Direct Identification of Enteroviruses in Cerebrospinal Fluid of Patients with Suspected Meningitis by Nested PCR Amplification
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
2.2. General Characteristic of Patients Included in the Study
2.3. RNA Extraction
2.4. Reverse Transcription (RT)
2.5. Nested PCR
| Primer | Position, nt | Resulted Amplicon, nt |
|---|---|---|
| 224 d | 1977–1996 a | 1856 |
| 131 d | 3832–3812 a | |
| HEVBS1695 e | 2375–2409 b | 1093 |
| HEVBR132 e | 3467–3432 b | |
| AK1 | 2298–2320 c | 1373 |
| AK3 | 3670–3645 c |
2.6. Sequencing and Sequences Analysis
2.7. Assay Sensitivity
3. Results
3.1. Comparative Analysis of the Existing and New Amplification Methods
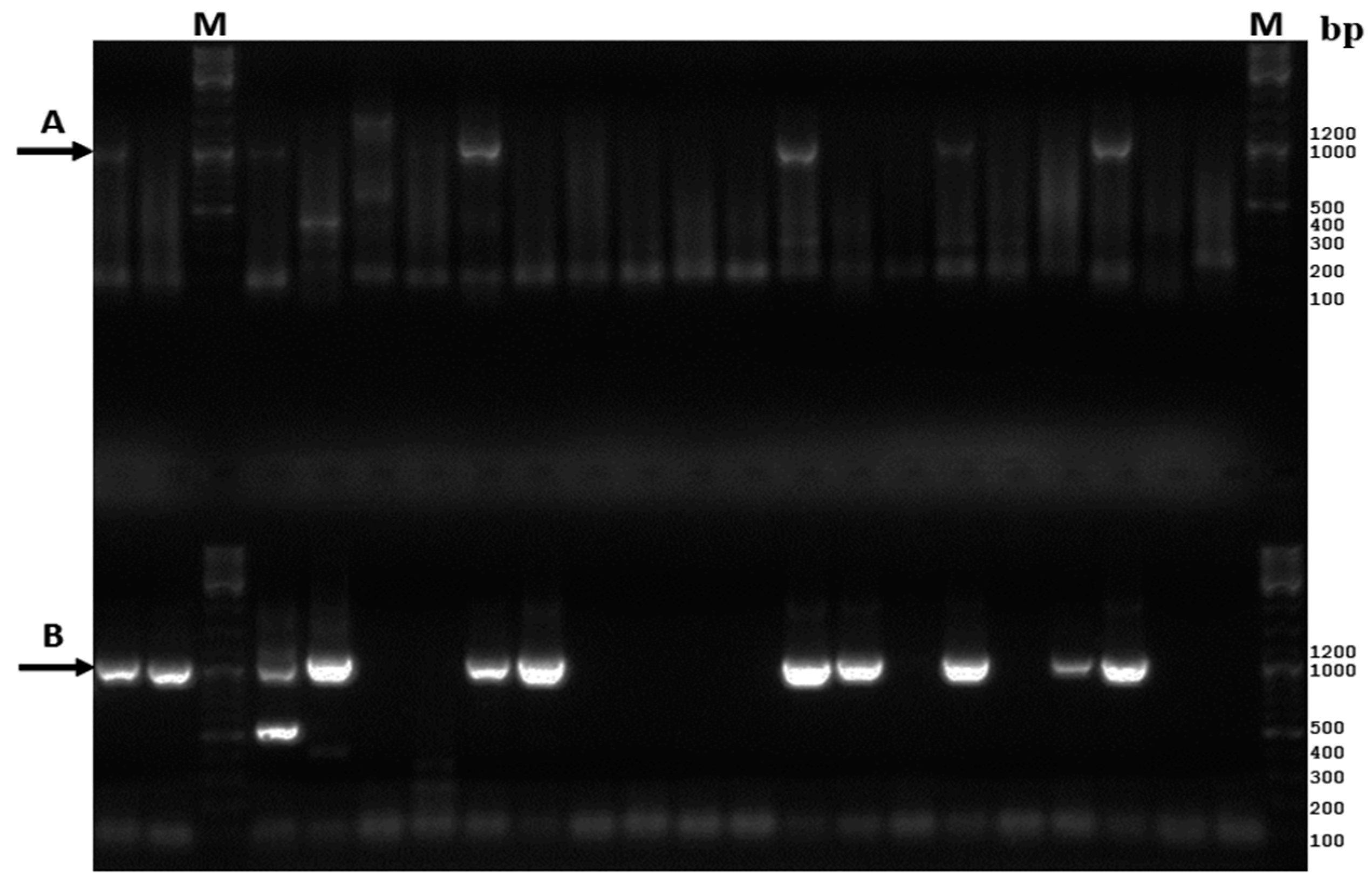
3.2. Assay Sensitivity
3.3. Retrospective Direct Genotyping of Enteroviruses Detected in CSF of Patients Hospitalized in 2012
| 2012 | 2013 | ||||||
|---|---|---|---|---|---|---|---|
| No. | CSF Specimen | EV Serotype | Accession No. | No. | CSF Specimen | EV Serotype | Accession No. |
| 1 | 257-CSF/RUS/Omsk/2012 | E-6 | KU133571 | 1 | 376-CSF/RUS/Omsk/2013 | E-11 | KU133616 |
| 2 | 259-CSF/RUS/Omsk/2012 | E-6 | KU133573 | 2 | 377-CSF/RUS/Omsk/2013 | E-11 | KU133617 |
| 3 | 266-CSF/RUS/Omsk/2012 | E-6 | KU133577 | 3 | 380-CSF/RUS/Omsk/2013 | E-11 | KU133618 |
| 4 | 268-CSF/RUS/Omsk/2012 | E-6 | KU133579 | 4 | 381-CSF/RUS/Omsk/2013 | E-11 | KU133619 |
| 5 | 271-CSF/RUS/Omsk/2012 | E-6 | KU133581 | 5 | 394-CSF/RUS/Omsk/2013 | E-11 | KU133626 |
| 6 | 272-CSF/RUS/Omsk/2012 | E-6 | KU133582 | 6 | 395-CSF/RUS/Omsk/2013 | E-11 | KU133627 |
| 7 | 274-CSF/RUS/Omsk/2012 | E-6 | KU133583 | 7 | 400-CSF/RUS/Omsk/2013 | E-11 | KU133629 |
| 8 | 320-CSF/RUS/Omsk/2012 | E-6 | KU133602 | 8 | 402-CSF/RUS/Omsk/2013 | E-11 | KU133630 |
| 9 | 321-CSF/RUS/Omsk/2012 | E-6 | KU133603 | 9 | 404-CSF/RUS/Omsk/2013 | E-11 | KU133632 |
| 10 | 322-CSF/RUS/Omsk/2012 | E-6 | KU133604 | 10 | 412-CSF/RUS/Omsk/2013 | E-11 | KU133637 |
| 11 | 324-CSF/RUS/Omsk/2012 | E-6 | KU133605 | 11 | 419-CSF/RUS/Omsk/2013 | E-11 | KU133639 |
| 12 | 329-CSF/RUS/Omsk/2012 | E-6 | KU133608 | 12 | 430-CSF/RUS/Omsk/2013 | E-11 | KU133643 |
| 13 | 332-CSF/RUS/Omsk/2012 | E-6 | KU133610 | 13 | 443-CSF/RUS/Omsk/2013 | E-11 | KU133647 |
| 14 | 338-CSF/RUS/Omsk/2012 | E-6 | KU133612 | 14 | 445-CSF/RUS/Omsk/2013 | E-11 | KU133648 |
| 15 | 340-CSF/RUS/Omsk/2012 | E-6 | KU133613 | 15 | 446-CSF/RUS/Omsk/2013 | E-11 | KU133649 |
| 16 | 341-CSF/RUS/Omsk/2012 | E-6 | KU133614 | 16 | 448-CSF/RUS/Omsk/2013 | E-11 | KU133650 |
| 17 | 261-CSF/RUS/Omsk/2012 | CV-A9 | KU133574 | 17 | 452-CSF/RUS/Omsk/2013 | E-11 | KU133653 |
