The 5′ and 3′ Untranslated Regions of the Flaviviral Genome
Abstract
:1. General Features of Flaviviruses
2. Flavivirus Untranslated Regions
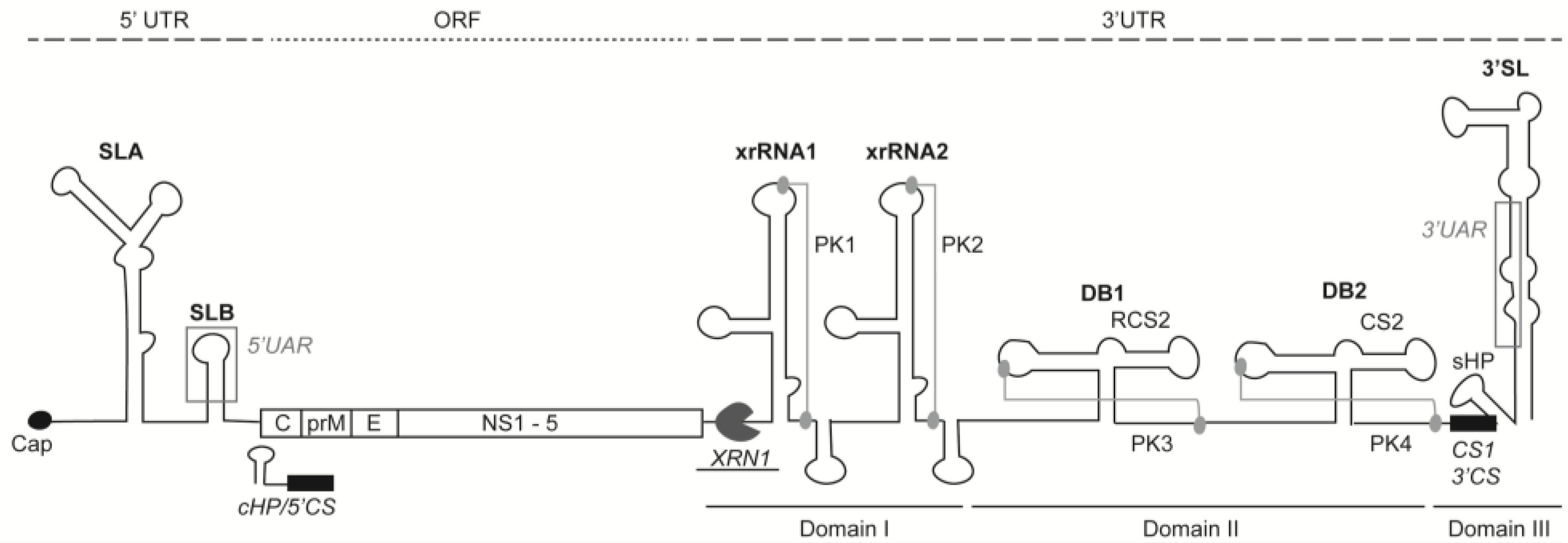
3. 5′ Untranslated Region
4. 3′ Untranslated Region
5. Generation of sfRNA
6. Vaccine Implications
7. Conclusions
Acknowledgments
Conflicts of Interest
References
- Moureau, G.; Cook, S.; Lemey, P.; Nougairede, A.; Forrester, N.L.; Khasnatinov, M.; Charrel, R.N.; Firth, A.E.; Gould, E.A.; de Lamballerie, X. New Insights into Flavivirus Evolution, Taxonomy and Biogeographic History, Extended by Analysis of Canonical and Alternative Coding Sequences. PLoS ONE 2015, 10, e0117849. [Google Scholar] [CrossRef] [PubMed]
- Blitvich, B.; Firth, A. Insect-Specific Flaviviruses: A Systematic Review of Their Discovery, Host Range, Mode of Transmission, Superinfection Exclusion Potential and Genomic Organization. Viruses 2015, 7, 1927–1959. [Google Scholar] [CrossRef] [PubMed]
- Chawla, T.; Chan, K.R.; Zhang, S.L.; Tan, H.C.; Lim, A.P.C.; Hanson, B.J.; Ooi, E.E. Dengue Virus Neutralization in Cells Expressing Fc Gamma Receptors. PLoS ONE 2013, 8, e65231. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; Kuhn, R.J.; Rossmann, M.G. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 2005, 3, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Lindenbach, B.D.; Rice, C.M. Molecular biology of flaviviruses. Adv. Virus Res. 2003, 59, 23–61. [Google Scholar] [PubMed]
- Bartholomeusz, A.I.; Wright, P.J. Synthesis of dengue virus RNA in vitro: Initiation and the involvement of proteins NS3 and NS5. Arch. Virol. 1993, 128, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Gebhard, L.G.; Filomatori, C.V.; Gamarnik, A.V. Functional RNA Elements in the Dengue Virus Genome. Viruses 2011, 3, 1739–1756. [Google Scholar] [CrossRef] [PubMed]
- Bidet, K.; Garcia-Blanco, M.A. Flaviviral RNAs: Weapons and targets in the war between virus and host. Biochem. J. 2014, 462, 215–230. [Google Scholar] [CrossRef] [PubMed]
- Corver, J.; Lenches, E.; Smith, K.; Robison, R.A.; Sando, T.; Strauss, E.G.; Strauss, J.H. Fine Mapping of a cis-Acting Sequence Element in Yellow Fever Virus RNA That Is Required for RNA Replication and Cyclization. J. Virol. 2003, 77, 2265–2270. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, D.E.; De Lella Ezcurra, A.L.; Fucito, S.; Gamarnik, A.V. Role of RNA structures present at the 3′UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology 2005, 339, 200–212. [Google Scholar] [CrossRef] [PubMed]
- Filomatori, C.V.; Lodeiro, M.F.; Alvarez, D.E.; Samsa, M.M.; Pietrasanta, L.; Gamarnik, A.V. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 2006, 20, 2238–2249. [Google Scholar] [CrossRef] [PubMed]
- Lodeiro, M.F.; Filomatori, C.V.; Gamarnik, A.V. Structural and functional studies of the promoter element for dengue virus RNA replication. J. Virol. 2009, 83, 993–1008. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Brinton, M.A. The 3′ stem loop of the West Nile virus genomic RNA can suppress translation of chimeric mRNAs. Virology 2001, 287, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Holden, K.L.; Stein, D.A.; Pierson, T.C.; Ahmed, A.A.; Clyde, K.; Iversen, P.L.; Harris, E. Inhibition of dengue virus translation and RNA synthesis by a morpholino oligomer targeted to the top of the terminal 3' stem-loop structure. Virology 2006, 344, 439–452. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Qin, C.; Jiang, T.; Li, X.; Zhao, H.; Liu, Z.; Deng, Y.; Liu, R.; Chen, S.; Yu, M.; et al. Translational regulation by the 3′ untranslated region of the dengue type 2 virus genome. Am. J. Trop. Med. Hyg. 2009, 81, 817–824. [Google Scholar] [CrossRef] [PubMed]
