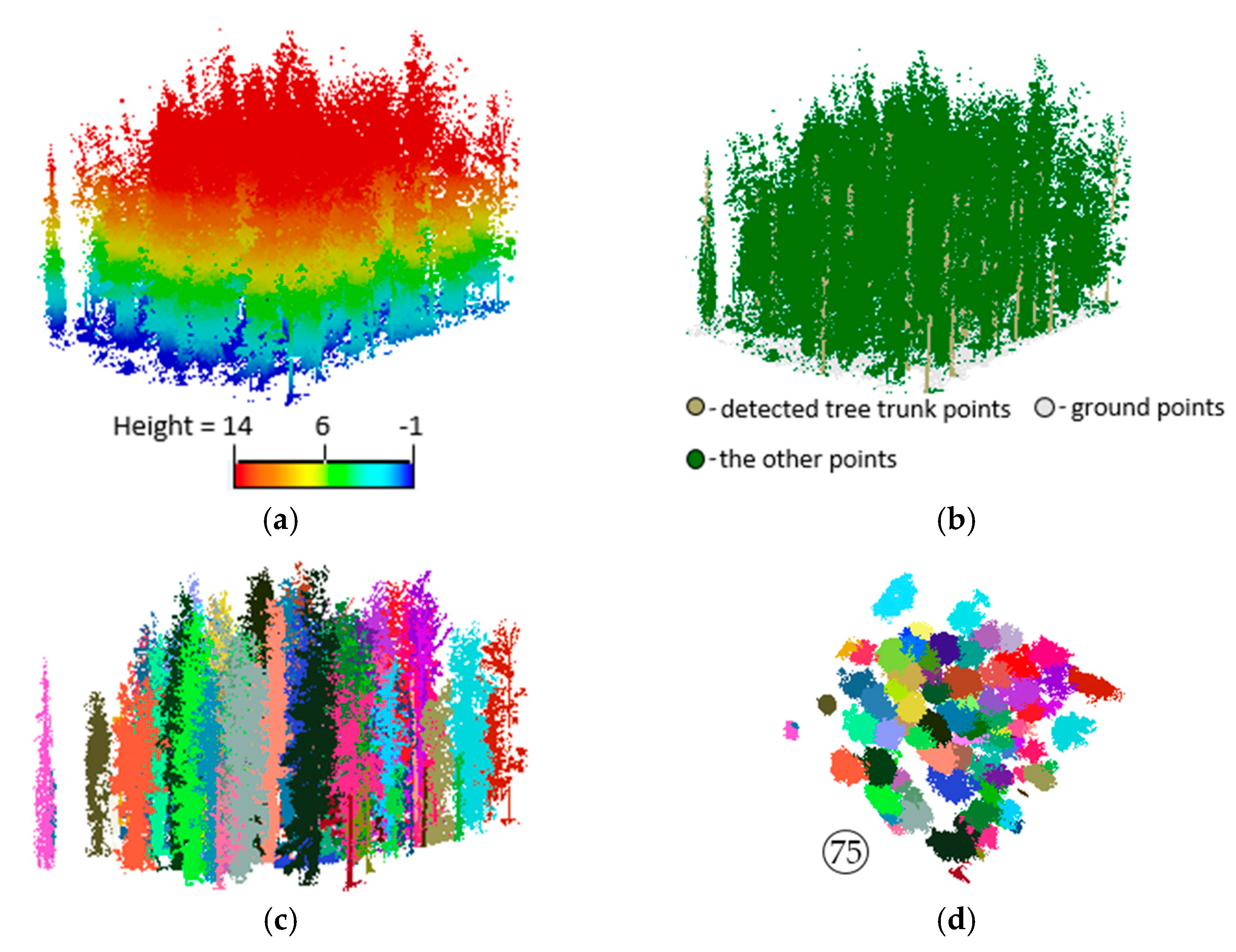
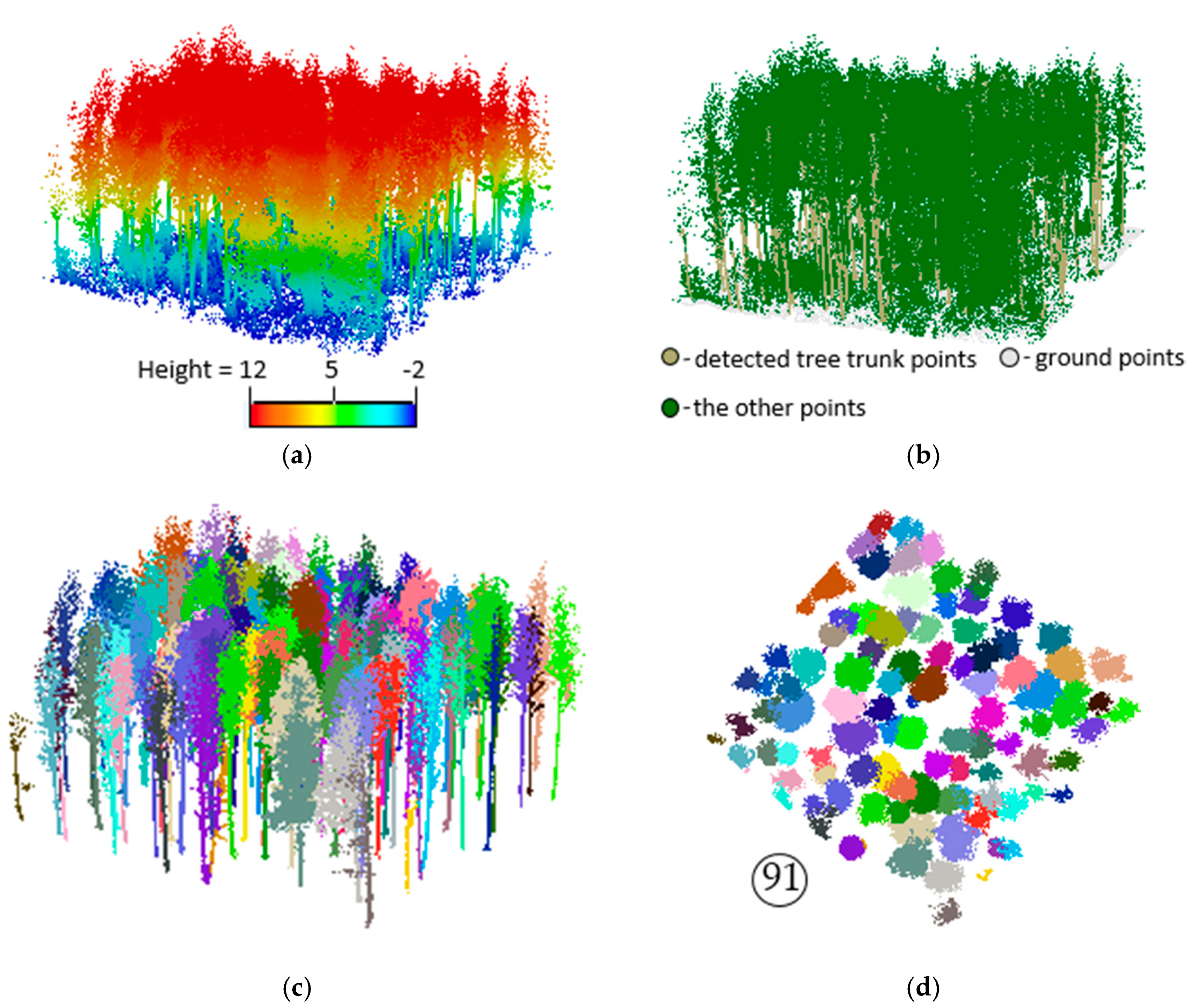
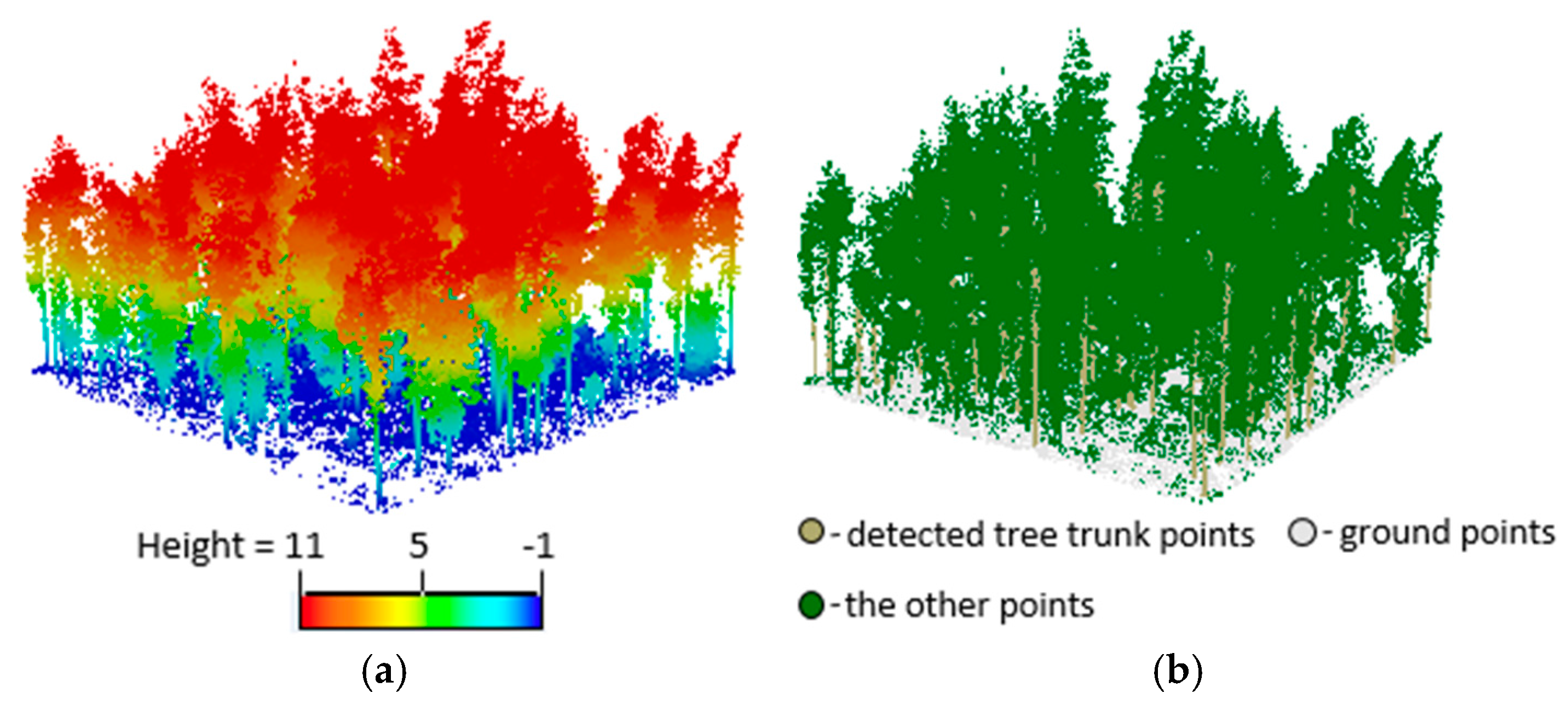
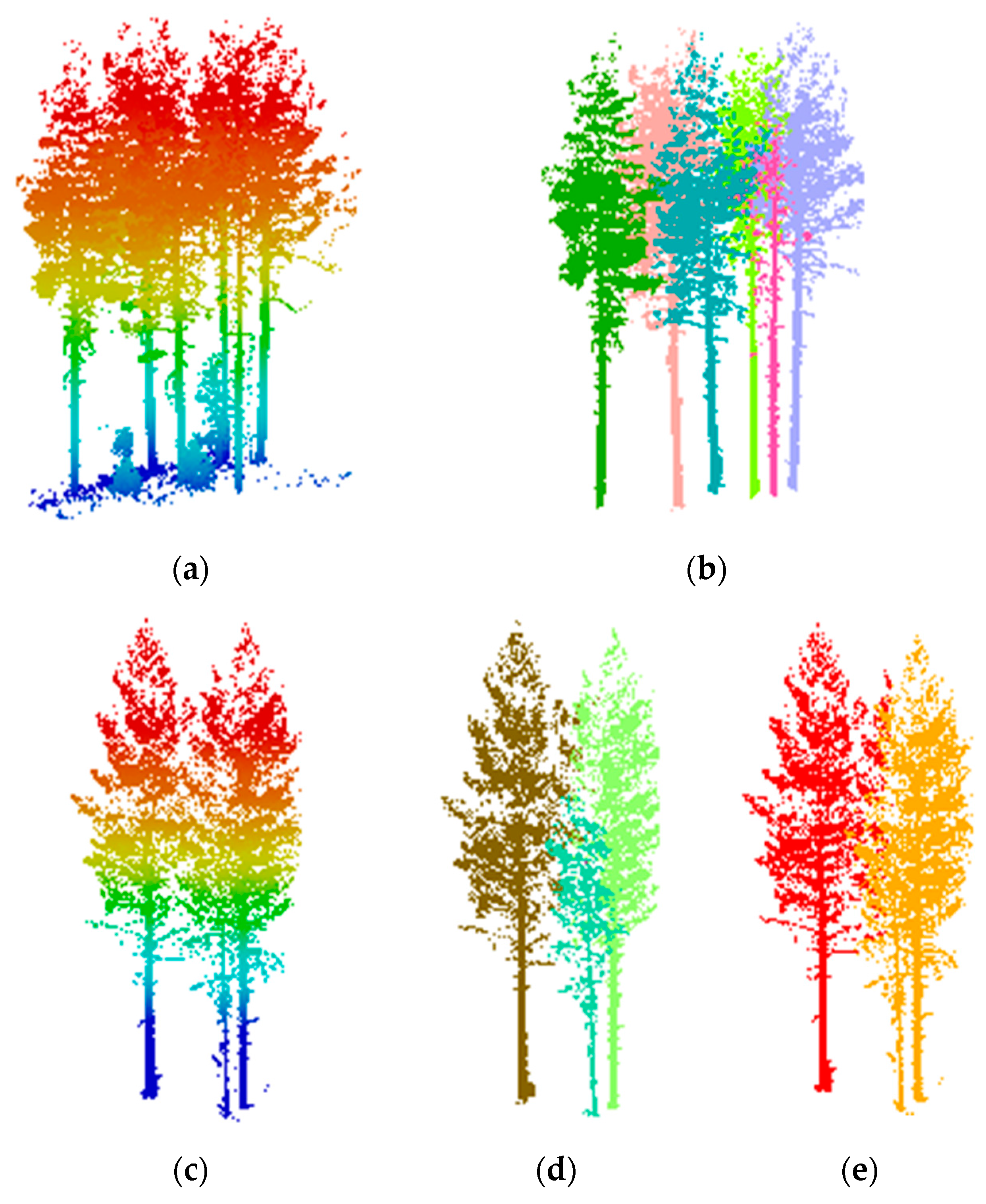
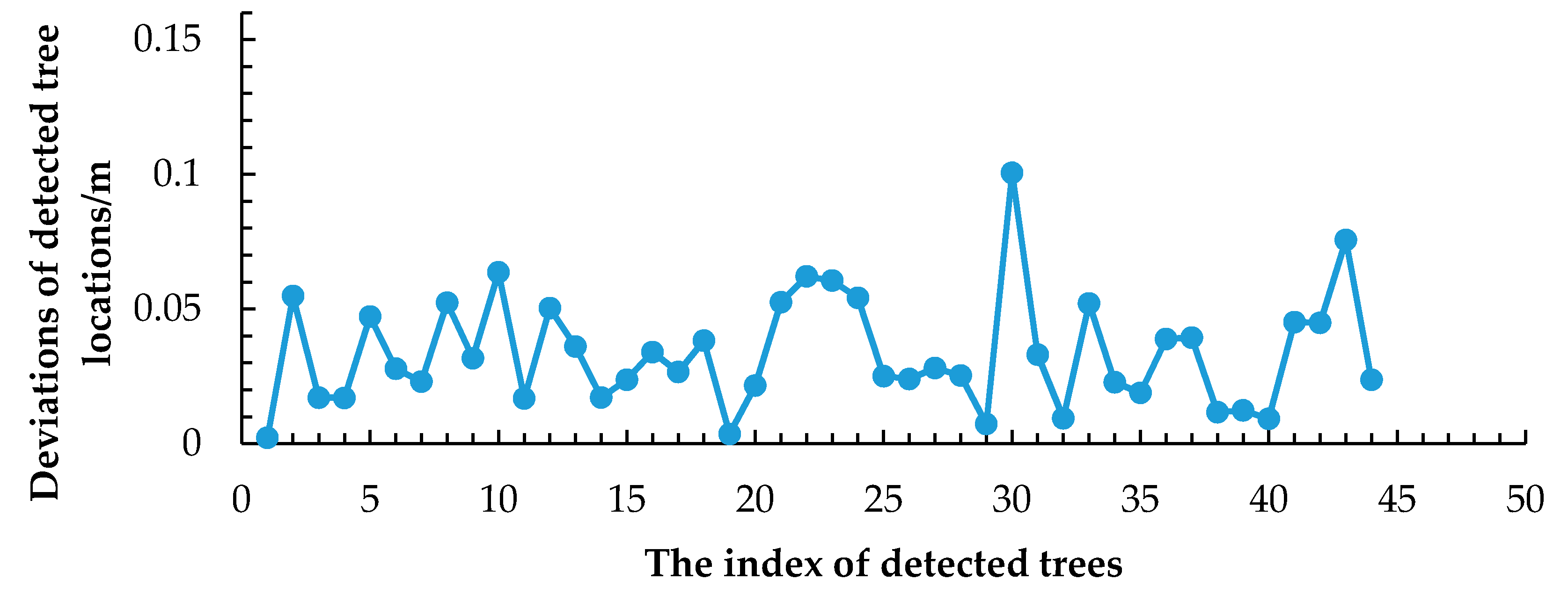
3.2. Extracting Results of Individual Trees
3.3. Accuracy Assessment and Evaluation of the Proposed Method
- TP (true positive): the number of trees detected with correct ground positions.
- FP (false positive): when the trunk is not detected and missed and the corresponding tree crown is allocated to the neighboring trees.
- FN (false negative): the number of ground truth trees undetected.
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix


References
- Hyyppä, J.; Kelle, O.; Lehikoinen, M.; Inkinen, M. A segmentation-based method to retrieve stem volume estimates from 3-D tree height models produced by laser scanners. IEEE Trans. Geosci. Remote Sens. 2001, 39, 969–975. [Google Scholar] [CrossRef]
