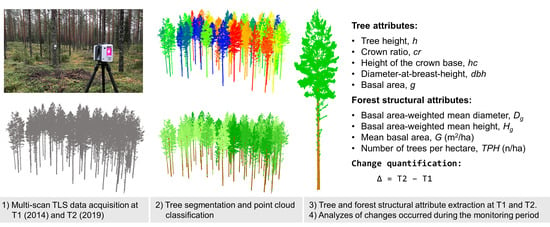
Structural Changes in Boreal Forests Can Be Quantified Using Terrestrial Laser Scanning
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Materials
2.2. Deriving Tree and Forest Structural Attributes from TLS Point Clouds
2.3. Quantifying Changes in Tree and Forest Structural Attributes Using Bi-Temporal TLS Data
2.4. Assessing The Performance of The TLS-Based Method in Quantifying Changes in Forest Structure
3. Results
3.1. Performance of Detecting Trees Using Bi-Temporal TLS Data
3.2. Performance of Characterizing Trees in Space Using TLS
3.3. Performance of Characterizing Tree Attributes in Time Using TLS
3.4. Performance of Characterizing Forest Structural Attributes in Space with TLS
3.5. Performance of Characterizing Forest Structural Attributes in Time Using TLS
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yrttimaa, T.; Saarinen, N.; Luoma, V.; Tanhuanpää, T.; Kankare, V.; Liang, X.; Hyyppä, J.; Holopainen, M.; Vastaranta, M. Detecting and characterizing downed dead wood using terrestrial laser scanning. ISPRS J. Photogramm. Remote Sens. 2019, 151, 76–90. [Google Scholar] [CrossRef]
- Saarinen, N.; Vastaranta, M.; Honkavaara, E.; Wulder, M.A.; White, J.C.; Litkey, P.; Holopainen, M.; Hyyppä, J. Using multi-source data to map and model the predisposition of forests to wind disturbance. Scand. J. For. Res. 2016, 31, 66–79. [Google Scholar] [CrossRef]
- Carvajal-Ramírez, F.; da Silva, J.R.M.; Agüera-Vega, F.; Martínez-Carricondo, P.; Serrano, J.; Moral, F.J. Evaluation of fire severity indices based on pre- and post-fire multispectral imagery sensed from UAV. Remote Sens. 2019, 11, 993. [Google Scholar] [CrossRef] [Green Version]
- Gupta, V.; Reinke, K.; Jones, S.; Wallace, L.; Holden, L. Assessing metrics for estimating fire induced change in the forest understorey structure using terrestrial laser scanning. Remote Sens. 2015, 7, 8180–8201. [Google Scholar] [CrossRef] [Green Version]
- Holopainen, M.; Vastaranta, M.; Hyyppä, J. Outlook for the next generation’s precision forestry in Finland. Forests 2014, 5, 1682–1694. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.; Hyyppä, J.; Kaartinen, H.; Holopainen, M.; Melkas, T. Detecting changes in forest structure over time with bi-temporal terrestrial laser scanning data. ISPRS Int. J. Geo-Inf. 2012, 1, 242–255. [Google Scholar] [CrossRef]
- Dassot, M.; Constant, T.; Fournier, M. The use of terrestrial LiDAR technology in forest science: Application fields, benefits and challenges. Ann. For. Sci. 2011, 68, 959–974. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.; Kankare, V.; Hyyppä, J.; Wang, Y.; Kukko, A.; Haggrén, H.; Yu, X.; Kaartinen, H.; Jaakkola, A.; Guan, F.; et al. Terrestrial laser scanning in forest inventories. ISPRS J. Photogramm. Remote Sens. 2016, 115, 63–77. [Google Scholar] [CrossRef]
- Newnham, G.J.; Armston, J.D.; Calders, K.; Disney, M.I.; Lovell, J.L.; Schaaf, C.B.; Strahler, A.H.; Mark Danson, F. Terrestrial laser scanning for plot-scale forest measurement. Curr. For. Rep. 2015, 1, 239–251. [Google Scholar] [CrossRef] [Green Version]