- Holden, K.L.; Harris, E. Enhancement of dengue virus translation: Role of the 3′ untranslated region and the terminal 3′ stem-loop domain. Virology 2004, 329, 119–133. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Falgout, B.; Markoff, L. Identification of specific nucleotide sequences within the conserved 3′-SL in the dengue type 2 virus genome required for replication. J. Virol. 1998, 72, 7510–7522. [Google Scholar] [PubMed]
- Yu, L.; Nomaguchi, M.; Padmanabhan, R.; Markoff, L. Specific requirements for elements of the 5′ and 3′ terminal regions in flavivirus RNA synthesis and viral replication. Virology 2008, 374, 170–185. [Google Scholar] [CrossRef] [PubMed]
- Clyde, K.; Barrera, J.; Harris, E. The capsid-coding region hairpin element (cHP) is a critical determinant of dengue virus and West Nile virus RNA synthesis. Virology 2008, 379, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Cahour, A.; Pletnev, A.; Vazielle-Falcoz, M.; Rosen, L.; Lai, C.J. Growth-restricted dengue virus mutants containing deletions in the 5′ noncoding region of the RNA genome. Virology 1995, 207, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Markoff, L. The topology of bulges in the long stem of the flavivirus 3′ stem-loop is a major determinant of RNA replication competence. J. Virol. 2005, 79, 2309–2324. [Google Scholar] [CrossRef] [PubMed]
- Clyde, K.; Harris, E. RNA secondary structure in the coding region of dengue virus type 2 directs translation start codon selection and is required for viral replication. J. Virol. 2006, 80, 2170–2182. [Google Scholar] [CrossRef] [PubMed]
- Villordo, S.M.; Alvarez, D.E.; Gamarnik, A.V. A balance between circular and linear forms of the dengue virus genome is crucial for viral replication. RNA 2010, 16, 2325–2335. [Google Scholar] [CrossRef] [PubMed]
- Villordo, S.M.; Gamarnik, A.V. Differential RNA Sequence Requirement for Dengue Virus Replication in Mosquito and Mammalian Cells. J. Virol. 2013, 87, 9365–9372. [Google Scholar] [CrossRef] [PubMed]
- Khromykh, A.A.; Meka, H.; Guyatt, K.J.; Westaway, E.G. Essential Role of Cyclization Sequences in Flavivirus RNA Replication. J. Virol. 2001, 75, 6719–6728. [Google Scholar] [CrossRef] [PubMed]
- Villordo, S.M.; Gamarnik, A.V. Genome cyclization as strategy for flavivirus RNA replication. Virus Res. 2009, 139, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Hahn, C.S.; Hahn, Y.S.; Rice, C.M.; Lee, E.; Dalgarno, L.; Strauss, E.G.; Strauss, J.H. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 1987, 198, 33–41. [Google Scholar] [CrossRef]
- Basu, M.; Brinton, M.A. West Nile virus (WNV) genome RNAs with up to three adjacent mutations that disrupt long distance 5′–3′ cyclization sequence basepairs are viable. Virology 2011, 412, 220–232. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, D.E.; Lodeiro, M.F.; Luduena, S.J.; Pietrasanta, L.I.; Gamarnik, A.V. Long-Range RNA-RNA Interactions Circularize the Dengue Virus Genome. J. Virol. 2005, 79, 6631–6643. [Google Scholar] [CrossRef] [PubMed]
- Khromykh, A.A.; Kondratieva, N.; Sgro, J.-Y.; Palmenberg, A.; Westaway, E.G. Significance in replication of the terminal nucleotides of the flavivirus genome. J. Virol. 2003, 77, 10623–10629. [Google Scholar] [CrossRef] [PubMed]
- Tilgner, M.; Deas, T.S.; Shi, P.-Y. The flavivirus-conserved penta-nucleotide in the 3′ stem-loop of the West Nile virus genome requires a specific sequence and structure for RNA synthesis, but not for viral translation. Virology 2005, 331, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Elghonemy, S.; Davis, W.G.; Brinton, M.A. The majority of the nucleotides in the top loop of the genomic 3′ terminal stem loop structure are cis-acting in a West Nile virus infectious clone. Virology 2005, 331, 238–246. [Google Scholar] [CrossRef] [PubMed]
