The Whitefly Bemisia tabaci Knottin-1 Gene Is Implicated in Regulating the Quantity of Tomato Yellow Leaf Curl Virus Ingested and Transmitted by the Insect
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plants and Whiteflies
2.2. Isolation of RNA from B. tabaci, Preparation of cDNA and Quantitative RT-PCR (qPCR)
2.3. Isolation of DNA from B. tabaci and Tomato Plants; Measurement of TYLCV Amounts
2.4. dsRNA Synthesis
2.5. Gene Silencing by Leaf-Mediated dsRNA Feeding
2.6. Fluorescence in Situ Hybridization (FISH)
2.7. Statistical Analysis of Data
3. Results
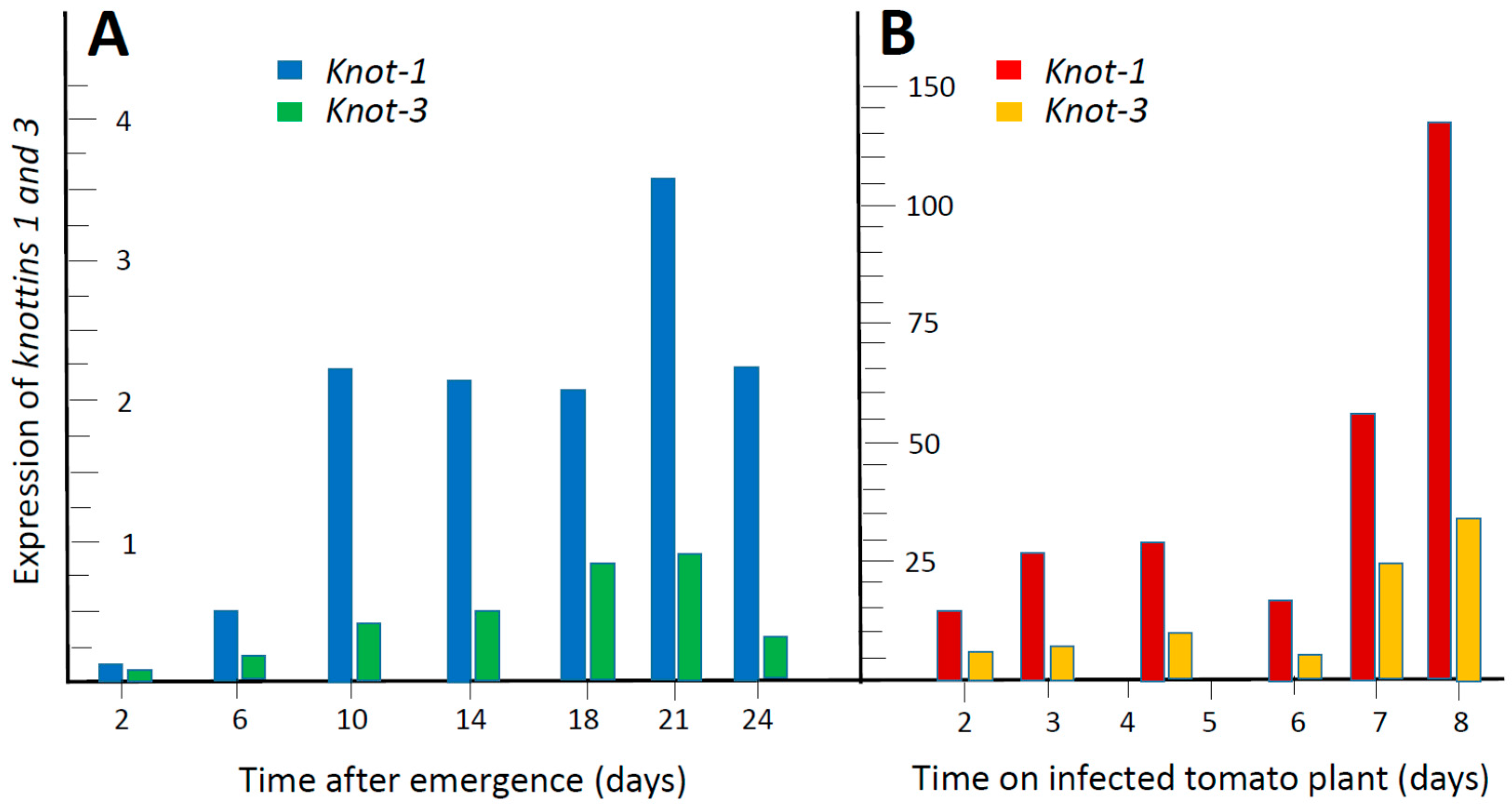
3.1. Out of the Four Whitefly Knottin Genes, knot-1 Shows the Highest Levels of Expression. It Is Upregulated upon Ingestion of TYLCV
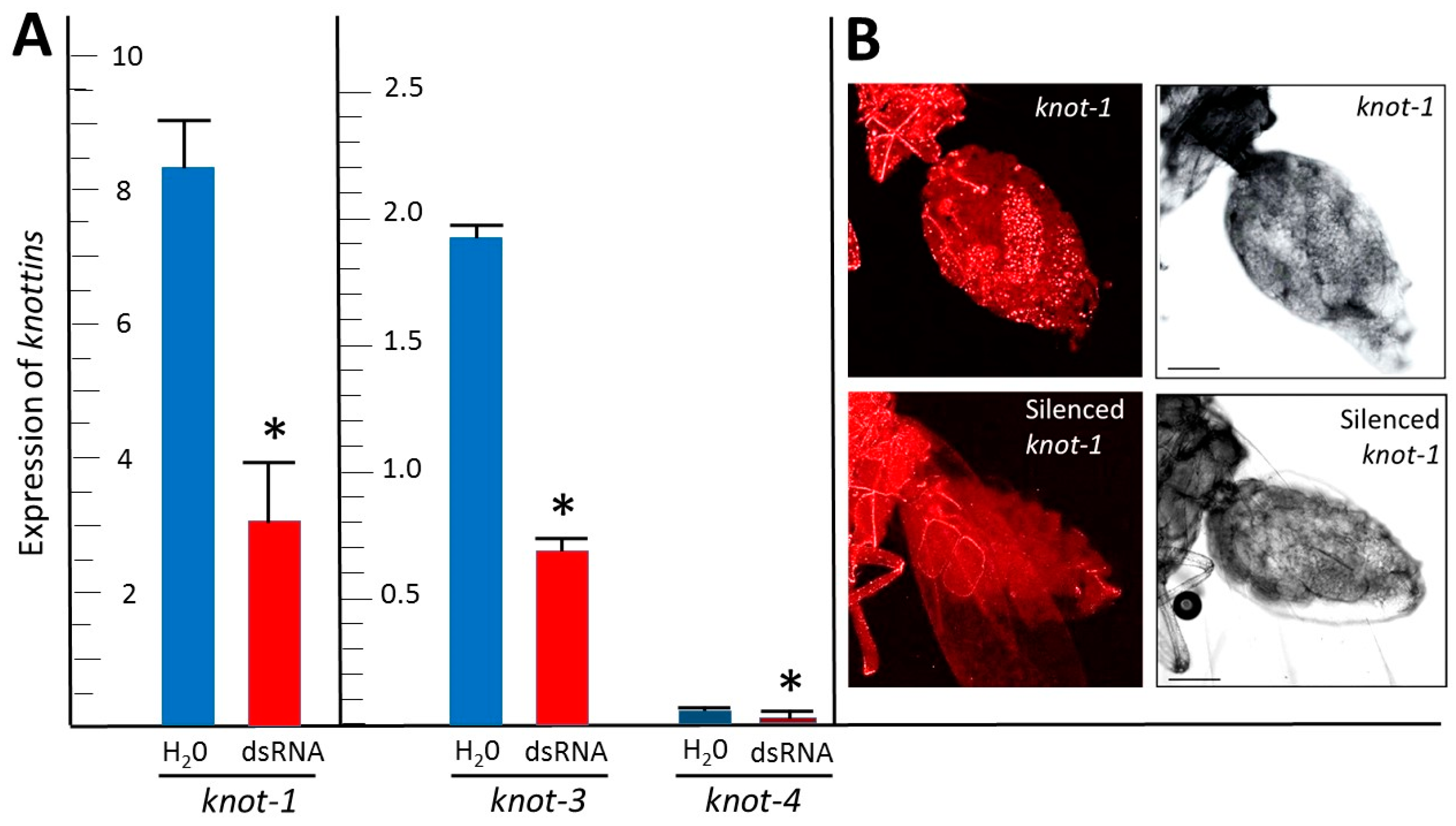
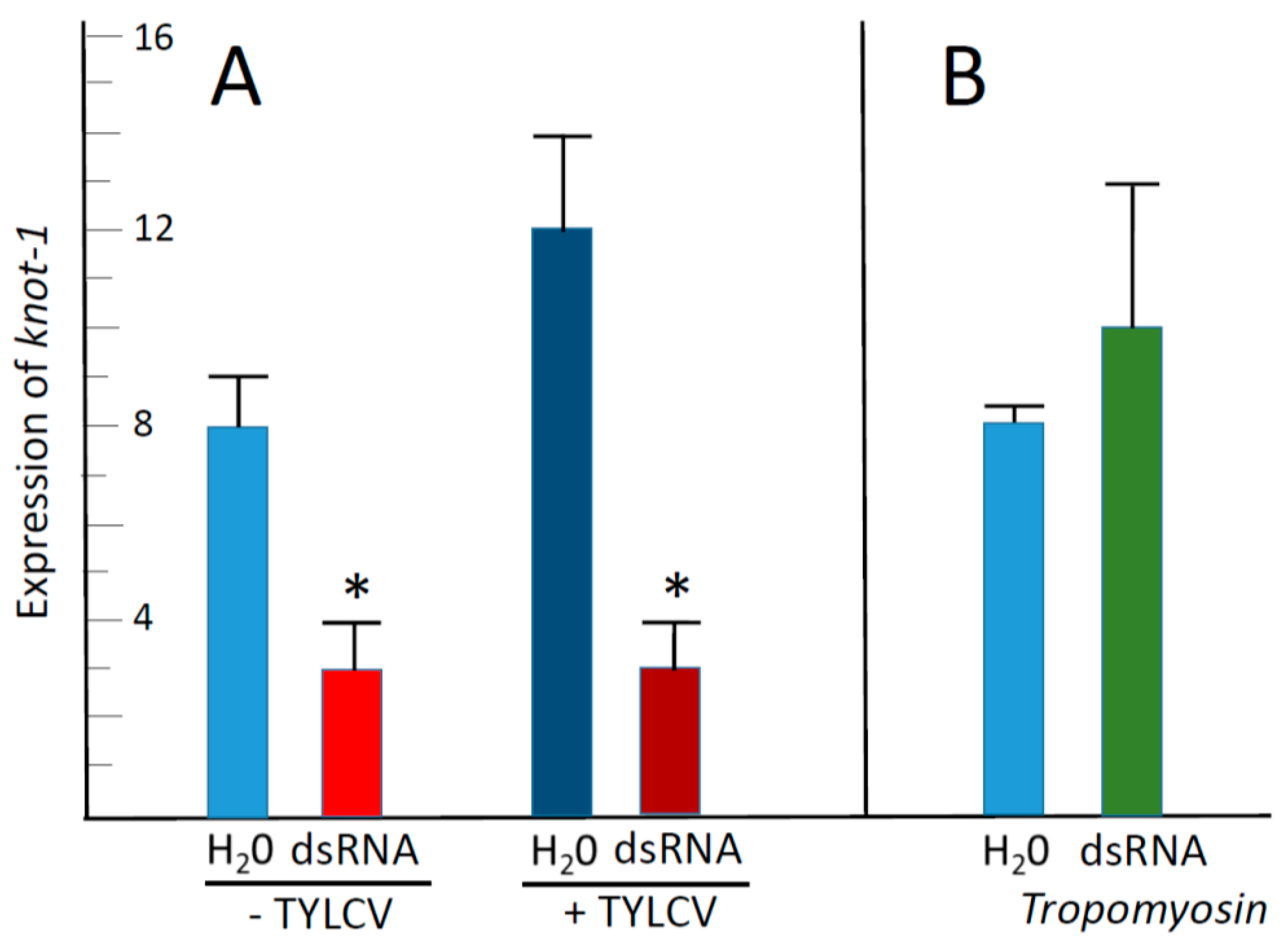
3.2. Feeding Whiteflies on Tomato Leaflets Bathing in dsRNA Targeted against knot-1 and knot-3 Downregulated These Genes by about Two Thirds
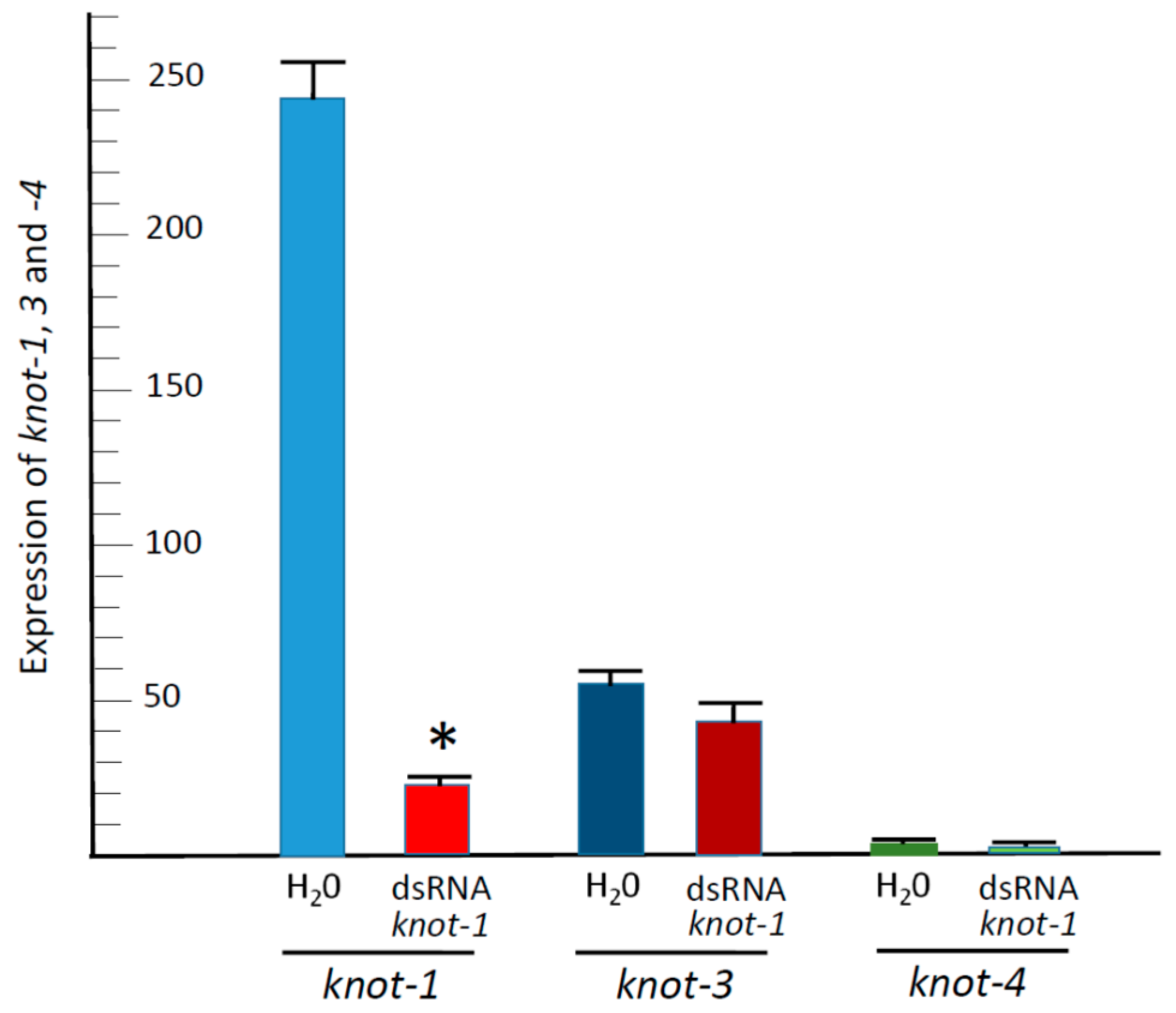
3.3. Silencing knot-1 Has no Effect on the Expression of the Other Whitefly Knottin Genes
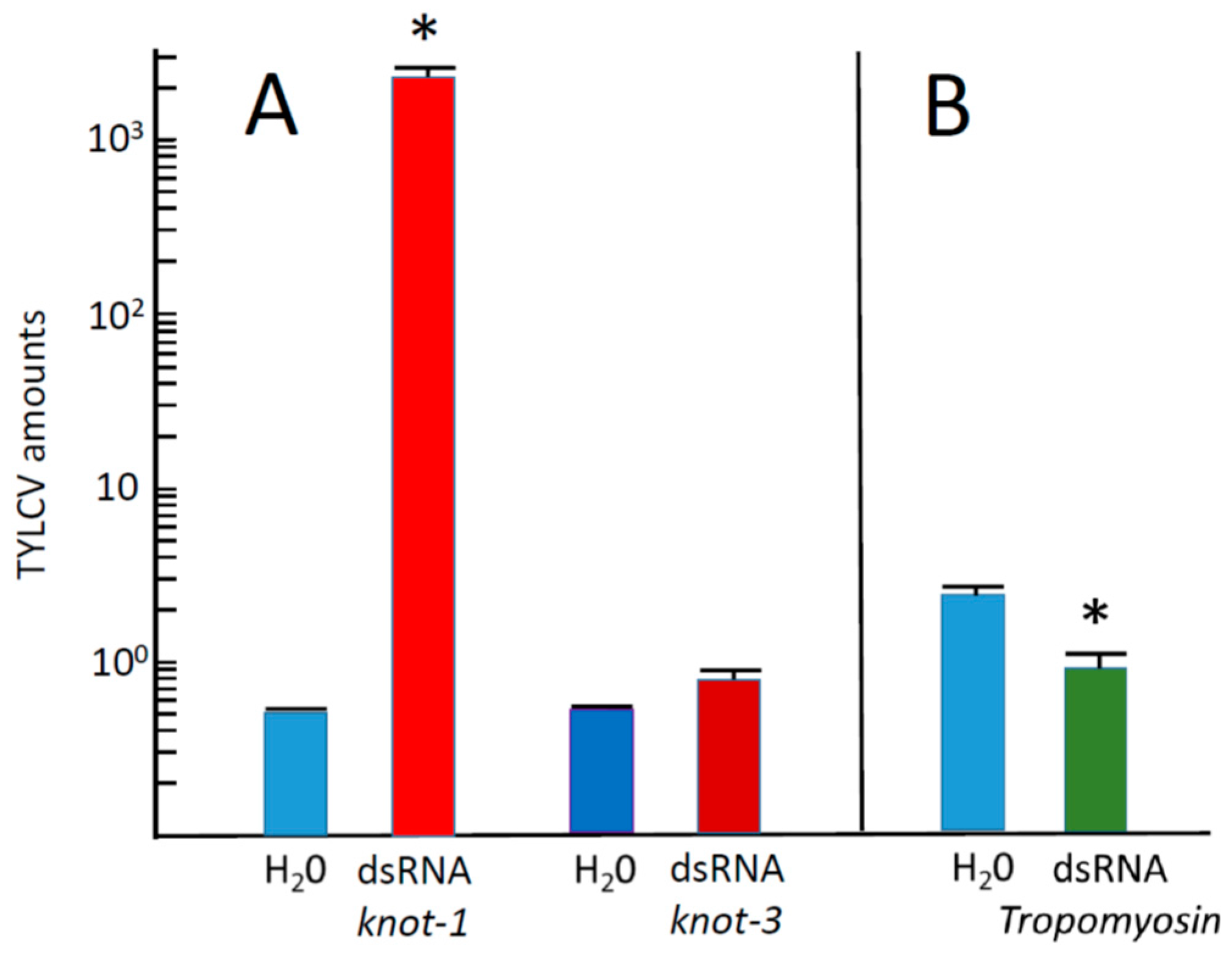
3.4. Whiteflies with Silencing knot-1 Contain Increased Amounts of TYLCV upon Feeding on Leaves of Infected Tomato Plants
3.5. Tomato Plants Inoculated by Viruliferous Whiteflies with Silenced knot-1 Presented Early Disease Symptoms and Contained Large Amounts of Virus
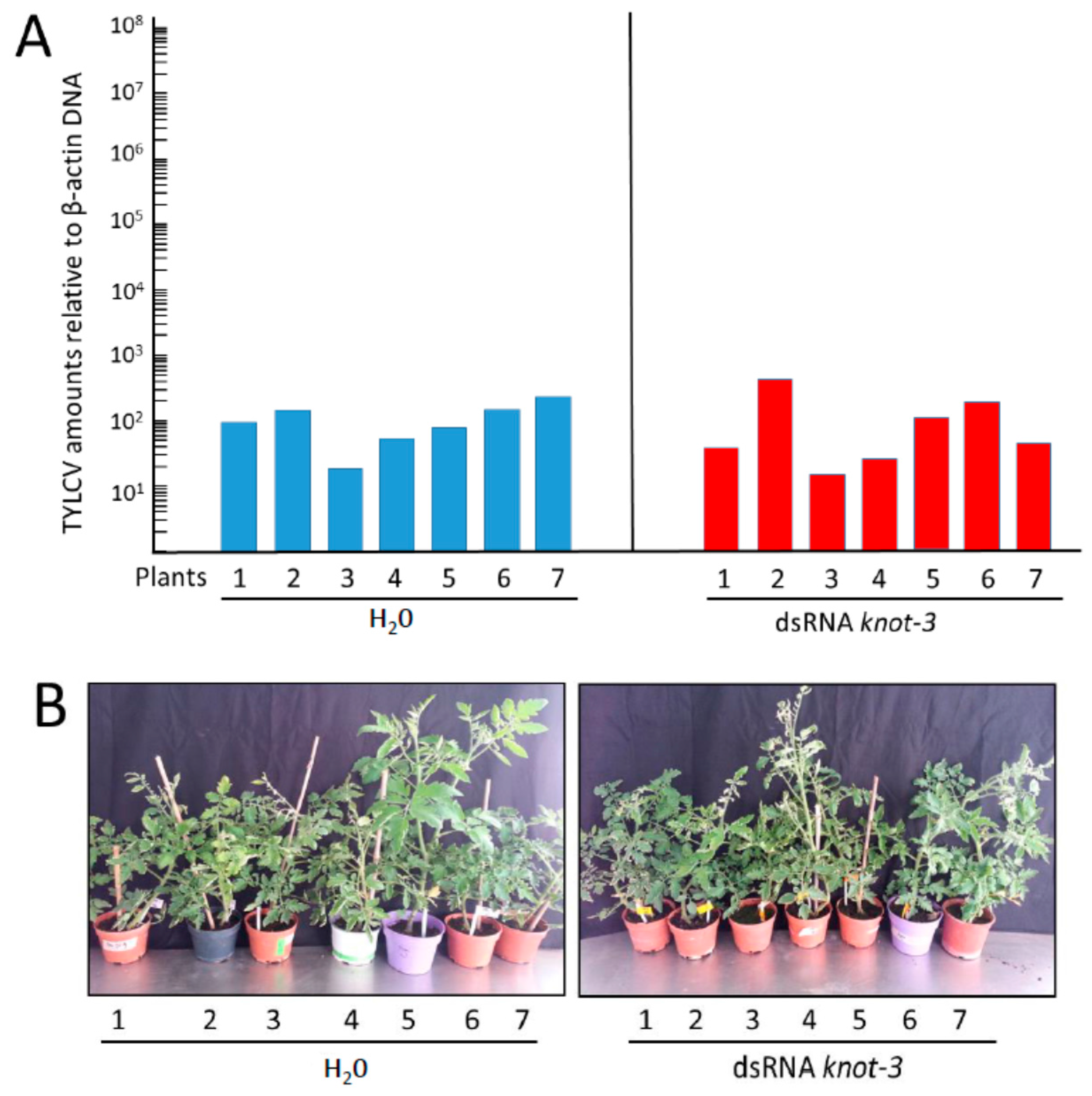
3.6. Inoculation of Tomato Plants by Whiteflies with Silenced knot-3 Significantly Modified Neither the Time of Symptom Appearance nor the Accumulation of TYLCV
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Stansley, P.A.; Naranjo, S.E. Bemisia: Bionomics and Management of a Global Pest; Springer: Dordrecht, The Netherlands, 2010. [Google Scholar]