- He, Q.; Chen, E.; An, R.; Li, Y. Above-ground biomass and biomass components estimation using LiDAR data in a coniferous forest. Forests 2013, 4, 984–1002. [Google Scholar] [CrossRef]
- Yang, X.; Strahler, A.H.; Schaaf, C.B.; Jupp, D.L.B.; Yao, T.; Zhao, F.; Wang, Z.; Culvenor, D.S.; Newnham, G.J.; Lovell, J.L.; et al. Three-dimensional forest reconstruction and structural parameter retrievals using a terrestrial full-waveform LiDAR instrument (Echidna®). Remote Sens. Environ. 2013, 135, 36–51. [Google Scholar] [CrossRef]
- Lin, Y.; Hyyppä, J.; Jaakkola, A.; Yu, X. Three-level frame and RD-schematic algorithm for automatic detection of individual trees from MLS point clouds. Int. J. Remote Sens. 2012, 33, 1701–1716. [Google Scholar] [CrossRef]
- Zhang, C.; Zhou, Y.; Qiu, F. Individual tree segmentation from LiDAR point clouds for urban forest inventory. Remote Sens. 2015, 7, 7892–7913. [Google Scholar] [CrossRef]
- Unger, D.R.; Hung, I.K.; Brooks, R.; Williams, H. Estimating number of trees, tree height and crown width using LiDAR data. GISci Remote Sens. 2014, 51, 227–238. [Google Scholar] [CrossRef]
- Srinivasan, S.; Popescu, S.C.; Eriksson, M.; Sheridan, R.D.; Ku, N.W. Terrestrial Laser Scanning as an effective tool to retrieve tree level height, crown width, and stem diameter. Remote Sens. 2015, 7, 1877–1896. [Google Scholar] [CrossRef]
- Henning, J.G.; Radtke, P.J. Detailed stem measurements of standing trees from ground-based scanning LiDAR. For. Sci. 2006, 52, 67–80. [Google Scholar]
- Liang, X.; Hyyppä, J.; Kankare, V.; Holopainen, M. Stem curve measurement using terrestrial laser scanning. In Proceedings of the 11th International Conference on LiDAR Applications for Assessing Forest Ecosystems, SilviLaser 2011, Hobart, Tasmania, Australia, 16–20 October 2011.
