1. Introduction
The publishing of both data and narratives on those data are changing radically. Linked Open Data and related semantic technologies allow for semantic publishing of data. We still need, however, to publish the narratives on that data and that style of publishing is in the process of change; one of those changes is the incorporation of semantics [
1,
2,
3]. The idea of semantic publishing is an attractive one for those who wish to consume papers electronically; it should enhance the richness of the computational component of papers [
2]. It promises a realisation of the vision of a next generation of the web, with papers becoming a critical part of a linked data environment [
1,
4], where the results and narratives become one.
The reality, however, is somewhat different. There are significant barriers to the acceptance of semantic publishing as a standard mechanism for academic publishing. The web was invented around 1990 as a lightweight mechanism for publication of documents. It has subsequently had a massive impact on society in general. It has, however, barely touched most scientific publishing; while most journals have a website, the publication process still revolves around the generation of papers, moving from Microsoft Word or
![Futureinternet 04 01004 i001]()
[
5], through to a final PDF which looks, feels and is something designed to be printed onto paper. Adding semantics into this environment is difficult or impossible; the content of the PDF has to be exposed and semantic content retrofitted or, in all likelihood, a complex process of author and publisher interaction has to be devised and followed. If semantic data publishing and semantic publishing of academic narratives are to work together, then academic publishing needs to change.
In this paper, we describe our attempts to take a commodity publication environment and modify it to bring in some of the formality required from academic publishing. We illustrate this with three exemplars—different kinds of knowledge that we wish to enhance. In the process, we add a small amount of semantics to the finished articles. Our key constraint is the desire to add value for all the human participants. Both authors and readers should see and recognise additional value, with the semantics a useful or necessary byproduct of the process, rather than the primary motivation. We characterise this process as our “three steps to heaven”, namely:
make life better for the machine to;
make life better for the author to;
make life better for the reader.
While requiring additional value for all of these participants is hard, and places significant limitations on the level of semantics that can be achieved, we believe that it does increase the likelihood that content will be generated in the first place, and represents an attempt to enable semantic publishing in a real-world workflow.
2. Knowledgeblog
The knowledgeblog project stemmed from the desire for a book describing the many aspects of ontology development, from the underlying formal semantics, to the practical technology layer and, finally, through to the knowledge domain [
6]. However, we have found the traditional book publishing process frustrating and unrewarding. While scientific authoring is difficult in its own right, our own experience suggests that the
publishing process is extremely hard work. This is particularly so for multi-author collected works that are often harder for the editor than writing a book “solo”. Finally, the expense and hard copy nature of academic books means that, again in our experience, few people read them.
This contrasts starkly with the web-first publication process that has become known as blogging. With any of a number of ready-made platforms, it is possible for authors with little or no technical skill to publish content to the web with ease. For knowledgeblog (“kblog”), we have taken one blogging engine, WordPress [
7], running on low-end hardware, and used it to develop a multi-author resource describing the use of ontologies in the life sciences (our main field of expertise). There are also kblogs on bioinformatics [
8] and the Taverna workflow environment [
9,
10]. We have previously described how we addressed some of the social aspects, including attribution, reviewing and immutability of articles [
6].
As well as delivering content, we are also using this framework to investigate semantic academic publishing, investigating how we can enhance the machine interpretability of the final paper, while living within the key constraint of making life (slightly) better for machine, author and reader without adding complexity for the human participants. Primarily here, we are interested only in the machine that mediate either the authoring or reading process; however, increased interpretability for these machines should also benefit others, including for instance those of librarians or publishers.
Scientific authors are relatively conservative. Most of them have well-established toolsets and workflows that they are relatively unwilling to change. For instance, within the kblog project, we have used workshops to start the process of content generation. For our initial meeting, we gave little guidance on authoring process to authors, as a result of which most attempted to use WordPress directly for authoring. The WordPress editing environment is, however, web-based, and was originally designed for editing short non-technical articles. It appeared to not work well for most scientists [
6].