- Silva, P.A.G.C.; Molenkamp, R.; Dalebout, T.J.; Charlier, N.; Neyts, J.H.; Spaan, W.J.M.; Bredenbeek, P.J. Conservation of the pentanucleotide motif at the top of the yellow fever virus 17D 3′ stem-loop structure is not required for replication. J. Gen. Virol. 2007, 88, 1738–1747. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, D.E.; Filomatori, C.V.; Gamarnik, A.V. Functional analysis of dengue virus cyclization sequences located at the 5′ and 3′UTRs. Virology 2008, 375, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Dong, H.; Stein, D.A.; Iversen, P.L.; Shi, P.-Y. West Nile virus genome cyclization and RNA replication require two pairs of long-distance RNA interactions. Virology 2008, 373, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Friebe, P.; Harris, E. Interplay of RNA Elements in the Dengue Virus 5′ and 3′ Ends Required for Viral RNA Replication. J. Virol. 2010, 84, 6103–6118. [Google Scholar] [CrossRef] [PubMed]
- Friebe, P.; Shi, P.Y.; Harris, E. The 5′ and 3′ Downstream AUG Region Elements Are Required for Mosquito-Borne Flavivirus RNA Replication. J. Virol. 2011, 85, 1900–1905. [Google Scholar] [CrossRef] [PubMed]
- Brinton, M.A.; Dispoto, J.H. Sequence and secondary structure analysis of the 5′-terminal region of flavivirus genome RNA. Virology 1988, 162, 290–299. [Google Scholar] [CrossRef]
- Polacek, C.; Friebe, P.; Harris, E. Poly(A)-binding protein binds to the non-polyadenylated 3′ untranslated region of dengue virus and modulates translation efficiency. J. Gen. Virol. 2009, 90, 687–692. [Google Scholar] [CrossRef] [PubMed]
- Wengler, G.; Gross, H.J. Studies on virus-specific nucleic acids synthesized in vertebrate and mosquito cells infected with flaviviruses. Virology 1978, 89, 423–437. [Google Scholar] [CrossRef]
- Cleaves, G.R.; Dubin, D.T. Methylation status of intracellular dengue type 2 40 S RNA. Virology 1979, 96, 159–165. [Google Scholar] [CrossRef]
- Edgil, D.; Polacek, C.; Harris, E. Dengue Virus Utilizes a Novel Strategy for Translation Initiation When Cap-Dependent Translation Is Inhibited. J. Virol. 2006, 80, 2976–2986. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-Y.; Li, X.-F.; Jiang, T.; Deng, Y.-Q.; Ye, Q.; Zhao, H.; Yu, J.-Y.; Qin, C.-F. Viral RNA switch mediates the dynamic control of flavivirus replicase recruitment by genome cyclization. eLife 2016, 5, e17636. [Google Scholar] [CrossRef] [PubMed]
- Carletti, T.; Zakaria, M.K.; Marcello, A. The host cell response to tick-borne encephalitis virus. Biochem. Biophys. Res. Commun. 2017, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kofler, R.M.; Hoenninger, V.M.; Thurner, C.; Mandl, C.W. Functional Analysis of the Tick-Borne Encephalitis Virus Cyclization Elements Indicates Major Differences between Mosquito-Borne and Tick-Borne Flaviviruses. J. Virol. 2006, 80, 4099–4113. [Google Scholar] [CrossRef] [PubMed]
- Rouha, H.; Hoenninger, V.M.; Thurner, C.; Mandl, C.W. Mutational analysis of three predicted 5′-proximal stem-loop structures in the genome of tick-borne encephalitis virus indicates different roles in RNA replication and translation. Virology 2011, 417, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Silva, R.L.A.; de Silva, A.M.; Harris, E.; MacDonald, G.H. Genetic analysis of Dengue 3 virus subtype III 5′ and 3′ non-coding regions. Virus Res. 2008, 135, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Mangada, M.N.; Igarashi, A. Sequences of terminal non-coding regions from four dengue-2 viruses isolated from patients exhibiting different disease severities. Virus Genes 1997, 14, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Kinney, R.M.; Butrapet, S.; Chang, G.J.; Tsuchiya, K.R.; Roehrig, J.T.; Bhamarapravati, N.; Gubler, D.J. Construction of infectious cDNA clones for dengue 2 virus: Strain 16681 and its attenuated vaccine derivative, strain PDK-53. Virology 1997, 230, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Butrapet, S.; Huang, C.Y.; Pierro, D.J.; Bhamarapravati, N.; Gubler, D.J.; Kinney, R.M. Attenuation markers of a candidate dengue type 2 vaccine virus, strain 16681 (PDK-53), are defined by mutations in the 5′ noncoding region and nonstructural proteins 1 and 3. J. Virol. 2000, 74, 3011–3019. [Google Scholar] [CrossRef] [PubMed]