| 18 | 263-CSF/RUS/Omsk/2012 | CV-A9 | KU133575 | 18 | 391-CSF/RUS/Omsk/2013 | EV-A71 | KU133624 |
| 19 | 270-CSF/RUS/Omsk/2012 | CV-A9 | KU133580 | 19 | 434-CSF/RUS/Omsk/2013 | EV-A71 | KU133644 |
| 20 | 296-CSF/RUS/Omsk/2012 | CV-A9 | KU133590 | 20 | 437-CSF/RUS/Omsk/2013 | EV-A71 | KU133645 |
| 21 | 297-CSF/RUS/Omsk/2012 | CV-A9 | KU133591 | 21 | 450-CSF/RUS/Omsk/2013 | EV-A71 | KU133651 |
| 22 | 299-CSF/RUS/Omsk/2012 | CV-A9 | KU133593 | 22 | 451-CSF/RUS/Omsk/2013 | EV-A71 | KU133652 |
| 23 | 302-CSF/RUS/Omsk/2012 | CV-A9 | KU133594 | 23 | 382-CSF/RUS/Omsk/2013 | CV-A9 | KU133620 |
| 24 | 309-CSF/RUS/Omsk/2012 | CV-A9 | KU133597 | 24 | 393-CSF/RUS/Omsk/2013 | CV-A9 | KU133625 |
| 25 | 312-CSF/RUS/Omsk/2012 | CV-A9 | KU133599 | 25 | 408-CSF/RUS/Omsk/2013 | CV-A9 | KU133635 |
| 26 | 313-CSF/RUS/Omsk/2012 | CV-A9 | KU133600 | 26 | 409-CSF/RUS/Omsk/2013 | CV-A9 | KU133636 |
| 27 | 325-CSF/RUS/Omsk/2012 | CV-A9 | KU133606 | 27 | 386-CSF/RUS/Omsk/2013 | E-7 | KU133622 |
| 28 | 256-CSF/RUS/Omsk/2012 | E-9 | KU133570 | 28 | 413-CSF/RUS/Omsk/2013 | E-7 | KU133638 |
| 29 | 267-CSF/RUS/Omsk/2012 | E-9 | KU133578 | 29 | 428-CSF/RUS/Omsk/2013 | E-7 | KU133641 |
| 30 | 287-CSF/RUS/Omsk/2012 | E-9 | KU133589 | 30 | 403-CSF/RUS/Omsk/2013 | E-30 | KU133631 |
| 31 | 307-CSF/RUS/Omsk/2012 | E-9 | KU133596 | 31 | 405-CSF/RUS/Omsk/2013 | E-30 | KU133633 |
| 32 | 342-CSF/RUS/Omsk/2012 | E-9 | KU133615 | 32 | 455-CSF/RUS/Omsk/2013 | E-30 | KU133654 |
| 33 | 258-CSF/RUS/Omsk/2012 | CV-B1 | KU133572 | 33 | 390-CSF/RUS/Omsk/2013 | CV-B2 | KU133623 |
| 34 | 277-CSF/RUS/Omsk/2012 | CV-B1 | KU133584 | 34 | 407-CSF/RUS/Omsk/2013 | CV-B2 | KU133634 |
| 35 | 298-CSF/RUS/Omsk/2012 | CV-B1 | KU133592 | 35 | 425-CSF/RUS/Omsk/2013 | CV-B2 | KU133640 |
| 36 | 265-CSF/RUS/Omsk/2012 | CV-B1 | KU133576 | 36 | 383-CSF/RUS/Omsk/2013 | E-18 | KU133621 |
| 37 | 328-CSF/RUS/Omsk/2012 | CV-B1 | KU133607 | 37 | 399-CSF/RUS/Omsk/2013 | E-18 | KU133628 |
| 38 | 279-CSF/RUS/Omsk/2012 | E-11 | KU133585 | 38 | 429-CSF/RUS/Omsk/2013 | CV-B4 | KU133642 |
| 39 | 282-CSF/RUS/Omsk/2012 | E-11 | KU133586 | 39 | 439-CSF/RUS/Omsk/2013 | CV-B5 | KU133646 |
| 40 | 318-CSF/RUS/Omsk/2012 | E-11 | KU133601 | – | – | – | – |
| 41 | 284-CSF/RUS/Omsk/2012 | E-18 | KU133587 | – | – | – | – |
| 42 | 330-CSF/RUS/Omsk/2012 | E-18 | KU133609 | – | – | – | – |
| 43 | 253-CSF/RUS/Omsk/2012 | E-32 | KU133569 | – | – | – | – |
| 44 | 286-CSF/RUS/Omsk/2012 | E-30 | KU133588 | – | – | – | – |
| 45 | 304-CSF/RUS/Omsk/2012 | E-14 | KU133595 | – | – | – | – |
| 46 | 310-CSF/RUS/Omsk/2012 | CV-B2 | KU133598 | – | – | – | – |