The requirements that authors have for such “scientific” articles are manifold. Many wish to be able to author while offline (particularly on trains or planes). Almost all scientific papers are multi-author, and some degree of collaboration is required. Collaboration is limited however, and more over articles are not usually changed once published, making Wikis inappropriate. Many scientists in the life sciences wish to author in Word because grant bodies and journals often produce templates as Word documents. Many wish to use
![Futureinternet 04 01004 i001]()
, because its idiomatic approach to programming documents is not replicable with anything else. Fortunately, it is possible to induce WordPress to accept content from many different authoring tools, including Word and
![Futureinternet 04 01004 i001]()
[
6].
As a result, during the kblog project, we have seen many different workflows in use, often highly idiosyncratic in nature. These include:
Word/Email Many authors write using MS Word and collaborate by emailing files around. This method has a low barrier to entry, but requires significant social processes to prevent conflicting versions, particularly as the number of authors increases.
Word/Dropbox For the tavernakblog, authors wrote in Word and collaborated with Dropbox [
11]. This method works reasonably well where many authors are involved; Dropbox detects conflicts, although it cannot prevent or merge them.
Asciidoc/Dropbox Used by the authors of this paper.Asciidoc [
12] is relatively simple, somewhat programmable and accessible. Unlike
![Futureinternet 04 01004 i001]()
which can be induced to produce HTML with effort, asciidoc is designed to do so.
Of these three approaches, probably the Word/Dropbox combination is the most generally used. Some older techniques, such as ASCII and versioning systems have disappeared in our experience.
From the readers perspective, a decision that we have made within knowledgeblog is to be “HTML-first”. The initial reasons for this were entirely practical; supporting multiple toolsets is hard, particularly if any degree of consistency is to be maintained; the generation of the HTML is at least partly controlled by the middleware—WordPress in kblog ’s case. As well as enabling consistency of presentation, it also, potentially, allows us to add additional knowledge; it makes semantic publication a possibility. However, we are aware that knowledgeblog currently scores rather badly on what we describe as the “bathtub test”; while exporting to PDF or printing out is possible, the presentation is not as “neat” as would be ideal. In this regard (and we hope only in this regard), the knowledgeblog experience is limited. However, increasingly, readers are happy and capable of interacting with material on the web without printouts.
From this background and aim, we have drawn the following requirements:
The author can, as much as possible, remain within familiar authoring environments;
The representation of the published work should remain extensible to, for instance, semantic enhancements;
The author and reader should be able to have the amount of “formal” academic publishing they need;
Support for semantic publishing should be gradual and offer advantages for author and reader at all stages.
We describe how we have achieved this with three exemplars, two of which are relatively general in use, and one more specific to biology. In each case, we have taken a slightly different approach, but have fulfilled our primary aim of making life better for machine, author and reader.
3. Representing Mathematics
The representation of mathematics is a common need in academic literature. Mathematical notation has grown from a requirement for a syntax that is highly expressive and relatively easy to write. It presents specific challenges because of its complexity, the difficulty of authoring and the difficulty of rendering, away from the chalkboard that is its natural home.
Support for mathematics has had a significant impact on academic publishing. It was, for example, the original motivation behind the development of
![Futureinternet 04 01004 i017]()
[
13], and it still one of the main reasons why authors wish to use it or its derivatives. This is to such an extent that much mathematics rendering on the web is driven by a
![Futureinternet 04 01004 i017]()
engine somewhere in the process. So MediaWiki (and therefore Wikipedia), Drupal and, of course, WordPress follow this route. The latter provides plugin support for
![Futureinternet 04 01004 i017]()
markup using the wp-latex plugin [
14]. Within kblog, we have developed a new plugin called mathjax-latex [
15]. From the kblog author’s perspective these two offer a similar interface—differences are, therefore, described later.