- Butrapet, S.; Kinney, R.M.; Huang, C.Y.-H. Determining genetic stabilities of chimeric dengue vaccine candidates based on dengue 2 PDK-53 virus by sequencing and quantitative TaqMAMA. J. Virol. Methods 2006, 131, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Brinton, M.A.; Fernandez, A.V.; Dispoto, J.H. The 3′-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology 1986, 153, 113–121. [Google Scholar] [CrossRef]
- Mandl, C.W.; Holzmann, H.; Kunz, C.; Heinz, F.X. Complete genomic sequence of Powassan virus: Evaluation of genetic elements in tick-borne versus mosquito-borne flaviviruses. Virology 1993, 194, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, J.L.; Brinton, M.A. BHK cell proteins that bind to the 3′ stem-loop structure of the West Nile virus genome RNA. J. Virol. 1995, 69, 5650–5658. [Google Scholar] [PubMed]
- Shi, P.Y.; Brinton, M.A.; Veal, J.M.; Zhong, Y.Y.; Wilson, W.D. Evidence for the existence of a pseudoknot structure at the 3′ terminus of the flavivirus genomic RNA. Biochemistry 1996, 35, 4222–4230. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, J.L.; Brinton, M.A. Translation elongation factor-1 alpha interacts with the 3′ stem-loop region of West Nile virus genomic RNA. J. Virol. 1997, 71, 6433–6444. [Google Scholar] [PubMed]
- Selisko, B.; Potisopon, S.; Agred, R.; Priet, S.; Varlet, I.; Thillier, Y.; Sallamand, C.; Debart, F.; Vasseur, J.-J.; Canard, B. Molecular Basis for Nucleotide Conservation at the Ends of the Dengue Virus Genome. PLoS Pathog. 2012, 8, e1002912. [Google Scholar] [CrossRef] [PubMed]
- Paranjape, S.M.; Harris, E. Y box-binding protein-1 binds to the dengue virus 3′-untranslated region and mediates antiviral effects. J. Biol. Chem. 2007, 282, 30497–30508. [Google Scholar] [CrossRef] [PubMed]
- Proutski, V.; Gritsun, T.S.; Gould, E.A.; Holmes, E.C. Biological consequences of deletions within the 3′-untranslated region of flaviviruses may be due to rearrangements of RNA secondary structure. Virus Res. 1999, 64, 107–123. [Google Scholar] [CrossRef]
- Asghar, N.; Lindblom, P.; Melik, W.; Lindqvist, R.; Haglund, M.; Forsberg, P.; Överby, A.K.; Andreassen, Å.; Lindgren, P.-E.; Johansson, M. Tick-Borne Encephalitis Virus Sequenced Directly from Questing and Blood-Feeding Ticks Reveals Quasispecies Variance. PLoS ONE 2014, 9, e103264. [Google Scholar] [CrossRef] [PubMed]
- Sakai, M.; Yoshii, K.; Sunden, Y.; Yokozawa, K.; Hirano, M.; Kariwa, H. Variable region of the 3′ UTR is a critical virulence factor in the Far-Eastern subtype of tick-borne encephalitis virus in a mouse model. J. Gen. Virol. 2014, 95, 823–835. [Google Scholar] [CrossRef] [PubMed]
- Villordo, S.M.; Filomatori, C.V.; Sánchez-Vargas, I.; Blair, C.D.; Gamarnik, A.V. Dengue Virus RNA Structure Specialization Facilitates Host Adaptation. PLoS Pathog. 2015, 11, e1004604. [Google Scholar] [CrossRef] [PubMed]
- Olsthoorn, R.C.; Bol, J.F. Sequence comparison and secondary structure analysis of the 3′ noncoding region of flavivirus genomes reveals multiple pseudoknots. RNA 2001, 7, 1370–1377. [Google Scholar] [PubMed]
- Romero, T.A.; Tumban, E.; Jun, J.; Lott, W.B.; Hanley, K.A. Secondary structure of dengue virus type 4 3′ untranslated region: Impact of deletion and substitution mutations. J. Gen. Virol. 2006, 87, 3291–3296. [Google Scholar] [CrossRef] [PubMed]
- Sztuba-Solinska, J.; Teramoto, T.; Rausch, J.W.; Shapiro, B.A.; Padmanabhan, R.; Le Grice, S.F.J. Structural complexity of Dengue virus untranslated regions: Cis-acting RNA motifs and pseudoknot interactions modulating functionality of the viral genome. Nucleic Acids Res. 2013, 41, 5075–5089. [Google Scholar] [CrossRef] [PubMed]