- Yrttimaa, T.; Saarinen, N.; Kankare, V.; Liang, X.; Hyyppä, J.; Holopainen, M.; Vastaranta, M. Investigating the feasibility of multi-scan terrestrial laser scanning to characterize tree communities in southern boreal forests. Remote Sens. 2019, 11, 1423. [Google Scholar] [CrossRef] [Green Version]
- Maas, H.-G.; Bienert, A.; Scheller, S.; Keane, E. Automatic forest inventory parameter determination from terrestrial laser scanner data. Int. J. Remote Sens. 2008, 29, 1579–1593. [Google Scholar] [CrossRef]
- Aschoff, T.; Thies, M.; Spiecker, H. Describing forest stands using terrestrial laser-scanning. In Proceedings of the 20th ISPRS World Congress, Istanbul, Turkey, 12–23 July 2004; pp. 237–241. [Google Scholar]
- Cabo, C.; Ordóñez, C.; López-Sánchez, C.A.; Armesto, J. Automatic dendrometry: Tree detection, tree height and diameter estimation using terrestrial laser scanning. Int. J. Appl. Earth Obs. Geoinf. 2018, 69, 164–174. [Google Scholar] [CrossRef]
- Zhang, W.; Wan, P.; Wang, T.; Cai, S.; Chen, Y.; Jin, X.; Yan, G. A novel approach for the detection of standing tree stems from plot-level terrestrial laser scanning data. Remote Sens. 2019, 11, 211. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.; Litkey, P.; Hyyppa, J.; Kaartinen, H.; Vastaranta, M.; Holopainen, M. Automatic stem mapping using single-scan terrestrial laser scanning. IEEE Trans. Geosci. Remote Sens. 2012, 50, 661–670. [Google Scholar] [CrossRef]
- Heinzel, J.; Huber, M. Detecting tree stems from volumetric TLS data in forest environments with rich understory. Remote Sens. 2016, 9, 9. [Google Scholar] [CrossRef] [Green Version]
- Raumonen, P.; Kaasalainen, M.; Åkerblom, M.; Kaasalainen, S.; Kaartinen, H.; Vastaranta, M.; Holopainen, M.; Disney, M.; Lewis, P. Fast automatic precision tree models from terrestrial laser scanner data. Remote Sens. 2013, 5, 491–520. [Google Scholar] [CrossRef] [Green Version]
- Hackenberg, J.; Morhart, C.; Sheppard, J.; Spiecker, H.; Disney, M. Highly accurate tree models derived from terrestrial laser scan data: A method description. Forests 2014, 5, 1069–1105. [Google Scholar] [CrossRef] [Green Version]
- Åkerblom, M.; Raumonen, P.; Kaasalainen, M.; Casella, E. Analysis of geometric primitives in quantitative structure models of tree stems. Remote Sens. 2015, 7, 4581–4603. [Google Scholar]
- Olofsson, K.; Holmgren, J. Single tree stem profile detection using terrestrial laser scanner data, flatness saliency features and curvature properties. Forests 2016, 7, 207. [Google Scholar] [CrossRef]
- Wilkes, P.; Lau, A.; Disney, M.; Calders, K.; Burt, A.; de Tanago, J.G.; Bartholomeus, H.; Brede, B.; Herold, M. Data acquisition considerations for terrestrial laser scanning of forest plots. Remote Sens. Environ. 2017, 196, 140–153. [Google Scholar] [CrossRef]
- Liang, X.; Hyyppä, J.; Kaartinen, H.; Lehtomäki, M.; Pyörälä, J.; Pfeifer, N.; Holopainen, M.; Brolly, G.; Francesco, P.; Hackenberg, J.; et al. International benchmarking of terrestrial laser scanning approaches for forest inventories. ISPRS J. Photogramm. Remote Sens. 2018, 144, 137–179. [Google Scholar] [CrossRef]