Authors write their mathematics directly as
![Futureinternet 04 01004 i017]()
using one of the four markup syntaxes. The most explicit (and therefore least likely to happen accidentally) is through the use of “shortcodes” [
16]. These are a HTML-like markup originating from some forum/bulletin board systems. In this form an equation would be entered as [latex]e=mc^2[/latex], which would be rendered as “
![Futureinternet 04 01004 i004]()
”. It is also possible to use three other syntaxes that are closer to math mode in
![Futureinternet 04 01004 i017]()
: $$e=mc^2$$, $latex e=mc^2$, or \[e=mc^2\].
From the authorial perspective, we have added significant value, as it is possible to use a variety of syntaxes, which are independent of the authoring engine. For example, a
![Futureinternet 04 01004 i017]()
-loving mathematician working with a Word-using biologist can still set their equations using
![Futureinternet 04 01004 i017]()
syntax; although Word will not render these at authoring time but, in practice, this causes few problems for such authors, who are experienced at reading
![Futureinternet 04 01004 i017]()
. Within an
![Futureinternet 04 01004 i001]()
workflow, equations can be both rendered locally with source compiled to PDF and published to WordPress.
There is also a W3C recommendation, MathML for the representation and presentation of mathematics. The kblog environment also supports this. In this case, the equivalent source appears as follows:
One problem with the MathML representation is obvious: it is very long-winded. A second issue, however, is that it is hard to integrate with existing workflows—on recognising an angle bracket, most of the publication workflows we have seen in use will turn it into the equivalent HTML entity. For some workflows (
![Futureinternet 04 01004 i001]()
, asciidoc) it is
possible, although not easy, to prevent this within the native syntax.
It is also possible to convert from Word’s native OMML (“equation editor”) XML representation to MathML, although this does not integrate with Word’s native blog publication workflow. Ironically, it is because MathML shares an XML based syntax with the final presentation format (HTML) that the problem arises. The shortcode syntax, for example, passes straight-through most of the publication frameworks to be consumed by the middleware. From a pragmatic point of view, therefore, supporting shortcodes and
![Futureinternet 04 01004 i017]()
-like syntaxes has considerable advantages.
For the reader, the use of mathjax-latex has significant advantages. The default mechanism within WordPress uses a math-mode-like syntax $latex e=mc^2$. This is rendered using a
![Futureinternet 04 01004 i017]()
engine into an image, which is then incorporated and linked using normal HTML capabilities. This representation is opaque and non-semantic; it has significant limitations for the reader. The images are not scalable—zooming in causes severe pixellation; the background to the mathematics is coloured inside the image and does not necessarily reflect the local style.
Kblog, however, uses the MathJax library [
17]. This has a number of significant advantages for the reader. First, where the browser supports them, MathJax can use native MathML support or web fonts to render the images that are scalable, attractive and standardized. Where they are not available, MathJax can fall back to bitmapped fonts. The reader can also access additional functionality: clicking on an equation will raise a zoomed-in popup, while the context menu allows access to a textual representation either as
![Futureinternet 04 01004 i017]()
or MathML irrespective of the form that the author used. This can be cut-and-paste for further use. Kblog uses the MathJax library [
17] to render the underlying
![Futureinternet 04 01004 i017]()
directly on the client.
Our use of MathJax provides no significant disadvantages to the middleware layers. It is implemented in JavaScript and runs in most environments. Although the library is fairly large (
![Futureinternet 04 01004 i008]()
100 Mb), it is available on a CDN and does not stress the server storage space. Most of this space comes from the bit-mapped fonts that are only downloaded on demand, which should not stress web clients either. It also obviates the need for a
![Futureinternet 04 01004 i017]()
installation that wp-latex may require (although this plugin can use an external server also).