- Manzano, M.; Reichert, E.D.; Polo, S.; Falgout, B.; Kasprzak, W.; Shapiro, B.A.; Padmanabhan, R. Identification of cis-acting elements in the 3′-untranslated region of the dengue virus type 2 RNA that modulate translation and replication. J. Biol. Chem. 2011, 286, 22521–22534. [Google Scholar] [CrossRef] [PubMed]
- Funk, A.; Truong, K.; Nagasaki, T.; Torres, S.; Floden, N.; Balmori Melian, E.; Edmonds, J.; Dong, H.; Shi, P.-Y.; Khromykh, A.A. RNA structures required for production of subgenomic flavivirus RNA. J. Virol. 2010, 84, 11407–11417. [Google Scholar] [CrossRef] [PubMed]
- Charlier, N.; Leyssen, P.; Pleij, C.W.A.; Lemey, P.; Billoir, F.; Van Laethem, K.; Vandamme, A.-M.; De Clercq, E.; de Lamballerie, X.; Neyts, J. Complete genome sequence of Montana Myotis leukoencephalitis virus, phylogenetic analysis and comparative study of the 3′ untranslated region of flaviviruses with no known vector. J. Gen. Virol. 2002, 83, 1875–1885. [Google Scholar] [CrossRef] [PubMed]
- Mutebi, J.P.; Rijnbrand, R.C.A.; Wang, H.; Ryman, K.D.; Wang, E.; Fulop, L.D.; Titball, R.; Barrett, A.D.T. Genetic Relationships and Evolution of Genotypes of Yellow Fever Virus and Other Members of the Yellow Fever Virus Group within the Flavivirus Genus Based on the 3′ Noncoding Region. J. Virol. 2004, 78, 9652–9665. [Google Scholar] [CrossRef] [PubMed]
- Gritsun, T.S.; Gould, E.A. Direct repeats in the 3′ untranslated regions of mosquito-borne flaviviruses: Possible implications for virus transmission. J. Gen. Virol. 2006, 87, 3297–3305. [Google Scholar] [CrossRef] [PubMed]
- Men, R.; Bray, M.; Clark, D.; Chanock, R.M.; Lai, C.J. Dengue type 4 virus mutants containing deletions in the 3′ noncoding region of the RNA genome: Analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J. Virol. 1996, 70, 3930–3937. [Google Scholar] [PubMed]
- Khromykh, A.A.; Westaway, E.G. Subgenomic replicons of the flavivirus Kunjin: Construction and applications. J. Virol. 1997, 71, 1497–1505. [Google Scholar] [PubMed]
- Bredenbeek, P.J.; Kooi, E.A.; Lindenbach, B.; Huijkman, N.; Rice, C.M.; Spaan, W.J.M. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. J. Gen. Virol. 2003, 84, 1261–1268. [Google Scholar] [CrossRef] [PubMed]
- Lo, M.K.; Tilgner, M.; Bernard, K.A.; Shi, P.-Y. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3′ untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J. Virol. 2003, 77, 10004–10014. [Google Scholar] [CrossRef] [PubMed]
- Gritsun, D.J.; Jones, I.M.; Gould, E.A.; Gritsun, T.S. Molecular Archaeology of Flaviviridae Untranslated Regions: Duplicated RNA Structures in the Replication Enhancer of Flaviviruses and Pestiviruses Emerged via Convergent Evolution. PLoS ONE 2014, 9, e92056. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, Y.; Kedersha, N.; Anderson, P.; Emara, M.; Swiderek, K.M.; Moreno, G.T.; Brinton, M.A. Cell proteins TIA-1 and TIAR interact with the 3′ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J. Virol. 2002, 76, 11989–12000. [Google Scholar] [CrossRef] [PubMed]
- Emara, M.M.; Brinton, M.A. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc. Natl. Acad. Sci. USA 2007, 104, 9041–9046. [Google Scholar] [CrossRef] [PubMed]
- Albornoz, A.; Carletti, T.; Corazza, G.; Marcello, A. The stress granule component TIA-1 binds tick-borne encephalitis virus RNA and is recruited to perinuclear sites of viral replication to inhibit viral translation. J. Virol. 2014, 88, 6611–6622. [Google Scholar] [CrossRef] [PubMed]
- Mandl, C.W.; Holzmann, H.; Meixner, T.; Rauscher, S.; Stadler, P.F.; Allison, S.L.; Heinz, F.X. Spontaneous and engineered deletions in the 3′ noncoding region of tick-borne encephalitis virus: Construction of highly attenuated mutants of a flavivirus. J. Virol. 1998, 72, 2132–2140. [Google Scholar] [PubMed]