- Moorthy, S.M.K.; Krishna Moorthy, S.M.; Calders, K.; Vicari, M.B.; Verbeeck, H. Improved supervised learning-based approach for leaf and wood classification from LiDAR point clouds of forests. IEEE Trans. Geosci. Remote Sens. 2020, 58, 3057–3070. [Google Scholar] [CrossRef] [Green Version]
- Åkerblom, M.; Raumonen, P.; Casella, E.; Disney, M.I.; Danson, F.M.; Gaulton, R.; Schofield, L.A.; Kaasalainen, M. Non-intersecting leaf insertion algorithm for tree structure models. Interface Focus 2018, 8, 20170045. [Google Scholar] [CrossRef] [PubMed]
- Vicari, M.B.; Disney, M.; Wilkes, P.; Burt, A.; Calders, K.; Woodgate, W. Leaf and wood classification framework for terrestrial LiDAR point clouds. Methods Ecol. Evol. 2019, 10, 680–694. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Zheng, G.; Eitel, J.U.H.; Monika Moskal, L.; He, W.; Huang, H. Improved salient feature-based approach for automatically separating photosynthetic and non-photosynthetic components within terrestrial LiDAR point cloud data of forest canopies. IEEE Trans. Geosci. Remote Sens. 2016, 54, 679–696. [Google Scholar] [CrossRef]
- Béland, M.; Baldocchi, D.D.; Widlowski, J.-L.; Fournier, R.A.; Verstraete, M.M. On seeing the wood from the leaves and the role of voxel size in determining leaf area distribution of forests with terrestrial LiDAR. Agric. For. Meteorol. 2014, 184, 82–97. [Google Scholar] [CrossRef]
- Zhu, X.; Skidmore, A.K.; Darvishzadeh, R.; Olaf Niemann, K.; Liu, J.; Shi, Y.; Wang, T. Foliar and woody materials discriminated using terrestrial LiDAR in a mixed natural forest. Int. J. Appl. Earth Obs. Geoinf. 2018, 64, 43–50. [Google Scholar] [CrossRef]
- Junttila, S.; Holopainen, M.; Vastaranta, M.; Lyytikäinen-Saarenmaa, P.; Kaartinen, H.; Hyyppä, J.; Hyyppä, H. The potential of dual-wavelength terrestrial LiDAR in early detection of Ips typographus (L.) infestation—Leaf water content as a proxy. Remote Sens. Environ. 2019, 231, 111264. [Google Scholar] [CrossRef]
- Srinivasan, S.; Popescu, S.C.; Eriksson, M.; Sheridan, R.D.; Ku, N.-W. Multi-temporal terrestrial laser scanning for modeling tree biomass change. For. Ecol. Manag. 2014, 318, 304–317. [Google Scholar] [CrossRef]
- Kaasalainen, S.; Krooks, A.; Liski, J.; Raumonen, P.; Kaartinen, H.; Kaasalainen, M.; Puttonen, E.; Anttila, K.; Mäkipää, R. Change detection of tree biomass with terrestrial laser scanning and quantitative structure modelling. Remote Sens. 2014, 6, 3906–3922. [Google Scholar] [CrossRef] [Green Version]
- Sheppard, J.; Morhart, C.; Hackenberg, J.; Spiecker, H. Terrestrial laser scanning as a tool for assessing tree growth. IFOREST 2017, 10, 172–179. [Google Scholar] [CrossRef]
- Hess, C.; Härdtle, W.; Kunz, M.; Fichtner, A.; von Oheimb, G. A high-resolution approach for the spatiotemporal analysis of forest canopy space using terrestrial laser scanning data. Ecol. Evol. 2018, 8, 6800–6811. [Google Scholar] [CrossRef] [PubMed]