At face value, mathjax-latex necessarily adds very little semantics to the math embedded within documents. The math could be represented as
Therefore, we have a heterogeneous representation for identical knowledge. However, in practice, the situation is much better than this. The author of the work created these equations and has then read them, transformed by MathJax into a rendered form. If MathJax has failed to translate them correctly, in line with the author’s intention, or if it has had some implications for the text in addition to setting the intended equations (say, if the
![Futureinternet 04 01004 i017]()
style markup appears accidentally elsewhere in the document), the author is likely to have seen this and fixed the problem. Someone wishing, for example, to extract all the mathematics as MathML from these documents computationally, therefore, knows:
that the document contains math as it imports MathJax;
that MathJax is capable of identifying this math correctly;
that equations can be transformed to MathML using MathJax. (This is assuming MathJax works correctly in general. The authors and readers are checking the rendered representation. It is possible that an equation would render correctly on screen, but be rendered to MathML inaccurately.)
Therefore, while our publication environment does not result directly in lower level of semantic heterogeneity, it does provide the data and the tools to enable the computational agent to make this transformation. While this is imperfect, it should help a bit. In short, we provide a practical mechanism to identify text that contains mathematics and a mechanism to transform this to a single standardised representation.
4. Representing References
Unlike mathematics, there is no standard mechanism for reference and in-text citation, but there are a large number of tools for authors such as BibTeX, Mendeley [
18] or EndNote. Because of this, the integration with existing toolsets is of primary importance, while the representation of the in-text citations is not, as it should be handled by the tool layer anyway.
Within kblog, we have developed a plugin called kcite [
19]. For the author, citations are inserted using the syntax:
The identifier used here is a digital object identifier (DOI), which is widely used within the publishing and library industry. Currently, kcite supports DOIs minted by either CrossRef [
19] or DataCite [
20] (unfortunately, DOIs
per se do not provide standardized metadata, however in practice, this means that we support the majority of DOIs in academic use). We also support identifiers from PubMed [
21], which covers most biomedical publications, and arXiv [
22], the physics (and other domains!) preprints archive, and we now have a system to support arbitrary URLs.
We have picked this “shortcode” format for similar reasons as described for math—it is relatively unambiguous, it is not XML based and passes through the HTML generation layer of most authoring tools unchanged, and it is explicitly supported in WordPress, bypassing the need for regular expressions and later parsing. However, it would be a little unwieldy from the perspective of the author. Nevertheless, in practice, it is relatively easy to integrate this with many reference managers. For example, tools such as Zotero [
23] and Mendeley use the Citation Style Language, and so can output kcite compliant citations with the following slightly elided code:
We do not yet support
![Futureinternet 04 01004 i001]()
/BibTeX citations, although we see no reason why a similar style file should not be supported. We do, however, support BibTeX-formatted files: the first author’s preferred editing/citation environment is based on these with Emacs, RefTeX (and Emacs package), and asciidoc. While this is undoubtedly a rather niche authoring environment, the (slightly elided) code shows that very little is needed to enable the tool chains to support kcite:
The key decision with kcite from the authorial perspective is to ignore the reference list itself and focus only on in-text citations, using public identifiers to references. This simplifies the tool integration process enormously, as this is the only data that needs to pass from the author’s bibliographic database onward. The key advantage for authors here is two-fold: they are not required to populate their reference metadata for themselves, and this metadata will update if it changes. Secondly, the identifiers are checked; if they are wrong, the authors will see this straightforwardly as the entire reference will be wrong. As a result, adding DOIs or other identifiers moves from becoming a burden for the author to becoming a specific advantage.
While supporting multiple forms of reference identifier (CrossRef DOI, DataCite DOI, arXiv and PubMed ID) provides a clear advantage to the author, it comes at considerable cost. While it is possible to get metadata about papers from all of these sources, there is little commonality between them. Moreover, resolving this metadata requires one outgoing HTTP request (in practice it is often more; for instance the DOI requests use 303 redirects) per reference, which might or might not be allowed by the browser security setting.
Hence, while the presentation of mathematics is performed largely on the client, for reference lists the kcite plugin performs metadata resolution and data integration on the server. A caching functionality is provided, storing this metadata in the WordPress database. The bibliographic metadata is finally transferred to the client encoded as JSON, using asynchronous callbacks to the server. While this provides the functionality needed, one negative impact is that Kcite makes heavy use of WordPress APIs (for caching, outgoing HTTP and so on), which directly makes it hard to port to other web frameworks.