| 47 | 333-CSF/RUS/Omsk/2012 | E-15 | KU133611 | – | – | – | – |
3.4. Retrospective Direct Genotyping of Enteroviruses Detected in CSFs of Patients Hospitalized in 2013
| Identified Serotype | No. and % (in Parentheses) of EV Serotype to All EVs Identified during the Corresponding Year | |
|---|---|---|
| 2012 | 2013 | |
| Coxsackievirus A9 | 11 (23.4) | 4 (10.3) |
| Coxsackievirus B1 | 5 (10.6) | 0 |
| Coxsackievirus B2 | 1 (2.1) | 3 (7.7) |
| Coxsackievirus B4 | 0 | 1 (2.6) |
| Coxsackievirus B5 | 0 | 1 (2.6) |
| Echovirus 6 | 16 (34.1) | 0 |
| Echovirus 7 | 0 | 3 (7.7) |
| Echovirus 9 | 5 (10.7) | 0 |
| Echovirus 11 | 3 (6.4) | 17 (43.5) |
| Echovirus 14 | 1 (2.1) | 0 |
| Echovirus 15 | 1 (2.1) | 0 |
| Echovirus 18 | 2 (4.3) | 2 (5.1) |
| Echovirus 30 | 1 (2.1) | 3 (7.7) |
| Echovirus 32 | 1 (2.1) | 0 |
| Enterovirus A71 | 0 | 5 (12.8) |
4. Discussion
5. Conclusions
Author Contributions
Conflics of Interest
References
- Knowles, N.; Hovi, T.; Hyypiä, T.; King, A.; Lindberg, A.; Pallansch, M.; Palmenberg, A.; Simmonds, P.; Skern, T.; Stanway, G.; et al. Family picornaviridae. In Virus Taxonomy. Ninth Report of the International Committee on Taxonomy of Viruses, 1st ed.; King, A., Adams, M., Carstens, E., Lefkowitz, E., Eds.; Elsevier Press: San Diego, CA, USA, 2012; Volume 1, pp. 855–880. [Google Scholar]
- Chevaliez, S.; Szendroi, A.; Caro, V.; Balanant, J.; Guillot, S.; Berencsi, G.; Delpeyroux, F. Molecular comparison of echovirus 11 strains circulating in Europe during an epidemic of multisystem hemorrhagic disease of infants indicates that evolution generally occurs by recombination. Virology 2004, 325, 56–70. [Google Scholar] [CrossRef] [PubMed]
- Lindberg, A.M.; Anderson, P.; Savolainen, M.N.; Mulders, M.N.; Hovi, T. Evolution of the genome of human enterovirus B: Incongruence between phylogenies of the VP1 and 3CD regions indicates frequent recombination within the species. J. Gen. Virol. 2003, 84, 1223–1235. [Google Scholar] [CrossRef] [PubMed]
- Mirand, A.; Henquell, C.; Archimbaud, C.; Peigue-Lafeuille, H.; Bailly, J. Emergence of echovirus 30 lineages is marked by serial genetic recombination events. J. Gen. Virol. 2007, 88, 166–176. [Google Scholar] [CrossRef] [PubMed]
- Oberste, M.S.; Maher, K.; Pallansch, M.A. Evidence for frequent recombination within species human enterovirus B based on complete genomic sequences of all thirty-seven serotypes. J. Virol. 2004, 78, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Oprisan, G.; Combiescu, M.; Guillot, S.; Caro, V.; Combiescu, A.; Delpeyroux, F.; Grainic, R. Natural genetic recombination between co-circulating heterotypic enteroviruses. J. Gen. Virol. 2002, 83, 2193–2200. [Google Scholar] [CrossRef] [PubMed]