- Strîmbu, V.F.; Strîmbu, B.M. A graph-based segmentation algorithm for tree crown extraction using airborne LiDAR data. ISPRS J. Photogramm. Remote Sens. 2015, 104, 30–43. [Google Scholar] [CrossRef]
- Mongus, D.; Zalik, B. An efficient approach to 3D single tree-crown delineation in LiDAR data. ISPRS J. Photogramm. Remote Sens. 2015, 108, 219–233. [Google Scholar] [CrossRef]
- Lee, H.; Slatton, K.C.; Roth, B.E.; Cropper, W.P., Jr. Adaptive clustering of airborne LiDAR data to segment individual tree crowns in managed pine forests. Int. J. Remote Sens. 2010, 31, 117–139. [Google Scholar] [CrossRef]
- Popescu, S.C.; Zhao, K. A voxel-based LiDAR method for estimating crown base height for deciduous and pine trees. Remote Sens. Environ. 2008, 112, 767–781. [Google Scholar] [CrossRef]
- Holmgren, J.; Persson, A. Identifying species of individual trees using airborne laser scanner. Remote Sens. Environ. 2004, 90, 415–423. [Google Scholar] [CrossRef]
- Estornell, J.; Velázquez-Martí, B.; López-Cortés, I.; Salazar, D.; Fernández-Sarría, A. Estimation of wood volume and height of olive tree plantations using airborne discrete-return LiDAR data. GISci Remote Sens. 2014, 51, 17–29. [Google Scholar] [CrossRef]
- Hecht, R.; Meinel, G.; Buchroithner, M.F. Estimation of urban green volume based on single-pulse LiDAR data. IEEE Trans. Geosci. Remote Sens. 2008, 46, 3832–3840. [Google Scholar] [CrossRef]
- Lee, S.J.; Kim, J.R.; Choi, Y.S. The extraction of forest CO2 storage capacity using high-resolution airborne LiDAR data. GISci Remote Sens. 2013, 50, 154–171. [Google Scholar]
- Lin, Y.; Jaakkola, A.; Hyyppä, J.; Kaartinen, H. From TLS to VLS: Biomass estimation at individual tree level. Remote Sens. 2010, 2, 1864–1879. [Google Scholar] [CrossRef]
- Olsoy, P.J.; Glenn, N.F.; Clark, P.E.; Derryberry, D.R. Above ground total and green biomass of dryland shrub derived from terrestrial laser scanning. ISPRS J. Photogramm. Remote Sens. 2014, 88, 166–173. [Google Scholar] [CrossRef]
- Seidel, D.; Albert, K.; Ammer, C.; Fehrmann, L.; Kleinn, C. Using terrestrial laser scanning to support biomass estimation in densely stocked young tree plantations. Int. J. Remote Sens. 2013, 34, 8699–8709. [Google Scholar] [CrossRef]
- Rahman, M.Z.A.; Gorte, B.G.H.; Bucksch, A.K. A new method for individual tree delineation and undergrowth removal from high resolution airborne LiDAR. In Proceedings of the ISPRS Workshop on Laser Scanning 2009, Paris, France, 1–2 September 2009.