Finally, this JSON is rendered using the citeproc-js library [
24] on the client. In our experience, this performs well, adding to the readers’ experience: in-text citations are initially shown as hyperlinks, rendering is rapid even on aging hardware, and finally in-text citations are linked both to the bibliography and directly through to the external source. Currently, the format of the reference list is fixed, however, citeproc-js is a generalised reference processor, driven by CSL [
25]. This makes it straightforward to change citation format, at the option of
thereader, rather than the author or the publisher. Both the in-text citation and the bibliography support outgoing links directing to the underlying resources (where the identifier allows—PubMed IDs redirect to PubMed). As these links have been used to gather metadata, they are likely to be correct. While these advantages are relatively small currently, we believe that the use of JavaScript rendering over a linked reference can be used to add further reader value in future.
For the computational agent wishing to consume bibliographic information, we have added significant value compared with the pre-formatted HTML reference list. First, all the information required to render the citation is present in the in-text citation, as a primary identifier, next to the text that the authors intended. A computational agent can therefore ignore the bibliography list itself entirely. These primary identifiers are, again, likely to be correct because the authors now need them to be correct for their own benefit. Currently, these identifiers are marked up with ad hoc microformat, although in future we may move to a more formal representation.
Should the computational agent wish, the (denormalised) bibliographic data used to render the bibliography is actually available in the underlying HTML as a JSON string. This is represented in a homogeneous format, although, of course, represents our (kcite’s) interpretation of the primary data.
A final, and subtle, advantage of kcite is that the authors can only use public metadata, and not their own. If they use the correct primary identifier, and still get an incorrect reference, it follows that the public metadata must be incorrect (or, we acknowledge, that kcite is broken!). Authors and readers therefore must ask the metadata providers to fix their metadata to the benefit of all. This form of data linking, therefore, can even help those who are not using it.
4.1. Microarray Data
Many publications require that papers discussing microarray experiments lodge their data in a publically available resource such as ArrayExpress [
26]. Authors do this by placing an ArrayExpress identifier, which has the form E-MEXP-1551. Currently, adding this identifier to a publication, as with adding the raw data to the repository, is no direct advantage to the author other than fulfilment of the publication requirement. Similarly, there is no existing support within most authoring environments for adding this form of reference.
For the knowledgeblog-arrayexpress plugin [
27], therefore, we have again used a shortcode representation, but allowed the author to automatically fill metadata, direct from ArrayExpress. Hence, a tag such as:
will be replaced with
Saccharomycescerevisiae, while:
will be replaced by “2010-02-24”. While the advantage here is small, it is significant. Hyperlinks to ArrayExpress are automatic and authors no longer need to look up detailed metadata. For the metadata that the authors are likely to know anyway (such as Species), the automatic lookup operates as a validation that their ArrayExpress ID is correct. As with references (see
Section 4), the use of an identifier becomes an
advantage rather than a burden to the authors.
For the reader there is currently less significant advantage, although there is some value to the author of the added correctness stemming from the ArrayExpress identifier. Nevertheless, knowledgeblog-arrayexpress is currently underdeveloped, and the added semantics that is now present could be used more extensively. The unambiguous knowledge that:
represents a species would allow us, for example, to link to the NCBI taxonomy database that contains data about species [
28].
Likewise, advantage for the computational agent from knowledgeblog-arrayexpress is currently limited; the identifiers are clearly marked up, and as the authors now care about them, they are likely to be correct. Again, however, knowledgeblog-arrayexpress is currently underdeveloped for the computational agent. The knowledge that is extracted from ArrayExpress could be presented within the HTML generated by knowledgeblog-arrayexpress, whether or not it is displayed to the reader for essentially no cost. By having an underlying shortcode representation, if we choose to add this functionality to knowledgeblog-arrayexpress, any posts that were written by using it would automatically update their HTML. For the text-mining bioinformatician, even the ability to unambiguously determine that a paper described or used a dataset relating to a specific species using standardised nomenclature (the standard nomenclature was only invented in 1753 and is still not used universally) would be a considerable boon.