- Kunz, M.; Fichtner, A.; Härdtle, W.; Raumonen, P.; Bruelheide, H.; von Oheimb, G. Neighbour species richness and local structural variability modulate aboveground allocation patterns and crown morphology of individual trees. Ecol. Lett. 2019, 22, 2130–2140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luoma, V.; Saarinen, N.; Kankare, V.; Tanhuanpää, T.; Kaartinen, H.; Kukko, A.; Holopainen, M.; Hyyppä, J.; Vastaranta, M. Examining changes in stem taper and volume growth with two-date 3D point clouds. Forests 2019, 10, 382. [Google Scholar] [CrossRef] [Green Version]
- Vastaranta, M.; Yrttimaa, T.; Saarinen, N.; Yu, X.; Karjalainen, M.; Nurminen, K.; Karila, K.; Kankare, V.; Luoma, V.; Pyörälä, J.; et al. Airborne laser scanning outperforms the alternative 3D techniques in capturing variation in tree height and forest density in southern boreal forests. Baltic For. 2018, 28, 268–277. [Google Scholar]
- Luoma, V.; Saarinen, N.; Wulder, M.; White, J.; Vastaranta, M.; Holopainen, M.; Hyyppä, J. Assessing precision in conventional field measurements of individual tree attributes. Forests 2017, 8, 38. [Google Scholar] [CrossRef] [Green Version]
- Ritter, T.; Schwarz, M.; Tockner, A.; Leisch, F.; Nothdurft, A. Automatic mapping of forest stands based on three-dimensional point clouds derived from terrestrial laser-scanning. Forests 2017, 8, 265. [Google Scholar] [CrossRef]
- Yrttimaa, T.; Saarinen, N.; Kankare, V.; Hynynen, J.; Huuskonen, S.; Holopainen, M.; Hyyppä, J.; Vastaranta, M. Performance of terrestrial laser scanning to characterize managed Scots pine (Pinus sylvestris L.) stands is dependent on forest structural variation. EarthArXiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Popescu, S.C.; Wynne, R.H. Seeing the trees in the forest. Photogramm. Eng. Remote Sens. 2004, 70, 589–604. [Google Scholar] [CrossRef] [Green Version]
- Meyer, F.; Beucher, S. Morphological segmentation. J. Visual Commun. Image Represent. 1990, 1, 21–46. [Google Scholar] [CrossRef]
- Saarinen, N.; Kankare, V.; Vastaranta, M.; Luoma, V.; Pyörälä, J.; Tanhuanpää, T.; Liang, X.; Kaartinen, H.; Kukko, A.; Jaakkola, A.; et al. Feasibility of terrestrial laser scanning for collecting stem volume information from single trees. ISPRS J. Photogramm. Remote Sens. 2017, 123, 140–158. [Google Scholar] [CrossRef]
- Abegg, M.; Kükenbrink, D.; Zell, J.; Schaepman, M.E.; Morsdorf, F. Terrestrial laser scanning for forest inventories—Tree diameter distribution and scanner location impact on occlusion. For. Trees Livelihoods 2017, 8, 184. [Google Scholar] [CrossRef] [Green Version]
- Gollob, C.; Ritter, T.; Wassermann, C.; Nothdurft, A. Influence of scanner position and plot size on the accuracy of tree detection and diameter estimation using terrestrial laser scanning on forest inventory plots. Remote Sens. 2019, 11, 1602. [Google Scholar] [CrossRef] [Green Version]
- Trochta, J.; Král, K.; Janík, D.; Adam, D. Arrangement of terrestrial laser scanner positions for area-wide stem mapping of natural forests. Can. J. For. Res. 2013, 43, 355–363. [Google Scholar] [CrossRef]
- Schneider, F.D.; Kükenbrink, D.; Schaepman, M.E.; Schimel, D.S.; Morsdorf, F. Quantifying 3D structure and occlusion in dense tropical and temperate forests using close-range LiDAR. Agric. For. Meteorol. 2019, 268, 249–257. [Google Scholar] [CrossRef]