- Dassot, M.; Constant, T.; Fournier, M. The use of terrestrial LiDAR technology in forest science: Application fields, benefits and challenges. Ann. For. Sci. 2011, 68, 959–974. [Google Scholar] [CrossRef]
- Yu, X.; Hyyppä, J.; Vastaranta, M.; Holopainen, M.; Viitala, R. Predicting individual tree attributes from airborne laser point clouds based on the random forests technique. ISPRS J. Photogramm. Remote Sens. 2011, 66, 28–37. [Google Scholar] [CrossRef]
- Alonzo, M.; Bookhagen, B.; Roberts, D.A. Urban tree species mapping using hyperspectral and LiDAR data fusion. Remote Sens. Environ. 2014, 148, 70–83. [Google Scholar] [CrossRef]
- Chen, Q.; Baldocchi, D.; Gong, P.; Kelly, M. Isolating individual trees in a Savanna woodland using small footprint LiDAR data. Photogramm. Eng. Remote Sens. 2006, 72, 923–932. [Google Scholar] [CrossRef]
- Tao, S.; Wu, F.; Guo, Q.; Wang, Y.; Li, W.; Xue, B.; Hu, X.; Li, P.; Tian, D.; Li, C.; et al. Segmenting tree crowns from terrestrial and mobile LiDAR data by exploring ecological theories. ISPRS J. Photogramm. Remote Sens. 2015, 110, 66–76. [Google Scholar] [CrossRef]
- Liang, X.; Litkey, P.; Hyyppä, J.; Kaartinen, H.; Vastaranta, M.; Holopainen, M. Automatic stem mapping using single-scan terrestrial laser scanning. IEEE Trans. Geosci. Remote Sens. 2012, 50, 661–670. [Google Scholar] [CrossRef]
- Arachchige, N.H. Automatic tree stem detection-a geometric feature based approach for MLS point clouds. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2013, 1, 109–114. [Google Scholar] [CrossRef]
- Xia, S.; Wang, C.; Pan, F.; Xi, X.; Zeng, H.; Liu, H. Detecting stems in dense and homogeneous forest using single-scan TLS. Forests 2015, 6, 3923–3945. [Google Scholar] [CrossRef]
- Lalonde, J.F.; Vandapel, N.; Huber, D.F.; Hebert, M. Natural terrain classification using three-dimensional ladar data for ground robot mobility. J. Field Robot. 2006, 23, 839–862. [Google Scholar] [CrossRef]
- Rutzinger, M.; Pratihast, A.K.; Elberink, S.O.; Vosselman, G. Detection and modelling of 3D trees from mobile laser scanning data. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2010, 38, 520–525. [Google Scholar]
- Olofsson, K.; Holmgren, J.; Olsson, H. Tree stem and height measurements using Terrestrial Laser Scanning and the RANSAC algorithm. Remote Sens. 2014, 6, 4323–4344. [Google Scholar] [CrossRef]
- Sirmacek, B.; Lindenbergh, R. Automatic classification of trees from laser scanning point clouds. In Proceedings of the International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences, ISPRS Geospatial Week 2015, La Grande Motte, France, 28 September–3 October 2015.
- Hernández, J.; Marcotegui, B. Point cloud segmentation towards urban ground modeling. In Proceedings of the 5th GRSS, ISPRS Joint Workshop on Remote Sensing and Data Fusion over Urban Areas, Shanghai, China, 20–22 May 2009.