5. Discussion
Our approach to semantic enrichment of articles is a measured and evolutionary approach. We are investigating how we can increase the amount of knowledge in academic articles presented in a computationally accessible form. However, we are doing so in an environment that does not require all the different aspects of authoring and publishing to be overturned, compared with the use of Wikis (semantic or otherwise) that provides a significantly different publishing paradigm. Moreover, we have followed a strong principle of semantic enhancement that offers advantages to both reader and author immediately. Hence, adding references as a DOI, or other identifier, “automagically” produces an in-text citation and a nicely formatted reference list. The reference list is no longer present in the article but is a visualisation over linked data. A happy by-product is that the article itself becomes a first class citizen of this linked data environment. As described, existing bibliographic tools can be used to make this process transparent to the author.
This approach, however, also has disadvantages. There are several semantic enhancements that we could but decided not to introduce to the knowledgeblog environment. The principles that we have adopted requires significant compromise. We offer here two examples.
First, there has been significant work by others on CiTO [
29]—an ontology that helps to describe the relationship between the citations and a paper. Kcite lays the groundwork for an easy and straightforward addition of CiTO tags surrounding each in-text citation. Doing so would enable increased machine understandability of a reference list. Potentially, we could use this to the advantage to the reader also: we could distinguish between reviews and primary research papers, highlight the authors’ previous work, emphasise older papers that are being refuted,
etc. However, to do this requires additional semantics from the author. Although these CiTO semantic enhancements would be easy to insert directly using the shortcode syntax, most authors will want to use their existing reference manager, which do not allow them to express this form of semantics. Even if it did, the author themselves gain little advantage from adding these semantics. There are advantages for the reader, but in this case not for both author and reader. As a result, we may add such support to kcite, but then—honestly speaking—be unlikely find the time to add such additional semantics when acting as content authors.
Second, our presentation of mathematics could be modified to automatically generate MathML from any included
![Futureinternet 04 01004 i017]()
markup. The transformation could be performed on the server, using MathJax, while MathML would still be rendered on the client in web fonts. This would mean that any embedded math would be discoverable because of the existence of MathML, which is a considerable advantage. However, neither the reader nor the author gain any advantage from doing this, while suffering from the longer load times and higher server load that would result from running JavaScript on the server. Moreover, they would pay this cost regardless of whether their content was actually being consumed computationally. As the situation now stands, the computational user needs to identify the insert of MathJax into the web page, and then transform the page using this library, none of which is standard. This is clearly a serious compromise, but necessary, as we feel.
Our support for microarrays offers the possibility of the most specific and increased level of semantics of all of our plugins. Knowledge about a species or a microarray experimental design can be precisely represented. However, almost by definition, this form of knowledge is fairly niche and only likely to be of relevance to a small community. However, we do note that the knowledgeblog process based on commodity technology does offer a publishing process that can be adapted, extended and specialised relatively easily in this way. Ultimately, the many small communities that make up the long tail of scientific publishing add up to one large one.
6. Conclusions
Semantic publishing is a desirable goal, but goals need to be realistic and achievable. To move towards semantic publishing in kblog , we have tried to put in place an approach that gives benefit to readers, authors and computational interpretation. As a result, at this stage, we have light semantic publishing, with small but definite benefits for all.
Semantics give meaning to entities. In kblog , we have sought benefit by “saying” within the kblog environment that entity x is either math, a citation or a microarray data entity reference. This is sufficient for the kblog infrastructure to “know what to do” with the entity in question. Knowing that some publishable entity is a “lump” of math tells the infrastructure how to handle that entity: the reader gets benefit as it looks like math, the author gets benefit by not having to do very much, and the infrastructure knows what to do. In addition, this approach leaves in hooks for doing more later.