- Wang, Y.; Lehtomäki, M.; Liang, X.; Pyörälä, J.; Kukko, A.; Jaakkola, A.; Liu, J.; Feng, Z.; Chen, R.; Hyyppä, J. Is field-measured tree height as reliable as believed—A comparison study of tree height estimates from field measurement, airborne laser scanning and terrestrial laser scanning in a boreal forest. ISPRS J. Photogramm. Remote Sens. 2019, 147, 132–145. [Google Scholar] [CrossRef]
- Pesonen, E. A new girth band for measuring stem diameter changes. Forestry 2004, 77, 431–439. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.; Hyyppa, J.; Kukko, A.; Kaartinen, H.; Jaakkola, A.; Yu, X. The use of a mobile laser scanning system for mapping large forest plots. IEEE Geosci. Remote Sens. Lett. 2014, 11, 1504–1508. [Google Scholar] [CrossRef]
- Kukko, A.; Kaartinen, H.; Hyyppä, J.; Chen, Y. Multiplatform mobile laser scanning: Usability and performance. Sensors 2012, 12, 11712–11733. [Google Scholar] [CrossRef] [Green Version]
- Hyyppä, E.; Kukko, A.; Kaijaluoto, R.; White, J.C.; Wulder, M.A.; Pyörälä, J.; Liang, X.; Yu, X.; Wang, Y.; Kaartinen, H.; et al. Accurate derivation of stem curve and volume using backpack mobile laser scanning. ISPRS J. Photogramm. Remote Sens. 2020, 161, 246–262. [Google Scholar] [CrossRef]
- Bauwens, S.; Bartholomeus, H.; Calders, K.; Lejeune, P. Forest inventory with terrestrial LiDAR: A comparison of static and hand-held mobile laser scanning. Forests 2016, 7, 127. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Liu, H.; Feng, Z.; Shen, C.; Chen, P. Applicability of personal laser scanning in forestry inventory. PLoS ONE 2019, 14, e0211392. [Google Scholar] [CrossRef] [PubMed]
- Kukko, A.; Kaijaluoto, R.; Kaartinen, H.; Lehtola, V.V.; Jaakkola, A.; Hyyppä, J. Graph SLAM correction for single scanner MLS forest data under boreal forest canopy. ISPRS J. Photogramm. Remote Sens. 2017, 132, 199–209. [Google Scholar] [CrossRef]
- Jaakkola, A.; Hyyppä, J.; Kukko, A.; Yu, X.; Kaartinen, H.; Lehtomäki, M.; Lin, Y. A low-cost multi-sensoral mobile mapping system and its feasibility for tree measurements. ISPRS J. Photogramm. Remote Sens. 2010, 65, 514–522. [Google Scholar] [CrossRef]
- Liu, K.; Shen, X.; Cao, L.; Wang, G.; Cao, F. Estimating forest structural attributes using UAV-LiDAR data in Ginkgo plantations. ISPRS J. Photogramm. Remote Sens. 2018, 146, 465–482. [Google Scholar] [CrossRef]
- Jaakkola, A.; Hyyppä, J.; Yu, X.; Kukko, A.; Kaartinen, H.; Liang, X.; Hyyppä, H.; Wang, Y. Autonomous collection of forest field reference—The outlook and a first step with UAV laser scanning. Remote Sens. 2017, 9, 785. [Google Scholar] [CrossRef] [Green Version]
- Hyyppä, E.; Hyyppä, J.; Hakala, T.; Kukko, A.; Wulder, M.A.; White, J.C.; Pyörälä, J.; Yu, X.; Wang, Y.; Virtanen, J.-P.; et al. Under-canopy UAV laser scanning for accurate forest field measurements. ISPRS J. Photogramm. Remote Sens. 2020, 164, 41–60. [Google Scholar] [CrossRef]
- Yrttimaa, T.; Saarinen, N.; Kankare, V.; Viljanen, N.; Hynynen, J.; Huuskonen, S.; Holopainen, M.; Hyyppä, J.; Honkavaara, E.; Vastaranta, M. Multisensorial close-range sensing generates benefits for characterization of managed Scots pine (Pinus sylvestris L.) stands. ISPRS Int. J. Geo-Inf. 2020, 9, 309. [Google Scholar] [CrossRef]




