Quantification of HEV RNA by Droplet Digital PCR
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. Analytical Performance of the RT-ddPCR Assay
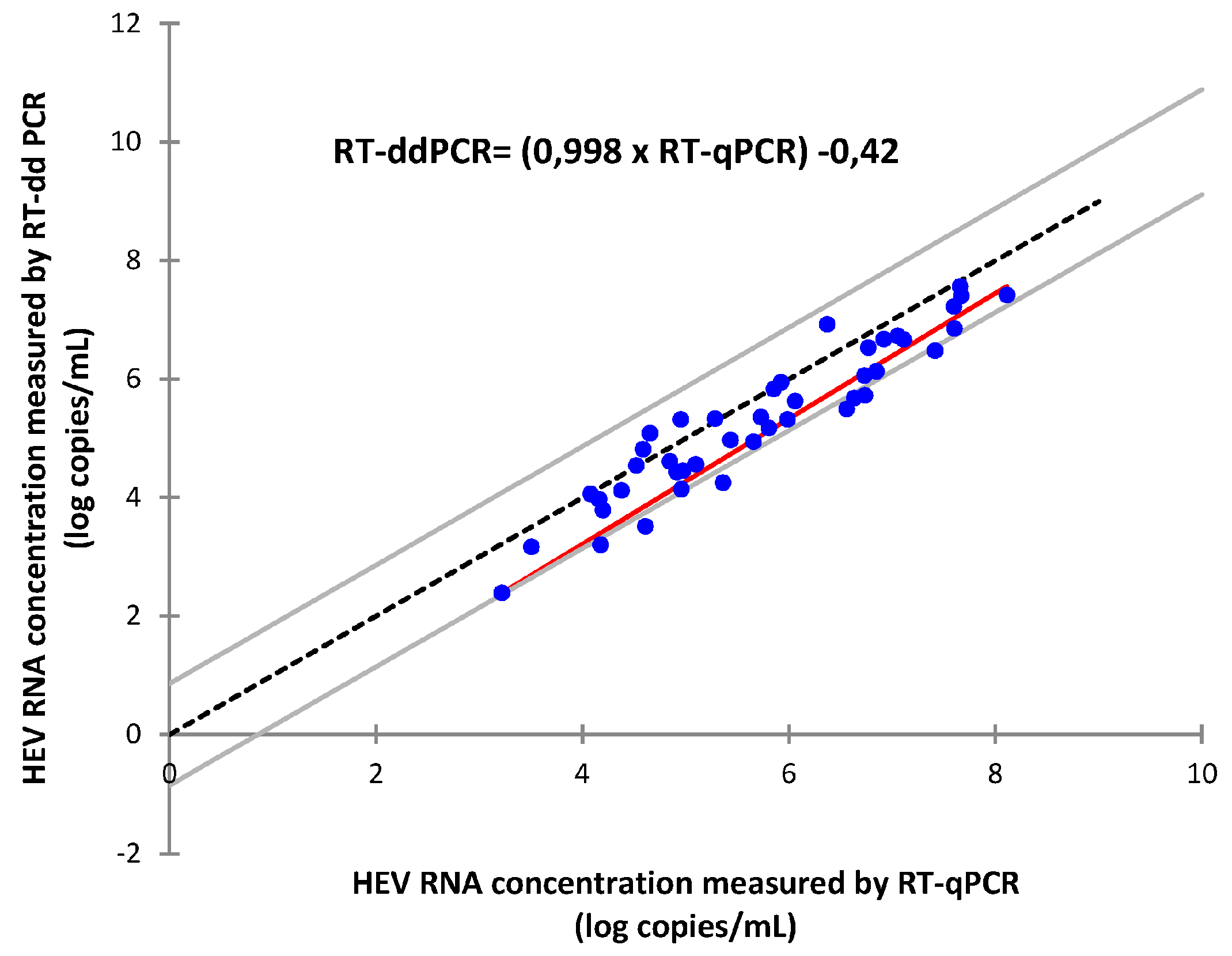
3.2. Comparison of HEV RNA Quantification between RT-ddPCR and RT-qPCR
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kamar, N.; Dalton, H.R.; Abravanel, F.; Izopet, J. Hepatitis E virus infection. Clin. Microbiol. Rev. 2014, 27, 116–138. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Li, C.; Hagedorn, C.H. Phylogenetic analysis of global hepatitis E virus sequences: genetic diversity, subtypes and zoonosis. Rev. Med. Virol. 2006, 16, 5–36. [Google Scholar] [CrossRef] [PubMed]
- Legrand-Abravanel, F.; Mansuy, J.-M.; Dubois, M.; Kamar, N.; Peron, J.-M.; Rostaing, L.; Izopet, J. Hepatitis E virus genotype 3 diversity, France. Emerg. Infect. Dis. 2009, 15, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.B.; Simmonds, P.; Jameel, S.; Emerson, S.U.; Harrison, T.J.; Meng, X.-J.; Okamoto, H.; Van der Poel, W.H.M.; Purdy, M.A.; International Committee on Taxonomy of Viruses Hepeviridae Study Group. Consensus proposals for classification of the family Hepeviridae. J. Gen. Virol. 2014, 95, 2223–2232. [Google Scholar] [CrossRef] [PubMed]
- Lhomme, S.; Abravanel, F.; Dubois, M.; Chapuy-Regaud, S.; Sandres-Saune, K.; Mansuy, J.-M.; Rostaing, L.; Kamar, N.; Izopet, J. Temporal evolution of the distribution of hepatitis E virus genotypes in Southwestern France. Infect. Genet. Evol. 2015, 35, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Kamar, N.; Bendall, R.; Legrand-Abravanel, F.; Xia, N.-S.; Ijaz, S.; Izopet, J.; Dalton, H.R. Hepatitis E. Lancet 2012, 379, 2477–2488. [Google Scholar] [CrossRef]
- Tavitian, S.; Péron, J.-M.; Huynh, A.; Mansuy, J.-M.; Ysebaert, L.; Huguet, F.; Vinel, J.-P.; Attal, M.; Izopet, J.; Récher, C. Hepatitis E virus excretion can be prolonged in patients with hematological malignancies. J. Clin. Virol. 2010, 49, 141–144. [Google Scholar] [CrossRef] [PubMed]
- Kamar, N.; Rostaing, L.; Abravanel, F.; Garrouste, C.; Lhomme, S.; Esposito, L.; Basse, G.; Cointault, O.; Ribes, D.; Nogier, M.B.; et al. Ribavirin therapy inhibits viral replication on patients with chronic hepatitis E virus infection. Gastroenterology 2010, 139, 1612–1618. [Google Scholar] [CrossRef] [PubMed]