- Cologna, R.; Rico-Hesse, R. American genotype structures decrease dengue virus output from human monocytes and dendritic cells. J. Virol. 2003, 77, 3929–3938. [Google Scholar] [CrossRef] [PubMed]
- Manokaran, G.; Finol, E.; Wang, C.; Gunaratne, J.; Bahl, J.; Ong, E.Z.; Tan, H.C.; Sessions, O.M.; Ward, A.M.; Gubler, D.J.; et al. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science 2015, 350, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Edgil, D.; Diamond, M.S.; Holden, K.L.; Paranjape, S.M.; Harris, E. Translation efficiency determines differences in cellular infection among dengue virus type 2 strains. Virology 2003, 317, 275–290. [Google Scholar] [CrossRef] [PubMed]
- Urosevic, N.; van Maanen, M.; Mansfield, J.P.; Mackenzie, J.S.; Shellam, G.R. Molecular characterization of virus-specific RNA produced in the brains of flavivirus-susceptible and -resistant mice after challenge with Murray Valley encephalitis virus. J. Gen. Virol. 1997, 78 Pt 1, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Pijlman, G.P.; Funk, A.; Kondratieva, N.; Leung, J.; Torres, S.; van der Aa, L.; Liu, W.J.; Palmenberg, A.C.; Shi, P.-Y.; Hall, R.A.; et al. A Highly Structured, Nuclease-Resistant, Noncoding RNA Produced by Flaviviruses Is Required for Pathogenicity. Cell Host Microbe 2008, 4, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Silva, P.A.G.C.; Pereira, C.F.; Dalebout, T.J.; Spaan, W.J.M.; Bredenbeek, P.J. An RNA pseudoknot is required for production of yellow fever virus subgenomic RNA by the host nuclease XRN1. J. Virol. 2010, 84, 11395–11406. [Google Scholar] [CrossRef] [PubMed]
- Chapman, E.G.; Costantino, D.A.; Rabe, J.L.; Moon, S.L.; Wilusz, J.; Nix, J.C.; Kieft, J.S. The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science 2014, 344, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, B.M.; Laurence, H.M.; Massey, A.R.; Costantino, D.A.; Xie, X.; Yang, Y.; Shi, P.-Y.; Nix, J.C.; Beckham, J.D.; Kieft, J.S. Zika virus produces noncoding RNAs using a multi-pseudoknot structure that confounds a cellular exonuclease. Science 2016, 354, 1148–1152. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.I.; Zabolotskaya, M.V.; Newbury, S.F. The 5″→3″ exoribonuclease XRN1/Pacman and its functions in cellular processes and development. Wiley Interdiscip. Rev. RNA 2012, 3, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Kieft, J.S.; Rabe, J.L.; Chapman, E.G. New hypotheses derived from the structure of a flaviviral Xrn1-resistant RNA: Conservation, folding, and host adaptation. RNA Biol. 2015, 12, 1169–1177. [Google Scholar] [CrossRef] [PubMed]
- Filomatori, C.V.; Carballeda, J.M.; Villordo, S.M.; Aguirre, S.; Pallarés, H.M.; Maestre, A.M.; Sánchez-Vargas, I.; Blair, C.D.; Fabri, C.; Morales, M.A.; et al. Dengue virus genomic variation associated with mosquito adaptation defines the pattern of viral non-coding RNAs and fitness in human cells. PLoS Pathog. 2017, 13, e1006265. [Google Scholar] [CrossRef] [PubMed]
- Chapman, E.G.; Moon, S.L.; Wilusz, J.; Kieft, J.S. RNA structures that resist degradation by XRN1 produce a pathogenic Dengue virus RNA. eLife 2014, 3, e01892. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.L.; Anderson, J.R.; Kumagai, Y.; Wilusz, C.J.; Akira, S.; Khromykh, A.A.; Wilusz, J. A noncoding RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease XRN1 and alters host mRNA stability. RNA 2012, 18, 2029–2040. [Google Scholar] [CrossRef] [PubMed]
- Schnettler, E.; Sterken, M.G.; Leung, J.Y.; Metz, S.W.; Geertsema, C.; Goldbach, R.W.; Vlak, J.M.; Kohl, A.; Khromykh, A.A.; Pijlman, G.P. Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and Mammalian cells. J. Virol. 2012, 86, 13486–13500. [Google Scholar] [CrossRef] [PubMed]