- Czosnek, H. Tomato Yellow Leaf Curl Virus Disease: Management, Molecular Biology, Breeding for Resistance; Springer: Dordrecht, The Netherlands, 2007. [Google Scholar]
- Brown, J.K.; Zerbuni, F.M.; Navas-Castillo, J.; Moriones, E.; Ramos-Sobrinho, R.; Silva, J.C.F.; Fiallo-Olivé, E.; Briddon, R.W.; Hernández-Zepeda, C.; Idris, A. Revision of Begomovirus taxonomy based on pairwise sequence comparisons. Arch. Virol. 2014, 160, 1593–1619. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Pendon, J.A.; Caňizares, M.C.; Moriones, E.; Bejarano, E.R.; Czosnek, H.; Navas-Castillo, J. Tomato yellow leaf curl viruses: Ménage a trios between the virus complex, the plant and the whitefly vector. Mol. Plant Pathol. 2010, 11, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Czosnek, H.; Ghanim, M.; Ghanim, M. Circulative pathway of begomoviruses in the whitefly vector Bemisia tabaci—Insights from studies with Tomato yellow leaf curl virus. Ann. Appl. Biol. 2002, 140, 215–231. [Google Scholar] [CrossRef]
- Wei, J.; Zhao, J.-J.; Zhang, T.; Li, F.-F.; Ghanim, M.; Zhou, X.-P.; Ye, G.-Y.; Liu, S.-S.; Wang, X.-W. Specific cells in the primary salivary glands of the whitefly Bemisia tabaci control retention and transmission of begomoviruses. J. Virol. 2014, 88, 13460–13468. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, S.; Hoshino, S. Effect of tomato yellow leaf curl disease on reproduction of Bemisia tabaci Q biotype (Hemiptera: Aleyrodidae) on tomato plants. Appl. Entomol. Zool. 2009, 44, 143–148. [Google Scholar] [CrossRef]
