Identification of HIV-1 Tat-Associated Proteins Contributing to HIV-1 Transcription and Latency
Abstract
:1. Introduction
2. Methods and Materials
2.1. Cells and Plasmids
2.2. Small Molecules
2.3. Viruses
2.4. Antibodies
2.5. shRNAs
2.6. PLATO
2.7. Luciferase Reporter Assays
2.8. Co-Immunoprecipitation (co-IP)
2.9. Real-Time Quantitative Polymerase Chain Reaction (qPCR)
3. Results
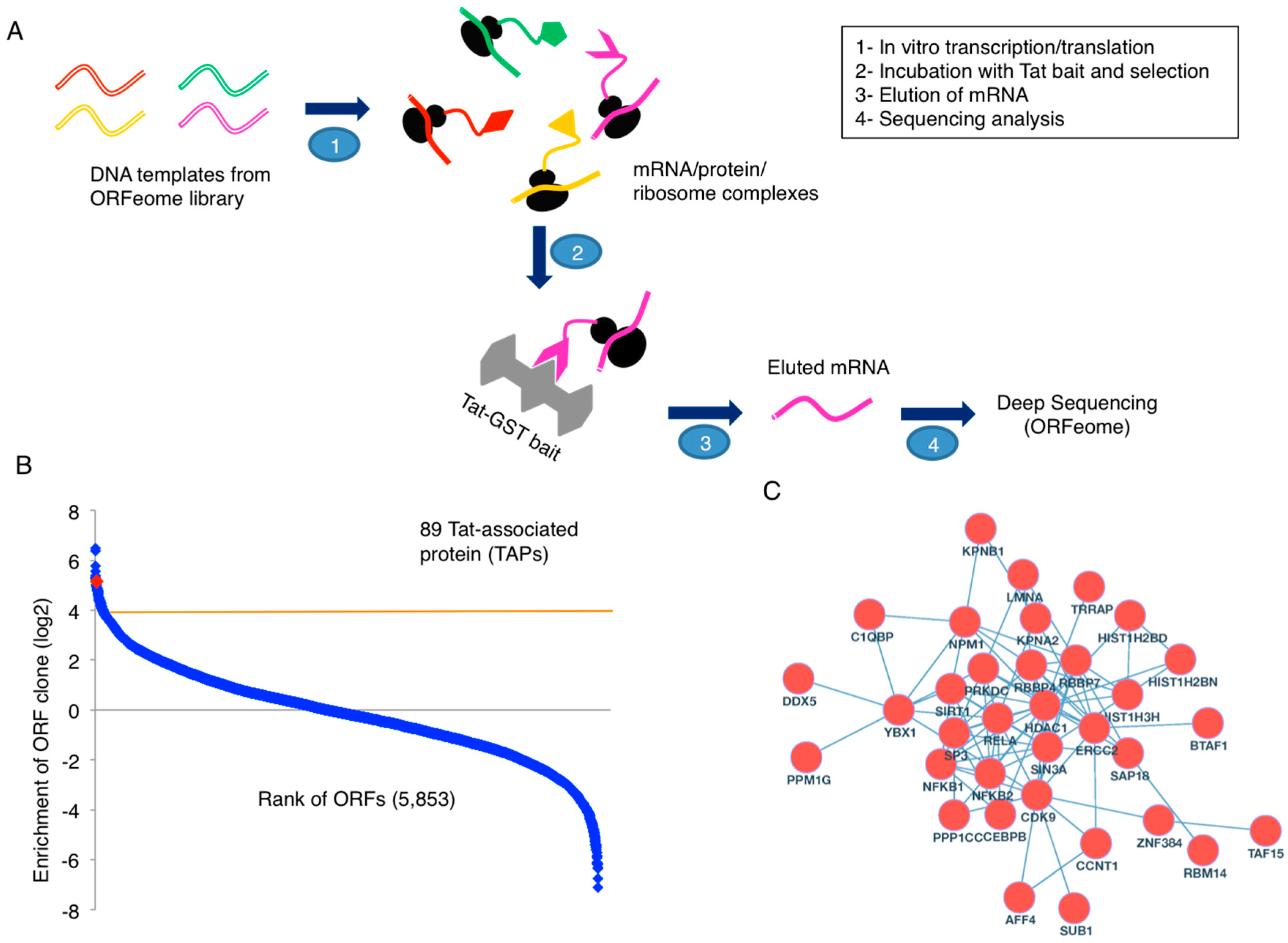
3.1. PLATO Analysis of Tat-Associated Proteins
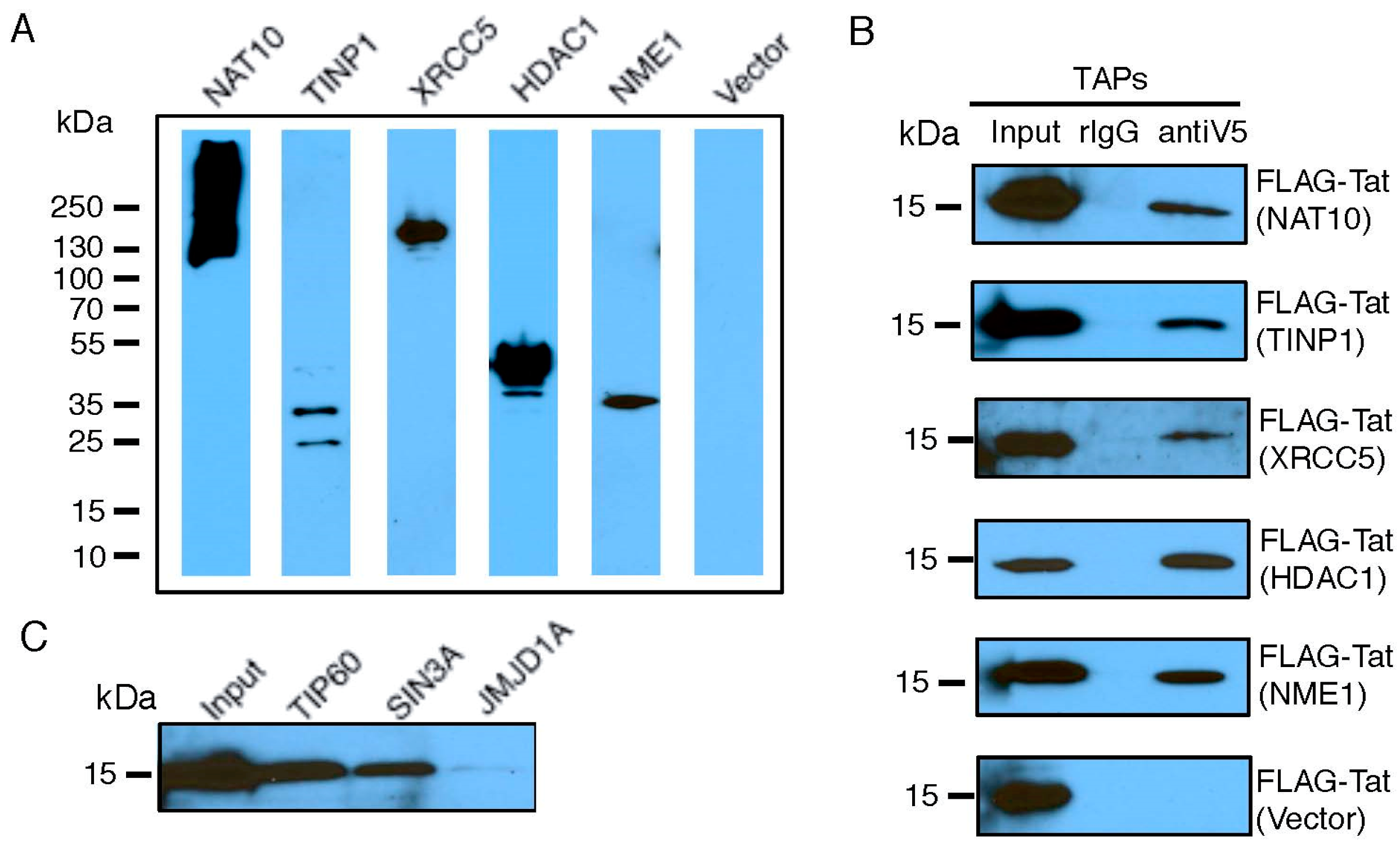
3.2. Validation of Tat-TAP Interactions
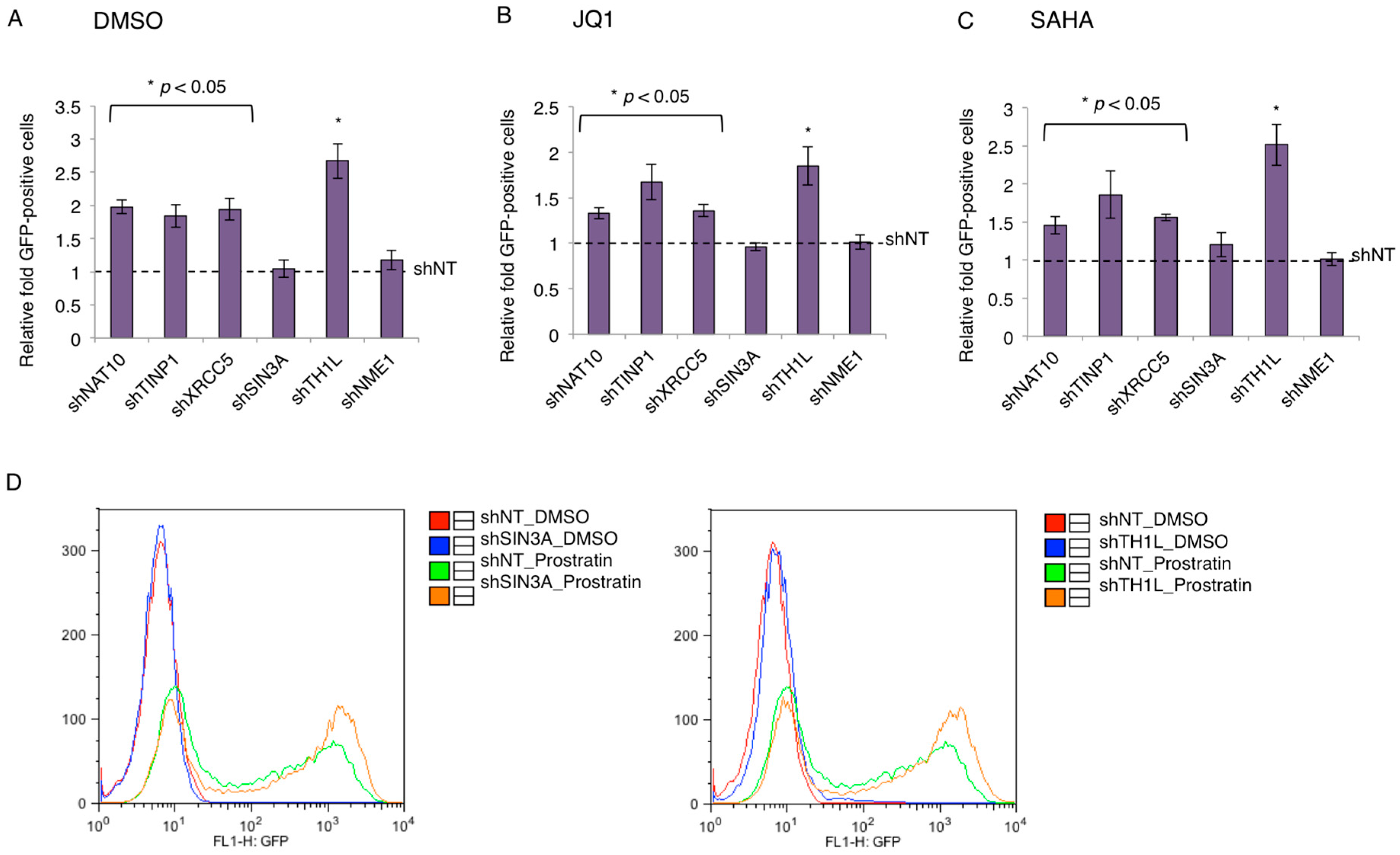
3.3. TAP-Mediated Suppression of HIV-1 LTR Promoter
3.4. Contribution of TAP to HIV-1 Latency
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Laspia, M.F.; Rice, A.P.; Mathews, M.B. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell 1989, 59, 283–292. [Google Scholar] [CrossRef]
- Brady, J.; Kashanchi, F. Tat gets the “green” light on transcription initiation. Retrovirology 2005, 2, 69. [Google Scholar] [CrossRef] [PubMed]
- He, N.; Liu, M.; Hsu, J.; Xue, Y.; Chou, S.; Burlingame, A.; Krogan, N.J.; Alber, T.; Zhou, Q. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol. Cell 2010, 38, 428–438. [Google Scholar] [CrossRef] [PubMed]
- Sobhian, B.; Laguette, N.; Yatim, A.; Nakamura, M.; Levy, Y.; Kiernan, R.; Benkirane, M. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol. Cell 2010, 38, 439–451. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Santoso, N.; Power, D.; Simpson, S.; Dieringer, M.; Miao, H.; Gurova, K.; Giam, C.Z.; Elledge, S.J.; Zhu, J. FACT Proteins, SUPT16H and SSRP1, Are Transcriptional Suppressors of HIV-1 and HTLV-1 That Facilitate Viral Latency. J. Biol. Chem. 2015, 290, 27297–27310. [Google Scholar] [CrossRef] [PubMed]