- Schuessler, A.; Funk, A.; Lazear, H.M.; Cooper, D.A.; Torres, S.; Daffis, S.; Jha, B.K.; Kumagai, Y.; Takeuchi, O.; Hertzog, P.; et al. West Nile virus noncoding subgenomic RNA contributes to viral evasion of the type I interferon-mediated antiviral response. J. Virol. 2012, 86, 5708–5718. [Google Scholar] [CrossRef] [PubMed]
- Chang, R.-Y.; Hsu, T.-W.; Chen, Y.-L.; Liu, S.-F.; Tsai, Y.-J.; Lin, Y.-T.; Chen, Y.-S.; Fan, Y.-H. Japanese encephalitis virus non-coding RNA inhibits activation of interferon by blocking nuclear translocation of interferon regulatory factor 3. Vet. Microbiol. 2013, 166, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Schnettler, E.; Tykalová, H.; Watson, M.; Sharma, M.; Sterken, M.G.; Obbard, D.J.; Lewis, S.H.; McFarlane, M.; Bell-Sakyi, L.; Barry, G.; et al. Induction and suppression of tick cell antiviral RNAi responses by tick-borne flaviviruses. Nucleic Acids Res. 2014, 42, 9436–9446. [Google Scholar] [CrossRef] [PubMed]
- Bidet, K.; Dadlani, D.; Garcia-Blanco, M.A. G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS Pathog. 2014, 10, e1004242. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.L.; Dodd, B.J.T.; Brackney, D.E.; Wilusz, C.J.; Ebel, G.D.; Wilusz, J. Flavivirus sfRNA suppresses antiviral RNA interference in cultured cells and mosquitoes and directly interacts with the RNAi machinery. Virology 2015, 485, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Göertz, G.P.; Fros, J.J.; Miesen, P.; Vogels, C.B.F.; van der Bent, M.L.; Geertsema, C.; Koenraadt, C.J.M.; van Rij, R.P.; van Oers, M.M.; Pijlman, G.P. Noncoding Subgenomic Flavivirus RNA is Processed by the Mosquito RNA Interference Machinery and Determines West Nile Virus Transmission by Culex pipiens Mosquitoes. J. Virol. 2016, 90, 10145–10159. [Google Scholar] [CrossRef] [PubMed]
- Myles, K.M.; Wiley, M.R.; Morazzani, E.M.; Adelman, Z.N. Alphavirus-derived small RNAs modulate pathogenesis in disease vector mosquitoes. Proc. Natl. Acad. Sci. USA 2008, 105, 19938–19943. [Google Scholar] [CrossRef] [PubMed]
- Cirimotich, C.M.; Scott, J.C.; Phillips, A.T.; Geiss, B.J.; Olson, K.E. Suppression of RNA interference increases alphavirus replication and virus-associated mortality in Aedes aegypti mosquitoes. BMC Microbiol. 2009, 9, 49. [Google Scholar] [CrossRef] [PubMed]
- Khoo, C.C.H.; Doty, J.B.; Heersink, M.S.; Olson, K.E.; Franz, A.W.E. Transgene-mediated suppression of the RNA interference pathway in Aedes aegypti interferes with gene silencing and enhances Sindbis virus and dengue virus type 2 replication. Insect Mol. Biol. 2013, 22, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.R.; Wang, X.; Saron, W.A.A.; Gan, E.S.; Tan, H.C.; Mok, D.Z.L.; Zhang, S.L.-X.; Lee, Y.H.; Liang, C.; Wijaya, L.; et al. Cross-reactive antibodies enhance live attenuated virus infection for increased immunogenicity. Nat. Microbiol. 2016, 1, 16164. [Google Scholar] [CrossRef] [PubMed]
- Durbin, A.P.; Karron, R.A.; Sun, W.; Vaughn, D.W.; Reynolds, M.J.; Perreault, J.R.; Thumar, B.; Men, R.; Lai, C.J.; Elkins, W.R.; et al. Attenuation and immunogenicity in humans of a live dengue virus type-4 vaccine candidate with a 30-nucleotide deletion in its 3′-untranslated region. Am. J. Trop. Med. Hyg. 2001, 65, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, S.S.; Falgout, B.; Hanley, K.A.; Blaney, J.E., Jr.; Markoff, L.; Murphy, B.R. A live, attenuated dengue virus type 1 vaccine candidate with a 30-nucleotide deletion in the 3′ untranslated region is highly attenuated and immunogenic in monkeys. J. Virol. 2003, 77, 1653–1657. [Google Scholar] [CrossRef] [PubMed]
- Blaney, J.E.; Hanson, C.T.; Hanley, K.A.; Murphy, B.R.; Whitehead, S.S. Vaccine candidates derived from a novel infectious cDNA clone of an American genotype dengue virus type 2. BMC Infect. Dis. 2004, 4, 39. [Google Scholar] [CrossRef] [PubMed]
- Blaney, J.E.; Hanson, C.T.; Firestone, C.-Y.; Hanley, K.A.; Murphy, B.R.; Whitehead, S.S. Genetically modified, live attenuated dengue virus type 3 vaccine candidates. Am. J. Trop. Med. Hyg. 2004, 71, 811–821. [Google Scholar] [PubMed]