- Jiu, M.; Zhou, X.P.; Tong, L.; Xu, J.; Yang, X.; Wan, F.H.; Liu, S.-S. Vector-virus mutualism accelerates population increase of an invasive whitefly. PLoS ONE 2007, 2, e182. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, G.; Czosnek, H. Long-term association of tomato yellow leaf curl virus (TYLCV) with its whitefly vector Bemisia tabaci: Effect on the insect transmission capacity, longevity and fecundity. J. Gen. Virol. 1997, 78, 2683–2689. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Chu, D.; Liu, B.; Shi, X.; Guo, L.; Xie, W.; Carrière, Y.; Li, X.; Zhang, Y. Differential effects of an exotic plant virus on its two closely related vectors. Sci. Rep. 2013, 3, 2230. [Google Scholar] [CrossRef] [PubMed]
- Leshkowitz, D.; Gazit, S.; Reuveni, E.; Ghanim, M.; Czosnek, H.; McKenzie, C.; Shatters, R.G., Jr.; Brown, J.K. Whitefly (Bemisia tabaci) genome project: Analysis of sequenced clones from egg, instar, and adult (viruliferous and non-viruliferous) cDNA libraries. BMC Genom. 2006, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Mahadav, A.; Gerling, D.; Gottlieb, Y.; Czosnek, H.; Ghanim, M. Gene expression in the whitefly Bemisia tabaci pupae in response to parasitization by the wasp Eretmocerus mundus. BMC Genom. 2000, 9, 342. [Google Scholar] [CrossRef] [PubMed]
- Luan, J.-B.; Li, J.-M.; Varela, N.; Wang, Y.-L.; Li, F.-F.; Bao, Y.-Y.; Zhang, C.-X.; Liu, S.-S.; Wang, X.-W. Global analysis of the transcriptional response of whitefly to Tomato Yellow leaf curl China virus reveals the relationship of coevolved adaptations. J. Virol. 2011, 85, 3330–3340. [Google Scholar] [CrossRef] [PubMed]
- Shatters, R.G., Jr.; McKenzie, C.L.; Boykin, L.M.; Gazit, S.; Sinisterra, X.; Weathersbee, A.A.; Brown, J.K.; Czosnek, H. A knottin-like putative antimicrobial gene family in the whitefly Bemisia tabaci biotype B: Cloning and transcript regulation. J. Insect Sci. 2008, 8, 4. [Google Scholar]