- Izmailova, E.; Bertley, F.M.; Huang, Q.; Makori, N.; Miller, C.J.; Young, R.A.; Aldovini, A. HIV-1 Tat reprograms immature dendritic cells to express chemoattractants for activated T cells and macrophages. Nat. Med. 2003, 9, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Peruzzi, F. The multiple functions of HIV-1 Tat: Proliferation versus apoptosis. Front. Biosci. 2006, 11, 708–717. [Google Scholar] [CrossRef] [PubMed]
- Debaisieux, S.; Rayne, F.; Yezid, H.; Beaumelle, B. The ins and outs of HIV-1 Tat. Traffic 2012, 13, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Shojania, S.; O’Neil, J.D. HIV-1 Tat is a natively unfolded protein: The solution conformation and dynamics of reduced HIV-1 Tat-(1–72) by NMR spectroscopy. J. Biol. Chem. 2006, 281, 8347–8356. [Google Scholar] [CrossRef] [PubMed]
- Tahirov, T.H.; Babayeva, N.D.; Varzavand, K.; Cooper, J.J.; Sedore, S.C.; Price, D.H. Crystal structure of HIV-1 Tat complexed with human P-TEFb. Nature 2010, 465, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Pumfery, A.; Deng, L.; Maddukuri, A.; de la Fuente, C.; Li, H.; Wade, J.D.; Lambert, P.; Kumar, A.; Kashanchi, F. Chromatin remodeling and modification during HIV-1 Tat-activated transcription. Curr. HIV Res. 2003, 1, 343–362. [Google Scholar] [CrossRef] [PubMed]
- Gautier, V.W.; Gu, L.; O’Donoghue, N.; Pennington, S.; Sheehy, N.; Hall, W.W. In vitro nuclear interactome of the HIV-1 Tat protein. Retrovirology 2009, 6, 47. [Google Scholar] [CrossRef] [PubMed]