| Tree/Forest Structural Attribute | Tree Species | Minimum (T1/T2) | Mean (T1/T2) | Maximum (T1/T2) | Standard Deviation (T1/T2) | |
|---|---|---|---|---|---|---|
| Tree Attributes | dbh (cm) | All trees | 5.0/5.1 | 16.9/17.9 | 59.9/64.0 | 10.2/10.5 |
| Scots pine | 5.2/5.1 | 20.3/21.3 | 59.7/60.1 | 10.3/10.6 | ||
| Norway spruce | 5.0/5.1 | 16.9/18.0 | 57.9/61.7 | 11.0/11.2 | ||
| Broadleaved | 5.1/5.1 | 14.9/15.7 | 59.9/64.0 | 8.1/8.7 | ||
| g (cm2) | All trees | 20.0/20.0 | 308.0/339.9 | 2818.0/3217.0 | 399.1/425.0 | |
| Scots pine | 21.2/20.8 | 407.7/445.5 | 2799.2/2836.9 | 468.2/487.1 | ||
| Norway spruce | 20.0/20.4 | 318.4/351.7 | 2633.0/2989.9 | 416.6/439.0 | ||
| Broadleaved | 20.0/20.0 | 225.7/251.8 | 2818.0/3217.0 | 291.4/331.7 | ||
| h (m) | All trees | 2.2/1.9 | 16.3/17.4 | 36.6/38.4 | 7.3/7.5 | |
| Scots pine | 5.0/5.0 | 17.8/19.2 | 34.5/37.2 | 5.5/5.8 | ||
| Norway spruce | 2.2/3.5 | 15.2/16.4 | 36.6/38.4 | 8.4/8.5 | ||
| Broadleaved | 2.2/1.9 | 17.0/17.7 | 32.5/35.8 | 6.0/6.6 | ||
| d-h-ratio | All trees | 0.42/0.41 | 1.03/1.02 | 4.70/5.00 | 0.31/0.33 | |
| Scots pine | 0.57/0.53 | 1.10/1.08 | 2.24/2.45 | 0.29/0.29 | ||
| Norway spruce | 0.73/0.65 | 1.09/1.08 | 2.86/2.15 | 0.21/0.19 | ||
| Broadleaved | 0.42/0.41 | 0.88/0.91 | 4.70/5.00 | 0.38/0.47 | ||
| hc (m) | Scots pine | 4.2/6.3 | 12.2/13.9 | 23.1/24.6 | 3.4/3.4 | |
| cr | Scots pine | 0.22/0.19 | 0.40/0.37 | 0.69/0.59 | 0.09/0.08 | |
| Forest Structural Attributes | Dg (cm) | All plots | 13.0/14.3 | 27.5/28.8 | 42.8/44.0 | 9.3/9.3 |
| Scots pine-dominated | 14.2/15.1 | 21.7/22.9 | 30.6/31.7 | 4.5/4.5 | ||
| Norway spruce-dominated | 19.7/21.2 | 34.8/36.1 | 42.8/44.0 | 6.9/6.9 | ||
| Mixed-species | 13.0/14.3 | 24.8/26.2 | 42.8/43.2 | 9.7/9.7 | ||
| Hg (m) | All plots | 12.4/13.7 | 22.3/23.6 | 31.6/32.4 | 4.9/4.9 | |
| Scots pine-dominated | 12.4/13.7 | 18.4 /20.0 | 23.1/25.3 | 3.2/3.4 | ||
| Norway spruce-dominated | 18.4/20.3 | 26.9/28.2 | 31.6/32.4 | 3.6/3.7 | ||
| Mixed-species | 15.9/17.3 | 20.8/21.9 | 27.3/27.8 | 3.6/3.5 | ||
| All plots | 15.3/17.2 | 31.6/34.5 | 51.5/56.8 | 10.5/11.0 | ||
| G (m2/ha) | Scots pine-dominated | 15.3/17.2 | 22.6/25.5 | 31.1/34.4 | 5.9/6.8 | |
| Norway spruce-dominated | 21.3/23.7 | 37.6/39.9 | 51.0/53.8 | 8.7/8.9 | ||
| Mixed-species | 16.2/17.5 | 32.4/35.9 | 51.5/56.8 | 10.6/11.7 | ||
| TPH (n/ha) | All plots | 368/368 | 1059/1045 | 3341/3236 | 706/711 | |
| Scots pine-dominated | 368/368 | 963/997 | 1894/2105 | 495/553 | ||
| Norway spruce-dominated | 395/368 | 635/605 | 1289/1289 | 289/301 | ||
| Mixed-species | 526/552 | 1522/1488 | 3341/3236 | 847/835 | ||
| Tree/Forest Structural Attribute | Abbreviation | Description/Formula | |
|---|---|---|---|
| Tree Attributes | diameter-at-breast-height (cm) | dbh | tree diameter measured at a 1.3-m height |
| basal area (cm2) | g | π ∗ dbh2/4 | |
| tree height (m) | h | vertical distance between ground and treetop | |
| diameter-height ratio | d-h-ratio | dbh/h | |