- Zhang, C.-R.; Zhang, S.; Xia, J.; Li, F.-F.; Xia, W.-Q.; Liu, S.-S.; Wang, X.-W. The immune strategy and stress response of the Mediterranean species of the Bemisia tabaci complex to an orally delivered bacterial pathogen. PLoS ONE 2014, 9, e94477. [Google Scholar] [CrossRef] [PubMed]
- Gracy, J.; Chiche, L. Structure and modeling of knottins, a promising molecular scaffold for drug discovery. Curr. Pharm. Des. 2011, 17, 4337–4350. [Google Scholar] [CrossRef] [PubMed]
- Gelly, J.C.; Gracy, J.; Kaas, Q.; Le-Nguyen, D.; Heitz, A.; Chiche, L. The KNOTTIN website and database: A new information system dedicated to the knottin scaffold. Nucleic Acids Res. 2004, 32, D156–D159. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.J.; Mahaffey, J.P.; Lorenzen, M.D.; Denell, R.E.; Mahaffey, J.W. Using RNAi to investigate orthologous homeotic gene function during development of distantly related insects. Evol. Dev. 1999, 1, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Tomoyasu, Y.; Denell, R.E. Larval RNAi in Tribolium (Coleoptera) for analyzing adult development. Dev. Genes Evol. 2004, 214, 575–578. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.M.; Ihle, K.E.; Fondrk, M.K.; Page, R.E., Jr.; Amdam, G.V. The gene vitellogenin has multiple coordinating effects on social organization. PLoS Biol. 2007, 5, e62. [Google Scholar] [CrossRef] [PubMed]