- Troyer, J.M.; Hanley, K.A.; Whitehead, S.S.; Strickman, D.; Karron, R.A.; Durbin, A.P.; Murphy, B.R. A live attenuated recombinant dengue-4 virus vaccine candidate with restricted capacity for dissemination in mosquitoes and lack of transmission from vaccinees to mosquitoes. Am. J. Trop. Med. Hyg. 2001, 65, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Blaney, J.E.; Sathe, N.S.; Goddard, L.; Hanson, C.T.; Romero, T.A.; Hanley, K.A.; Murphy, B.R.; Whitehead, S.S. Dengue virus type 3 vaccine candidates generated by introduction of deletions in the 3′ untranslated region (3′-UTR) or by exchange of the DENV-3 3′-UTR with that of DENV-4. Vaccine 2008, 26, 817–828. [Google Scholar] [CrossRef] [PubMed]
- Blaney, J.E.; Matro, J.M.; Murphy, B.R.; Whitehead, S.S. Recombinant, live-attenuated tetravalent dengue virus vaccine formulations induce a balanced, broad, and protective neutralizing antibody response against each of the four serotypes in rhesus monkeys. J. Virol. 2005, 79, 5516–5528. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, B.D.; Whitehead, S.S.; Pierce, K.K.; Tibery, C.M.; Grier, P.L.; Hynes, N.A.; Larsson, C.J.; Sabundayo, B.P.; Talaat, K.R.; Janiak, A.; et al. The live attenuated dengue vaccine TV003 elicits complete protection against dengue in a human challenge model. Sci. Transl. Med. 2016, 8, 330ra36. [Google Scholar] [CrossRef] [PubMed]
- Shan, C.; Muruato, A.E.; Nunes, B.T.D.; Luo, H.; Xie, X.; Medeiros, D.B.A.; Wakamiya, M.; Tesh, R.B.; Barrett, A.D.; Wang, T.; et al. A live-attenuated Zika virus vaccine candidate induces sterilizing immunity in mouse models. Nat. Med. 2017, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Osorio, J.E.; Huang, C.Y.-H.; Kinney, R.M.; Stinchcomb, D.T. Development of DENVax: A chimeric dengue-2 PDK-53-based tetravalent vaccine for protection against dengue fever. Vaccine 2011, 29, 7251–7260. [Google Scholar] [CrossRef] [PubMed]
- Chambers, T.J.; Liang, Y.; Droll, D.A.; Schlesinger, J.J.; Davidson, A.D.; Wright, P.J.; Jiang, X. Yellow Fever Virus/Dengue-2 Virus and Yellow Fever Virus/Dengue-4 Virus Chimeras: Biological Characterization, Immunogenicity, and Protection against Dengue Encephalitis in the Mouse Model. J. Virol. 2003, 77, 3655–3668. [Google Scholar] [CrossRef] [PubMed]
- Engel, A.R.; Rumyantsev, A.A.; Maximova, O.A.; Speicher, J.M.; Heiss, B.; Murphy, B.R.; Pletnev, A.G. The neurovirulence and neuroinvasiveness of chimeric tick-borne encephalitis/dengue virus can be attenuated by introducing defined mutations into the envelope and NS5 protein genes and the 3′ non-coding region of the genome. Virology 2010, 405, 243–252. [Google Scholar] [CrossRef] [PubMed]
| Downstream of Stop Codon: | ||||
|---|---|---|---|---|
| 1st SL | 2nd SL | 1st DB | 2nd DB | Reference |
| SL II | SL IV | [62] | ||
| DEN-SLI | DEN-SLII | [90] | ||
| xrRNA1 | xrRNA2 | xrRNA3 | xrRNA4 | [84,92] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ng, W.C.; Soto-Acosta, R.; Bradrick, S.S.; Garcia-Blanco, M.A.; Ooi, E.E. The 5′ and 3′ Untranslated Regions of the Flaviviral Genome. Viruses 2017, 9, 137. https://doi.org/10.3390/v9060137
Ng WC, Soto-Acosta R, Bradrick SS, Garcia-Blanco MA, Ooi EE. The 5′ and 3′ Untranslated Regions of the Flaviviral Genome. Viruses. 2017; 9(6):137. https://doi.org/10.3390/v9060137
Chicago/Turabian StyleNg, Wy Ching, Ruben Soto-Acosta, Shelton S. Bradrick, Mariano A. Garcia-Blanco, and Eng Eong Ooi. 2017. "The 5′ and 3′ Untranslated Regions of the Flaviviral Genome" Viruses 9, no. 6: 137. https://doi.org/10.3390/v9060137
APA StyleNg, W. C., Soto-Acosta, R., Bradrick, S. S., Garcia-Blanco, M. A., & Ooi, E. E. (2017). The 5′ and 3′ Untranslated Regions of the Flaviviral Genome. Viruses, 9(6), 137. https://doi.org/10.3390/v9060137