- Kamar, N.; Izopet, J.; Tripon, S.; Bismuth, M.; Hillaire, S.; Dumortier, J.; Radenne, S.; Coilly, A.; Garrigue, V.; D’Alteroche, L.; et al. Ribavirin for chronic hepatitis E virus infection in transplant recipients. N. Engl. J. Med. 2014, 370, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Baylis, S.A.; Hanschmann, K.-M.; Blümel, J.; Nübling, C.M. Standardization of hepatitis E virus (HEV) nucleic acid amplification technique-based assays: an initial study to evaluate a panel of HEV strains and investigate laboratory performance. J. Clin. Microbiol. 2011, 49, 1234–1239. [Google Scholar] [CrossRef] [PubMed]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, L.B.; Coleman, V.A.; Hindson, C.M.; Herrmann, J.; Hindson, B.J.; Bhat, S.; Emslie, K.R. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal. Chem. 2012, 84, 1003–1011. [Google Scholar] [CrossRef] [PubMed]
- Abravanel, F.; Sandres-Saune, K.; Lhomme, S.; Dubois, M.; Mansuy, J.-M.; Izopet, J. Genotype 3 diversity and quantification of hepatitis E virus RNA. J. Clin. Microbiol. 2012, 50, 897–902. [Google Scholar] [CrossRef] [PubMed]
- Baylis, S.A.; Blümel, J.; Mizusawa, S.; Matsubayashi, K.; Sakata, H.; Okada, Y.; Nübling, C.M.; Hanschmann, K.-M. World Health Organization International Standard to Harmonize Assays for Detection of Hepatitis E Virus RNA. Emerg. Infect. Dis. 2013, 19, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Abravanel, F.; Chapuy-Regaud, S.; Lhomme, S.; Dubois, M.; Peron, J.-M.; Alric, L.; Rostaing, L.; Kamar, N.; Izopet, J. Performance of two commercial assays for detecting hepatitis E virus RNA in acute or chronic infections. J. Clin. Microbiol. 2013, 51, 1913–1916. [Google Scholar] [CrossRef] [PubMed]
- Jothikumar, N.; Cromeans, T.L.; Robertson, B.H.; Meng, X.J.; Hill, V.R. A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J. Virol. Methods 2006, 131, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Hayden, R.T.; Gu, Z.; Ingersoll, J.; Abdul-Ali, D.; Shi, L.; Pounds, S.; Caliendo, A.M. Comparison of droplet digital PCR to real-time PCR for quantitative detection of cytomegalovirus. J. Clin. Microbiol. 2013, 51, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Henrich, T.J.; Gallien, S.; Li, J.Z.; Pereyra, F.; Kuritzkes, D.R. Low-level detection and quantitation of cellular HIV-1 DNA and 2-LTR circles using droplet digital PCR. J. Virol. Methods 2012, 186, 68–72. [Google Scholar] [CrossRef] [PubMed]