It is not necessarily easy to find compelling examples that give advantages for all steps. Adding in CiTO attributes to citations, for instance, has obvious advantages for the reader, but not the author. However, advantages may be indirect; richer reader semantics may attract more readers and thus more citations—which the authors appreciate as much as the act of publishing itself. It is, however, difficult to imagine how such advantages can be conveyed to the author at the point of writing. It is easy to see the advantages of semantic publishing for readers, but as a community, we need to pay attention to advantages to the authors too. Without these “carrots”, we will only have “sticks”—and authors, particularly technically skilled ones, are highly adept at working around the sticks.
 [5], through to a final PDF which looks, feels and is something designed to be printed onto paper. Adding semantics into this environment is difficult or impossible; the content of the PDF has to be exposed and semantic content retrofitted or, in all likelihood, a complex process of author and publisher interaction has to be devised and followed. If semantic data publishing and semantic publishing of academic narratives are to work together, then academic publishing needs to change.
[5], through to a final PDF which looks, feels and is something designed to be printed onto paper. Adding semantics into this environment is difficult or impossible; the content of the PDF has to be exposed and semantic content retrofitted or, in all likelihood, a complex process of author and publisher interaction has to be devised and followed. If semantic data publishing and semantic publishing of academic narratives are to work together, then academic publishing needs to change. , because its idiomatic approach to programming documents is not replicable with anything else. Fortunately, it is possible to induce WordPress to accept content from many different authoring tools, including Word and
, because its idiomatic approach to programming documents is not replicable with anything else. Fortunately, it is possible to induce WordPress to accept content from many different authoring tools, including Word and  [6].
[6].which can be induced to produce HTML with effort, asciidoc is designed to do so.
 [13], and it still one of the main reasons why authors wish to use it or its derivatives. This is to such an extent that much mathematics rendering on the web is driven by a
[13], and it still one of the main reasons why authors wish to use it or its derivatives. This is to such an extent that much mathematics rendering on the web is driven by a  engine somewhere in the process. So MediaWiki (and therefore Wikipedia), Drupal and, of course, WordPress follow this route. The latter provides plugin support for
engine somewhere in the process. So MediaWiki (and therefore Wikipedia), Drupal and, of course, WordPress follow this route. The latter provides plugin support for  markup using the wp-latex plugin [14]. Within kblog, we have developed a new plugin called mathjax-latex [15]. From the kblog author’s perspective these two offer a similar interface—differences are, therefore, described later.
markup using the wp-latex plugin [14]. Within kblog, we have developed a new plugin called mathjax-latex [15]. From the kblog author’s perspective these two offer a similar interface—differences are, therefore, described later. using one of the four markup syntaxes. The most explicit (and therefore least likely to happen accidentally) is through the use of “shortcodes” [16]. These are a HTML-like markup originating from some forum/bulletin board systems. In this form an equation would be entered as [latex]e=mc^2[/latex], which would be rendered as “
using one of the four markup syntaxes. The most explicit (and therefore least likely to happen accidentally) is through the use of “shortcodes” [16]. These are a HTML-like markup originating from some forum/bulletin board systems. In this form an equation would be entered as [latex]e=mc^2[/latex], which would be rendered as “  ”. It is also possible to use three other syntaxes that are closer to math mode in
”. It is also possible to use three other syntaxes that are closer to math mode in  : $$e=mc^2$$, $latex e=mc^2$, or \[e=mc^2\].
: $$e=mc^2$$, $latex e=mc^2$, or \[e=mc^2\]. -loving mathematician working with a Word-using biologist can still set their equations using
-loving mathematician working with a Word-using biologist can still set their equations using  syntax; although Word will not render these at authoring time but, in practice, this causes few problems for such authors, who are experienced at reading
syntax; although Word will not render these at authoring time but, in practice, this causes few problems for such authors, who are experienced at reading  . Within an
. Within an  workflow, equations can be both rendered locally with source compiled to PDF and published to WordPress.
workflow, equations can be both rendered locally with source compiled to PDF and published to WordPress.