- Huvenne, H.; Smagghe, G. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: A review. J. Insect Physiol. 2010, 56, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Coleman, A.D.; Wouters, R.H.M.; Mugford, S.T.; Hogenhout, S.A. Persistence and transgenerational effect of plant-mediated RNAi in aphids. J. Exp. Bot. 2015, 66, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Christiaens, O.; Swevers, L.; Smagghe, G. dsRNA degradation in the pea aphid (Acyrthosiphon pisum) associated with lack of response in RNAi feeding and injection assay. Peptides 2014, 53, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Ghanim, M.; Kontsedalov, S.; Czosnek, H. Tissue-specific gene silencing by RNA interference in the whitefly Bemisia tabaci (Gennadius). Insect Biochem. Mol. Biol. 2007, 37, 732–738. [Google Scholar] [CrossRef] [PubMed]
- Luan, J.-B.; Ghanim, M.; Liu, S.-S.; Czosnek, H. Silencing the ecdysone (synthesis and signaling) pathway genes disrupts nymphal development in the whitefly. Insect Biochem. Mol. Biol. 2013, 43, 740–746. [Google Scholar] [CrossRef] [PubMed]
- De Barro, P.J.; Liu, S.-S.; Boykin, L.M.; Dinsdale, A.B. Bemisia tabaci: A Statement of Species Status. Ann. Rev. Entomol. 2011, 56, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Shahjahan, R.M.; Roger, K.J.H.; Leopold, R.A.; Devault, J.D. Lower incubation temperature increases yield of insect genomic DNA isolated by the CTAB method. Biotechniques 1995, 19, 332–339. [Google Scholar] [PubMed]
- Götz, M.; Popovski, S.; Kollenberg, M.; Gorovits, R.; Brown, J.K.; Cicero, J.M.; Czosnek, H.; Winter, S.; Ghanim, M. Implication of Bemisia tabaci heat shock protein 70 in begomovirus-whitefly interactions. J. Virol. 2012, 86, 13241–13252. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.; Nitzany, F.E. Transmission and host range of the tomato yellow leaf curl virus. Phytopathology 1966, 56, 1127–1131. [Google Scholar]