- Jager, S.; Cimermancic, P.; Gulbahce, N.; Johnson, J.R.; McGovern, K.E.; Clarke, S.C.; Shales, M.; Mercenne, G.; Pache, L.; Li, K.; et al. Global landscape of HIV-human protein complexes. Nature 2012, 481, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Tacheny, A.; Michel, S.; Dieu, M.; Payen, L.; Arnould, T.; Renard, P. Unbiased proteomic analysis of proteins interacting with the HIV-1 5'LTR sequence: Role of the transcription factor Meis. Nucleic Acids Res. 2012, 40, e168. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Larman, H.B.; Gao, G.; Somwar, R.; Zhang, Z.; Laserson, U.; Ciccia, A.; Pavlova, N.; Church, G.; Zhang, W.; et al. Protein interaction discovery using parallel analysis of translated ORFs (PLATO). Nat. Biotechnol. 2013, 31, 331–334. [Google Scholar] [CrossRef] [PubMed]
- Larman, H.B.; Liang, A.C.; Elledge, S.J.; Zhu, J. Discovery of protein interactions using parallel analysis of translated ORFs (PLATO). Nat. Protoc. 2014, 9, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Gaiha, G.D.; John, S.P.; Pertel, T.; Chin, C.R.; Gao, G.; Qu, H.; Walker, B.D.; Elledge, S.J.; Brass, A.L. Reactivation of latent HIV-1 by inhibition of BRD4. Cell Rep. 2012, 2, 807–816. [Google Scholar] [CrossRef] [PubMed]
- Power, D.; Santoso, N.; Dieringer, M.; Yu, J.; Huang, H.; Simpson, S.; Seth, I.; Miao, H.; Zhu, J. IFI44 suppresses HIV-1 LTR promoter activity and facilitates its latency. Virology 2015, 481, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Rialdi, A.; Campisi, L.; Zhao, N.; Lagda, A.C.; Pietzsch, C.; Ho, J.S.; Martinez-Gil, L.; Fenouil, R.; Chen, X.; Edwards, M.; et al. Topoisomerase 1 inhibition suppresses inflammatory genes and protects from death by inflammation. Science 2016, 352, aad7993. [Google Scholar] [CrossRef] [PubMed]