- Santti, J.; Hyypiä, T.; Kinnunen, L.; Salminen, M. Evidence of recombination among enteroviruses. J. Virol. 1999, 73, 8741–8749. [Google Scholar] [PubMed]
- Simmonds, P.; Welch, J. Frequency and dynamics of recombination within different species of human enteroviruses. J. Virol. 2006, 80, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Harris, K.G.; Coyne, C.B. Enter at your own risk: How enteroviruses navigate the dangerous world of pattern recognition receptor signaling. Cytokine 2013, 63, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.E.; Davies, H.D. Aseptic meningitis. Curr. Opin. Infect. Dis. 2007, 20, 272–277. [Google Scholar] [CrossRef] [PubMed]
- Kelly, T.A.; O'Lorcain, P.; Moran, J.; Garvey, P.; McKeown, P.; Connell, J.; Cotter, S. Underreporting of viral encephalitis and viral meningitis, Ireland, 2005–2008. Emerg. Infect. Dis. 2013, 19, 1428–1436. [Google Scholar] [CrossRef] [PubMed]
- Nathanson, N.; Kew, O.M. From emergence to eradication: The epidemiology of poliomyelitis deconstructed. Am. J. Epidemiol. 2010, 172, 1213–1229. [Google Scholar] [CrossRef] [PubMed]
- Andréoletti, L.; Lévêque, N.; Boulagnon, C.; Brasselet, C.; Fornes, P. Viral causes of human myocarditis. Arch. Cardiovasc. Dis. 2009, 102, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Kühl, U.; Schultheiss, H.P. Viral myocarditis. Swiss. Med. Wkly. 2014, 144, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hober, D.; Sauter, P. Pathogenesis of type 1 diabetes mellitus: Interplay between enterovirus and host. Nat. Rev. Endocrinol. 2010, 6, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Rantakallio, P.; Jones, P.; Moring, J.; von Wendt, L. Association between central nervous system infections during childhood and adult onset schizophrenia and other psychoses: A 28-year follow-up. Int. J. Epidemiol. 1997, 26, 837–843. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.C.; Chi, C.S.; Chiu, Y.T.; Hsu, S.L.; Hwang, B.; Jan, S.L.; Chan, P.Y.; Huang, F.L.; Chang, Y. Cardiac complications of enterovirus rhombencephalitis. Arch. Dis. Child. 2004, 89, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Kung, C.M.; King, C.C.; Lee, C.N.; Huang, L.M.; Lee, P.I.; Kao, C.L. Differences in replication capacity between enterovirus 71 isolates obtained from patients with encephalitis and those obtained from patients with herpangina in Taiwan. J. Med. Virol. 2007, 79, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Solomon, T.; Lewthwaite, P.; Perera, D.; Cardosa, M.J.; McMinn, P.; Ooi, M.H. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet. Infect. Dis. 2010, 10, 778–790. [Google Scholar] [CrossRef]