- Mokhtari, C.; Marchadier, E.; Haïm-Boukobza, S.; Jeblaoui, A.; Tessé, S.; Savary, J.; Roque-Afonso, A.M. Comparison of real-time RT-PCR assays for hepatitis E virus RNA detection. J. Clin. Virol. 2013, 58, 36–40. [Google Scholar] [CrossRef] [PubMed]

| Plasma Dilution (HEV RNA Copies/mL) | Replicate 1 Replicate 2 Replicate 3 (HEV RNA Log Copies/mL) | Mean (HEV RNA Log Copies/mL) | SD | ||
|---|---|---|---|---|---|
| Genotype 1 | |||||
| 1,000,000 | 5.90 | 5.90 | 5.91 | 5.90 | 0 |
| 100,000 | 5 | 5 | 4.99 | 5 | 0.01 |
| 10,000 | 3.99 | 4.12 | 4.05 | 4.05 | 0.06 |
| 1000 | 2.01 | 3.05 | 3.01 | 2.69 | 0.59 |
| 500 | 2.44 | 2.61 | 2.61 | 2.55 | 0.10 |
| 200 | 2.68 | 2.47 | 2.01 | 2.39 | 0.34 |
| Genotype 3 | |||||
| 1,000,000 | 6.05 | 6 | 5.99 | 6.01 | 0.03 |
| 100,000 | 5 | 5.01 | 5.02 | 5.01 | 0.01 |
| 10,000 | 4.02 | 4.04 | 4.03 | 4.03 | 0.01 |
| 1000 | 3.01 | 3.03 | 3.12 | 3.07 | 0.06 |
| 500 | 2.79 | 2.26 | 2.62 | 2.56 | 0.27 |
| 200 | 1.96 | 1.96 | 2.67 | 2.20 | 0.41 |
| Genotype 4 | |||||
| 1,000,000 | 5.51 | 5.50 | 5.51 | 5.51 | 0 |
| 100,000 | 4.62 | 4.60 | 4.60 | 4.61 | 0.02 |
| 10,000 | 3.67 | 3.59 | 4.44 | 3.90 | 0.47 |
| 1000 | 2.79 | 2.85 | 2.74 | 2.79 | 0.06 |
| 500 | 2.31 | 2.33 | 2.09 | 2.24 | 0.14 |
| 200 | 0 | 0 | 2.05 | NA | NA |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nicot, F.; Cazabat, M.; Lhomme, S.; Marion, O.; Sauné, K.; Chiabrando, J.; Dubois, M.; Kamar, N.; Abravanel, F.; Izopet, J. Quantification of HEV RNA by Droplet Digital PCR. Viruses 2016, 8, 233. https://doi.org/10.3390/v8080233
Nicot F, Cazabat M, Lhomme S, Marion O, Sauné K, Chiabrando J, Dubois M, Kamar N, Abravanel F, Izopet J. Quantification of HEV RNA by Droplet Digital PCR. Viruses. 2016; 8(8):233. https://doi.org/10.3390/v8080233
Chicago/Turabian StyleNicot, Florence, Michelle Cazabat, Sébastien Lhomme, Olivier Marion, Karine Sauné, Julie Chiabrando, Martine Dubois, Nassim Kamar, Florence Abravanel, and Jacques Izopet. 2016. "Quantification of HEV RNA by Droplet Digital PCR" Viruses 8, no. 8: 233. https://doi.org/10.3390/v8080233