- von Mering, C.; Jensen, L.J.; Snel, B.; Hooper, S.D.; Krupp, M.; Foglierini, M.; Jouffre, N.; Huynen, M.A.; Bork, P. STRING: Known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005, 33, D433–D437. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef] [PubMed]
- Chou, S.; Upton, H.; Bao, K.; Schulze-Gahmen, U.; Samelson, A.J.; He, N.; Nowak, A.; Lu, H.; Krogan, N.J.; Zhou, Q.; et al. HIV-1 Tat recruits transcription elongation factors dispersed along a flexible AFF4 scaffold. Proc. Natl. Acad. Sci. USA 2013, 110, E123–E131. [Google Scholar] [CrossRef] [PubMed]
- Ott, M.; Geyer, M.; Zhou, Q. The control of HIV transcription: Keeping RNA polymerase II on track. Cell Host Microbe 2011, 10, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Davoli, T.; Perriera, J.M.; Chin, C.R.; Gaiha, G.D.; John, S.P.; Sigiollot, F.D.; Gao, G.; Xu, Q.; Qu, H.; et al. Comprehensive identification of host modulators of HIV-1 replication using multiple orthologous RNAi reagents. Cell Rep. 2014, 9, 752–766. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.H.; Haller, K.; Peloponese, J.M., Jr.; Jeang, K.T. Histone acetyltransferase hALP and nuclear membrane protein hsSUN1 function in de-condensation of mitotic chromosomes. J. Biol. Chem. 2007, 282, 27447–27458. [Google Scholar] [CrossRef] [PubMed]
- Alsarraj, J.; Faraji, F.; Geiger, T.R.; Mattaini, K.R.; Williams, M.; Wu, J.; Ha, N.H.; Merlino, T.; Walker, R.C.; Bosley, A.D.; et al. BRD4 short isoform interacts with RRP1B, SIPA1 and components of the LINC complex at the inner face of the nuclear membrane. PLoS ONE 2013, 8, e80746. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ma, X.; Shi, T.; Song, Q.; Zhao, H.; Ma, D. NSA2, a novel nucleolus protein regulates cell proliferation and cell cycle. Biochem. Biophys. Res. Commun. 2010, 391, 651–658. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Song, A.P.; Zhao, F.; Hu, Y.M.; Hua, M. A novel human TINP1 gene promotes cell proliferation through inhibition of p53 and p21 expression. Oncol. Rep. 2013, 30, 1848–1852. [Google Scholar] [CrossRef] [PubMed]
- Willis, D.M.; Loewy, A.P.; Charlton-Kachigian, N.; Shao, J.S.; Ornitz, D.M.; Towler, D.A. Regulation of osteocalcin gene expression by a novel Ku antigen transcription factor complex. J. Biol. Chem. 2002, 277, 37280–37291. [Google Scholar] [CrossRef] [PubMed]
- Li, G.C.; Yang, S.H.; Kim, D.; Nussenzweig, A.; Ouyang, H.; Wei, J.; Burgman, P.; Li, L. Suppression of heat-induced hsp70 expression by the 70-kDa subunit of the human Ku autoantigen. Proc. Natl. Acad. Sci. USA 1995, 92, 4512–4516. [Google Scholar] [CrossRef] [PubMed]
- Grzenda, A.; Lomberk, G.; Zhang, J.S.; Urrutia, R. Sin3: Master scaffold and transcriptional corepressor. Biochim. Biophys. Acta 2009, 1789, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Jeanson, L.; Mouscadet, J.F. Ku represses the HIV-1 transcription: Identification of a putative Ku binding site homologous to the mouse mammary tumor virus NRE1 sequence in the HIV-1 long terminal repeat. J. Biol. Chem. 2002, 277, 4918–4924. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarski, W.; Khan, S.A. Lupus autoantigen Ku protein binds HIV-1 TAR RNA in vitro. Biochem. Biophys. Res. Commun. 1993, 196, 935–942. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.A.; Chen, L.F.; Kwon, H.; Ruiz-Jarabo, C.M.; Verdin, E.; Greene, W.C. NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 2006, 25, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.; Espeseth, A.; Hazuda, D.J.; Margolis, D.M. c-Myc and Sp1 contribute to proviral latency by recruiting histone deacetylase 1 to the human immunodeficiency virus type 1 promoter. J. Virol. 2007, 81, 10914–10923. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, M.; Karn, J. CBF-1 promotes transcriptional silencing during the establishment of HIV-1 latency. EMBO J. 2007, 26, 4985–4995. [Google Scholar] [CrossRef] [PubMed]
- Leth, S.; Nymann, R.; Jorgensen, S.; Olesen, R.; Rasmussen, T.A.; Ostergaard, L.; Denton, P.W.; Tolstrup, M.; Sogaard, O.S. HIV-1 transcriptional activity during frequent longitudinal sampling in aviremic patients on antiretroviral therapy. AIDS 2016, 30, 713–721. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jean, M.J.; Power, D.; Kong, W.; Huang, H.; Santoso, N.; Zhu, J. Identification of HIV-1 Tat-Associated Proteins Contributing to HIV-1 Transcription and Latency. Viruses 2017, 9, 67. https://doi.org/10.3390/v9040067
Jean MJ, Power D, Kong W, Huang H, Santoso N, Zhu J. Identification of HIV-1 Tat-Associated Proteins Contributing to HIV-1 Transcription and Latency. Viruses. 2017; 9(4):67. https://doi.org/10.3390/v9040067
Chicago/Turabian StyleJean, Maxime Junior, Derek Power, Weili Kong, Huachao Huang, Netty Santoso, and Jian Zhu. 2017. "Identification of HIV-1 Tat-Associated Proteins Contributing to HIV-1 Transcription and Latency" Viruses 9, no. 4: 67. https://doi.org/10.3390/v9040067