- Scott, J.G.; Michel, K.; Bartholomay, L.C.; Siegfried, B.D.; Hunter, W.B.; Smagghe, G.; Zhu, K.Y.; Douglas, A.E. Towards the elements of successful insect RNAi. J. Insect Physiol. 2013, 59, 1212–1221. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Cai, Z.; Song, J.; Wu, Z.; Zhou, S. dsRNA uptake and persistence account for tissue-dependent susceptibility to RNA interference in the migratory locust, Locusta migratoria. Insect Mol. Biol. 2014, 23, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Asokan, R.; Rebijith, K.B.; Roopa, H.K.; Kumar, N.K. Non-invasive delivery of dsGST is lethal to the sweet potato whitefly, Bemisia tabaci (G.) (Hemiptera: Aleyrodidae). Appl. Biochem. Biotechnol. 2015, 175, 2288–2299. [Google Scholar] [CrossRef] [PubMed]
- Jia, S.; Wan, P.-J.; Zhou, L.-T.; Mu, L.-L.; Li, G.-Q. RNA interference-mediated silencing of a Halloween gene spookier affects nymph performance in the small brown planthopper Laodelphax striatellus. Insect Sci. 2015, 22, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Killiny, N.; Hajeri, S.; Tiwari, S.; Gowda, S.; Stelinski, L.L. Double-stranded RNA uptake through topical application, mediates silencing of five CYP4 genes and suppresses insecticide resistance in Diaphorina citri. PLoS ONE 2014, 9, e110536. [Google Scholar] [CrossRef] [PubMed]
- Thakur, N.; Upadhyay, S.K.; Verma, P.C.; Chandrashekar, K.; Tuli, R.; Singh, P.K. Enhanced whitefly resistance in transgenic tobacco plants expressing double stranded RNA of v-ATPase A gene. PLoS ONE 2014, 9, e87235. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.R.; Rajam, M.V. Targeting chitinase gene of Helicoverpa armigera by host-induced RNA interference confers insect resistance in tobacco and tomato. Plant Mol. Biol. 2016, 90, 281–292. [Google Scholar]