 , asciidoc) it is possible, although not easy, to prevent this within the native syntax.
, asciidoc) it is possible, although not easy, to prevent this within the native syntax. -like syntaxes has considerable advantages.
-like syntaxes has considerable advantages. engine into an image, which is then incorporated and linked using normal HTML capabilities. This representation is opaque and non-semantic; it has significant limitations for the reader. The images are not scalable—zooming in causes severe pixellation; the background to the mathematics is coloured inside the image and does not necessarily reflect the local style.
engine into an image, which is then incorporated and linked using normal HTML capabilities. This representation is opaque and non-semantic; it has significant limitations for the reader. The images are not scalable—zooming in causes severe pixellation; the background to the mathematics is coloured inside the image and does not necessarily reflect the local style. or MathML irrespective of the form that the author used. This can be cut-and-paste for further use. Kblog uses the MathJax library [17] to render the underlying
or MathML irrespective of the form that the author used. This can be cut-and-paste for further use. Kblog uses the MathJax library [17] to render the underlying  directly on the client.
directly on the client. 100 Mb), it is available on a CDN and does not stress the server storage space. Most of this space comes from the bit-mapped fonts that are only downloaded on demand, which should not stress web clients either. It also obviates the need for a
100 Mb), it is available on a CDN and does not stress the server storage space. Most of this space comes from the bit-mapped fonts that are only downloaded on demand, which should not stress web clients either. It also obviates the need for a  installation that wp-latex may require (although this plugin can use an external server also).
installation that wp-latex may require (although this plugin can use an external server also).
 style markup appears accidentally elsewhere in the document), the author is likely to have seen this and fixed the problem. Someone wishing, for example, to extract all the mathematics as MathML from these documents computationally, therefore, knows:
style markup appears accidentally elsewhere in the document), the author is likely to have seen this and fixed the problem. Someone wishing, for example, to extract all the mathematics as MathML from these documents computationally, therefore, knows: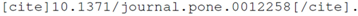

 /BibTeX citations, although we see no reason why a similar style file should not be supported. We do, however, support BibTeX-formatted files: the first author’s preferred editing/citation environment is based on these with Emacs, RefTeX (and Emacs package), and asciidoc. While this is undoubtedly a rather niche authoring environment, the (slightly elided) code shows that very little is needed to enable the tool chains to support kcite:
/BibTeX citations, although we see no reason why a similar style file should not be supported. We do, however, support BibTeX-formatted files: the first author’s preferred editing/citation environment is based on these with Emacs, RefTeX (and Emacs package), and asciidoc. While this is undoubtedly a rather niche authoring environment, the (slightly elided) code shows that very little is needed to enable the tool chains to support kcite:

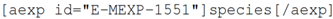
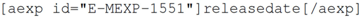
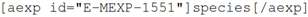
 markup. The transformation could be performed on the server, using MathJax, while MathML would still be rendered on the client in web fonts. This would mean that any embedded math would be discoverable because of the existence of MathML, which is a considerable advantage. However, neither the reader nor the author gain any advantage from doing this, while suffering from the longer load times and higher server load that would result from running JavaScript on the server. Moreover, they would pay this cost regardless of whether their content was actually being consumed computationally. As the situation now stands, the computational user needs to identify the insert of MathJax into the web page, and then transform the page using this library, none of which is standard. This is clearly a serious compromise, but necessary, as we feel.
markup. The transformation could be performed on the server, using MathJax, while MathML would still be rendered on the client in web fonts. This would mean that any embedded math would be discoverable because of the existence of MathML, which is a considerable advantage. However, neither the reader nor the author gain any advantage from doing this, while suffering from the longer load times and higher server load that would result from running JavaScript on the server. Moreover, they would pay this cost regardless of whether their content was actually being consumed computationally. As the situation now stands, the computational user needs to identify the insert of MathJax into the web page, and then transform the page using this library, none of which is standard. This is clearly a serious compromise, but necessary, as we feel.