| height of the crown base (m) | hc | height of the lowest living branches | |
| crown ratio | cr | (h − hc)/h | |
| Forest Structural Attributes | basal area-weighted mean diameter (cm) | Dg | |
| basal area-weighted mean height (m) | Hg | ||
| mean basal area (m2/ha) | G | ||
| number of trees per hectare (n/ha) | TPH | n/A | |
| Tree Species | Nref | NTLS | Completeness: Total Stem Number | Completeness: Total Basal Area |
|---|---|---|---|---|
| All trees | 1280 | 795 | 62.1% | 84.5% |
| Scots pine | 270 | 227 | 84.1% | 91.3% |
| Norway spruce | 649 | 366 | 56.4% | 85.7% |
| Broadleaved | 361 | 202 | 56.0% | 73.3% |
| Tree Attribute | Tree Species | Bias | RMSE | ||||
|---|---|---|---|---|---|---|---|
| T1 | T2 | Δ | T1 | T2 | Δ | ||
| dbh (cm) | All trees | −0.05 (−0.3%) | 0.04 (0.2%) | 0.10 (8.3%) | 1.18 (5.7%) | 0.90 (4.1%) | 1.13 (97.4%) |
| Scots pine | −0.43 (−2.0%) | −0.32 (−1.4%) | 0.12 (10.1%) | 1.11 (5.2%) | 0.99 (4.5%) | 0.99 (83.9%) | |
| Norway spruce | 0.26 (1.2%) | 0.30 (1.3%) | 0.04 (3.8%) | 1.26 (5.7%) | 0.77 (3.3%) | 1.22 (103.5%) | |
| Broadleaved | −0.19 (−1.1%) | −0.03 (−0.15%) | 0.17 (15.0%) | 1.11 (6.3%) | 0.99 (5.27%) | 1.12 (100.4%) | |
| g (cm2) | All trees | −7.65 (−1.9%) | −3.39 (−0.7%) | 4.26 (10.2%) | 49.28 (11.9%) | 47.59 (10.4%) | 49.40 (118.4%) |
| Scots pine | −21.47 (−5.1%) | −18.48 (−3.9%) | 2.99 (7.0%) | 52.82 (12.5%) | 60.81 (13.0%) | 44.14 (102.8%) | |
| Norway spruce | 1.75 (0.4%) | 7.38 (1.4%) | 5.64 (12.9%) | 50.51 (10.6%) | 32.35 (6.2%) | 55.49 (126.6%) | |
| Broadleaved | −9.16 (−3.1%) | −5.96 (−1.8%) | 3.21 (8.7%) | 42.42 (14.5%) | 53.51 (16.3%) | 42.98 (117.1%) | |
| h (m) | All trees | −1.33 (−6.9%) | −0.74 (−3.6%) | 0.59 (42.1%) | 4.37 (22.5%) | 4.10 (19.7%) | 3.53 (251.6%) |
| Scots pine | −0.89 (−4.8%) | −0.47 (−2.3%) | 0.43 (27.3%) | 2.52 (13.7%) | 2.42 (12.1%) | 1.91 (122.8%) | |
| Norway spruce | −0.51 (−2.6%) | 0.39 (1.8%) | 0.90 (72.3%) | 3.99 (20.1%) | 3.03 (14.4%) | 3.43 (274.6%) | |
| Broadleaved | −3.31 (−16.8%) | −3.11 (−14.6%) | 0.21 (13.8%) | 6.26 (31.7%) | 6.56 (30.8%) | 4.85 (321.5%) | |
| d-h-ratio | All trees | 0.10 (9.7%) | 0.07 (6.3%) | −0.04 (226.6%) | 0.35 (33.6%) | 0.30 (29.5%) | 0.33 (2028.6%) |
| Scots pine | 0.05 (4.6%) | 0.03 (2.6%) | −0.02 (78.3%) | 0.28 (24.9%) | 0.24 (21.6%) | 0.28 (942.6%) | |
| Norway spruce | 0.06 (5.3%) | −0.01 (0.1%) | −0.06 (563.8%) | 0.26 (23.6%) | 0.16 (14.8%) | 0.22 (2123.3%) | |
| Broadleaved | 0.24 (26.9%) | 0.23 (26.2%) | −0.01 (88.6%) | 0.53 (59.8%) | 0.51 (57.7%) | 0.49 (4516.2%) | |
| hc (m) | Scots pine | −1.70 (−16.1%) | −1.36 (−10.9%) | 0.34 (20.7%) | 2.71 (25.7%) | 2.55 (20.4%) | 1.97 (120.6%) |
| cr | Scots pine | 0.05 (11.7%) | 0.04 (10.3%) | −0.01 (36.2%) | 0.10 (22.8%) | 0.09 (22.5%) | 0.09 (318.1%) |
| Tree Species | dbh | g | h | d-h-Ratio | hc | cr |
|---|---|---|---|---|---|---|
| All trees | <0.000 | <0.000 | <0.000 | <0.000 | <0.000 | <0.000 |
| Scots pine | <0.000 | <0.000 | <0.000 | <0.000 | <0.000 | <0.000 |
| Norway spruce | <0.000 | <0.000 | <0.000 | <0.000 | - | - |
| Broadleaved tree | <0.000 | <0.000 | <0.000 | 0.003 | - | - |
| Forest Structural Attribute | Main Tree Species | Bias | RMSE | ||||