- Camargo, R.A.; Herai, R.H.; Santos, L.N.; Bento, F.M.M.; Lima, J.E.; Marques-Souza, H.; Figueira, A. De novo transcriptome assembly and analysis to identify potential gene targets for RNAi-mediated control of the tomato leafminer (Tuta absoluta). BMC Genom. 2015, 16, 635. [Google Scholar] [CrossRef] [PubMed]
- Chouabe, C.; Eyraud, V.; Da Silva, P.; Rahioui, I.; Royer, C.; Soulage, C.; Bonvallet, R.; Huss, M.; Gressent, F. New mode of action for a knottin protein bioinsecticide Pea albumin 1 subunit b (PA1b) is the first peptidic inhibitor of V-ATPase. J. Biol. Chem. 2011, 286, 36291–36296. [Google Scholar] [CrossRef] [PubMed]
- Bende, N.J.; Dziemborowicz, S.; Herzig, V.; Ramanujam, V.; Brown, G.W.; Bosmans, F.; Nicholson, G.M.; King, G.F.; Mobli, M. The insecticidal spider toxin SFI1 is a knottin peptide that blocks the pore of insect voltage-gated sodium channels via a large β-hairpin loop. FEBS J. 2015, 282, 904–920. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.-S.; Lee, J.; Hwang, B.; Nam, S.-H.; Yun, E.-Y.; Kim, S.-R.; Lee, D.G. Isolation and characterization of Psacotheasin, a novel knottin-type antimicrobial peptide, from Psacothea hilaris. J. Microbiol. Biotech. 2010, 20, 708–711. [Google Scholar] [CrossRef]
- Czosnek, H.; Ghanim, M.; Rubinstein, G.; Morin, S.; Fridman, V.; Zeidan, M. Whiteflies: Vectors–or victims?–of geminiviruses. Adv. Virus Res. 2001, 57, 291–322. [Google Scholar] [PubMed]
- Czosnek, H.; Ghanim, M. Back to basics: Are begomoviruses whitefly pathogens? J. Integr. Agric. 2012, 11, 225–234. [Google Scholar] [CrossRef]