- Boykov, Y.; Funka-Lea, G. Graph cuts and efficient ND image segmentation. Int. J. Comput. Vis. 2006, 70, 109–131. [Google Scholar] [CrossRef]
- Boykov, Y.Y.; Jolly, M.P. Interactive graph cuts for optimal boundary & region segmentation of objects in ND images. In Proceedings of the International Conference on Computer Vision, Vancouver, BC, Canada, 7–14 July 2001.
- Golovinskiy, A.; Funkhouser, T. Minimum cut based segmentation of point clouds. In Proceedings of the 2009 IEEE 12th International Conference Computer Vision Workshops (ICCV Workshops), Kyoto, Japan, 27 September–4 October 2009.
- Oesau, S.; Lafarge, F.; Alliez, P. Indoor scene reconstruction using feature sensitive primitive extraction and graph-cut. ISPRS J. Photogramm. Remote Sens. 2014, 90, 68–82. [Google Scholar] [CrossRef]
- Lindenbergh, R C.; Berthold, D.; Sirmacek, B.; Wang, J.; Ebersbach, D. Automated large scale parameter extraction of road-side trees sampled by a laser mobile mapping system. In Proceedings of the International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences, ISPRS Geospatial Week 2015, La Grande Motte, France, 28 September–3 October 2015.





| Plot | Plot 1 | Plot 2 | Plot 3 | Plot 4 | Plot 5 |
|---|---|---|---|---|---|
| Acquisition time | Summer, 2014 | ||||
| Instrument | Leica HDS6100 | ||||
| Field of view | 360° × 310° | ||||
| Beam divergence | 0.22 mrad | ||||
| Increment | 0.036° (horizontal)/0.036° (vertical) | ||||
| Plot size | 32 m × 32 m | ||||
| Number of points (million) | 47.18 | 68.11 | 32.87 | 50.42 | 69.12 |
| Point density (points/cm2) | 4.9 | 7.3 | 3.5 | 5.2 | 7.7 |
| Number of trees | 52 | 80 | 95 | 67 | 59 |
| Parameter | Descriptor | Value |
|---|---|---|
| Hslice | Thickness of horizontal slice | 0.2 m |
| rc_min | Min radius of cylinder in trunk point extraction | 0.05 m |
| rc_max | Max radius of cylinder in trunk point extraction | 0.5 m |
| σ | The average point span in Bp,q calculation | 1.0 |
| k | Constant value to determine the buffer zone as the input point cloud of the hierarchical minimum cut | 1.25 |
| Weight(p,S) | Weight of edges’ t-links connecting input points to terminal S | 0.8 |
| Plot | TP | Reference Trees | FP | FN | Recall | Precision | F-Measure |
|---|---|---|---|---|---|---|---|
| 1 | 47 | 52 | 5 | 5 | 90.38% | 90.38% | 90.38% |
| 2 | 75 | 80 | 7 | 5 | 93.75% | 91.46% | 92.59% |
| 3 | 91 | 95 | 9 | 4 | 95.79% | 91.00% | 93.33% |
| 4 | 62 | 67 | 6 | 5 | 92.54% | 91.18% | 91.85% |
| 5 | 47 | 59 | 7 | 12 | 79.66% | 87.04% | 83.19% |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, B.; Dai, W.; Dong, Z.; Liu, Y. Automatic Forest Mapping at Individual Tree Levels from Terrestrial Laser Scanning Point Clouds with a Hierarchical Minimum Cut Method. Remote Sens. 2016, 8, 372. https://doi.org/10.3390/rs8050372
Yang B, Dai W, Dong Z, Liu Y. Automatic Forest Mapping at Individual Tree Levels from Terrestrial Laser Scanning Point Clouds with a Hierarchical Minimum Cut Method. Remote Sensing. 2016; 8(5):372. https://doi.org/10.3390/rs8050372
Chicago/Turabian StyleYang, Bisheng, Wenxia Dai, Zhen Dong, and Yang Liu. 2016. "Automatic Forest Mapping at Individual Tree Levels from Terrestrial Laser Scanning Point Clouds with a Hierarchical Minimum Cut Method" Remote Sensing 8, no. 5: 372. https://doi.org/10.3390/rs8050372