|---|---|---|---|---|---|---|---|
| T1 | T2 | Δ | T1 | T2 | Δ | ||
| Dg (cm) | All plots | 0.08 (0.3%) | 0.25 (0.9%) | 0.17 (13.2%) | 1.42 (5.2%) | 1.72 (6.0%) | 0.93 (72.6%) |
| Scots pine-dominated | 0.08 (0.4%) | 0.55 (2.4%) | 0.47 (40.5%) | 0.59 (2.7%) | 0.77 (3.4%) | 0.60 (52.5%) | |
| Norway spruce-dominated | −0.26 (−0.7%) | −0.15 (−0.4%) | 0.10 (7.8%) | 0.87 (2.5%) | 1.06 (2.9%) | 1.13 (86.4%) | |
| Mixed-species | 0.39 (1.6%) | 0.41 (1.6%) | 0.02 (1.4%) | 2.09 (8.4%) | 2.51 (9.6%) | 0.91 (67.9%) | |
| Hg (m) | All plots | −1.73 (−7.8%) | −0.50 (−2.1%) | 1.24 (96.3%) | 2.51 (11.2%) | 1.85 (7.9%) | 1.64 (127.8%) |
| Scots pine-dominated | −0.35 (−1.9%) | 0.20 (1.0%) | 0.55 (35.3%) | 0.53 (2.9%) | 0.70 (3.5%) | 0.81 (51.9%) | |
| Norway spruce-dominated | −2.88 (−10.7%) | −0.83 (−2.9%) | 2.05 (165.0%) | 3.12 (11.6%) | 1.09 (3.9%) | 2.26 (182.2%) | |
| Mixed-species | −1.67 (−8.0%) | −0.69 (−3.1%) | 0.98 (86.6%) | 2.72 (13.1%) | 2.76 (12.6%) | 1.39 (122.9%) | |
| G (m2/ha) | All plots | −6.49 (−20.5%) | −6.60 (19.1%) | −0.11 (−3.7%) | 8.52 (26.9%) | 9.27 (26.9%) | 1.84 (64.2%) |
| Scots pine-dominated | −5.32 (−23.6%) | −5.56 (−21.8%) | −0.23 (−8.1%) | 7.03 (31.1%) | 8.05 (31.6%) | 1.40 (48.9%) | |
| Norway spruce-dominated | −3.84 (−10.2%) | −3.24 (−8.1%) | 0.59 (26.4%) | 5.09 (13.5%) | 4.23 (10.6%) | 1.48 (66.4%) | |
| Mixed-species | −9.79 (−30.2%) | −10.46 (−29.2%) | −0.67 (−19.3%) | 11.50 (35.5%) | 12.82 (35.7%) | 2.34 (68.0%) | |
| TPH (n/ha) | All plots | −373 (−35.2%) | −292 (−27.9%) | −81 (−570%) | 620 (58.5%) | 515 (49.3%) | 143 (1008.7%) |
| Scots pine-dominated | −337 (−35.0%) | −268 (−26.9%) | −68 (−200.0%) | 486 (50.5%) | 428 (42.9%) | 130 (381.5%) | |
| Norway spruce-dominated | −91 (−14.3%) | −47 (−7.7%) | −45 (−146.7%) | 117 (18.5%) | 65 (10.7%) | 74 (245.1%) | |
| Mixed-species | −659 (−43.3%) | −536 (−36.0) | −124 (−366.7%) | 914 (60.0%) | 753 (50.6%) | 193 (569.3%) | |
| Main Tree species | Dg | Hg | G | TPH |
|---|---|---|---|---|
| All plots | <0.000 | <0.000 | <0.000 | 0.105 |
| Scots pine-dominated | <0.000 | <0.000 | <0.000 | 0.014 |
| Norway spruce-dominated | <0.000 | <0.000 | <0.000 | 0.775 |
| Mixed-species | <0.000 | <0.000 | <0.000 | 0.984 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yrttimaa, T.; Luoma, V.; Saarinen, N.; Kankare, V.; Junttila, S.; Holopainen, M.; Hyyppä, J.; Vastaranta, M. Structural Changes in Boreal Forests Can Be Quantified Using Terrestrial Laser Scanning. Remote Sens. 2020, 12, 2672. https://doi.org/10.3390/rs12172672
Yrttimaa T, Luoma V, Saarinen N, Kankare V, Junttila S, Holopainen M, Hyyppä J, Vastaranta M. Structural Changes in Boreal Forests Can Be Quantified Using Terrestrial Laser Scanning. Remote Sensing. 2020; 12(17):2672. https://doi.org/10.3390/rs12172672
Chicago/Turabian StyleYrttimaa, Tuomas, Ville Luoma, Ninni Saarinen, Ville Kankare, Samuli Junttila, Markus Holopainen, Juha Hyyppä, and Mikko Vastaranta. 2020. "Structural Changes in Boreal Forests Can Be Quantified Using Terrestrial Laser Scanning" Remote Sensing 12, no. 17: 2672. https://doi.org/10.3390/rs12172672