- Li, J.M.; Ruan, Y.M.; Li, F.F.; Liu, S.S.; Wang, X.W. Gene expression profiling of the whitefly (Bemisia tabaci) Middle East—Asia Minor 1 feeding on healthy and Tomato yellow leaf curl China virus-infected tobacco. Insect Sci. 2011, 18, 11–22. [Google Scholar] [CrossRef]
- Pakkianathan, B.C.; Kontsedalov, S.; Lebedev, G.; Mahadav, A.; Zeidan, M.; Czosnek, H.; Ghanim, M. Replication of Tomato yellow leaf curl in its whitefly vector Bemisia tabaci. J. Virol. 2015, 89, 9791–9803. [Google Scholar] [CrossRef] [PubMed]


© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hariton Shalev, A.; Sobol, I.; Ghanim, M.; Liu, S.-S.; Czosnek, H. The Whitefly Bemisia tabaci Knottin-1 Gene Is Implicated in Regulating the Quantity of Tomato Yellow Leaf Curl Virus Ingested and Transmitted by the Insect. Viruses 2016, 8, 205. https://doi.org/10.3390/v8070205
Hariton Shalev A, Sobol I, Ghanim M, Liu S-S, Czosnek H. The Whitefly Bemisia tabaci Knottin-1 Gene Is Implicated in Regulating the Quantity of Tomato Yellow Leaf Curl Virus Ingested and Transmitted by the Insect. Viruses. 2016; 8(7):205. https://doi.org/10.3390/v8070205
Chicago/Turabian StyleHariton Shalev, Aliza, Iris Sobol, Murad Ghanim, Shu-Sheng Liu, and Henryk Czosnek. 2016. "The Whitefly Bemisia tabaci Knottin-1 Gene Is Implicated in Regulating the Quantity of Tomato Yellow Leaf Curl Virus Ingested and Transmitted by the Insect" Viruses 8, no. 7: 205. https://doi.org/10.3390/v8070205
APA StyleHariton Shalev, A., Sobol, I., Ghanim, M., Liu, S.-S., & Czosnek, H. (2016). The Whitefly Bemisia tabaci Knottin-1 Gene Is Implicated in Regulating the Quantity of Tomato Yellow Leaf Curl Virus Ingested and Transmitted by the Insect. Viruses, 8(7), 205. https://doi.org/10.3390/v8070205