- Tijsma, A.; Franco, D.; Tucker, S.; Hilgenfeld, R.; Froeyen, M.; Leyssen, P.; Neyts, J. The capsid binder Vapendavir and the novel protease inhibitor SG85 inhibit enterovirus 71 replication. Antimicrob. Agents Chemother. 2014, 58, 6990–6992. [Google Scholar] [CrossRef] [PubMed]
- Volle, R.; Bailly, J.-L.; Mirand, A.; Pepeira, B.; Marque-Juillet, S.; Chambon, M.; Regagnon, C.; Brebion, A.; Henquell, C.; Peigue-Lafeuille, H.; et al. Variations in cerebrospinal fluid viral loads among enterovirus genotypes in patients hospitalized with laboratory-confirmed meningitis due to enterovirus. J. Infect. Dis. 2014, 210, 576–584. [Google Scholar] [CrossRef] [PubMed]
- Mirand, A.; Henquell, C.; Archimbaud, C.; Chambon, M.; Charbonne, F.; Peigue-Lafeuille, H.; Bailly, J.-L. Prospective identification of enteroviruses involved in meningitis in 2006 through direct genotyping in celebrospinal fluid. J. Clin. Microbiol. 2008, 46, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Nix, W.A.; Oberste, M.S.; Pallansch, M.A. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all interovirus serotypes from original clinical specimens. J. Clin. Microbiol. 2006, 44, 2698–2704. [Google Scholar] [CrossRef] [PubMed]
- Casas, I.; Palacios, G.F.; Trallero, G.; Cisterna, D.; Freire, M.C.; Tennorio, A. Molecular characterization of human enteroviruses in clinical samples: Comparison between VP2,VP1, and RNA polymerase region using RT nested PCR assays and direct sequencing of products. J. Med. Virol. 2001, 65, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Oberste, M.S.; Penaranda, S.; Maher, K.; Pallansch, M.A. Complete genome sequences of all members of the species Human enterovirus A. J. Gen. Virol. 2004, 85, 1957–1607. [Google Scholar] [CrossRef] [PubMed]
- Basic Local Alignment Search Tool. Available online: http://blast. ncbi.nlm.nih.gov/Blast.cgi (accessed on 11 August 2015).
- Rose, T.M. CODEHOP-mediated PCR—A powerful technique for the identification and characterization of viral genomes. Virol. J. 2005, 15, 2–20. [Google Scholar]
- Khetsuriani, N.; LaMonte-Fowlkes, A.; Oberste, M.S.; Pallansch, M.A. Enterovirus surveillance-Unated States, 1970–2005. Morb. Mortal. Wkly. Rep. 2006, 55, 1–20. [Google Scholar]
- Bharti, O.K. Can non-polio enteroviruses be tamed with a vaccine to minimize paralysis caused by them? World J. Vacc. 2015, 5, 54–59. [Google Scholar] [CrossRef]
- Dhole, T.N.; Ayyagari, A.; Chowdhary, R.; Shakya, A.K.; Shrivastav, N.; Datta, T.; Prakash, V. Non-polio enteroviruses in acute flaccid paralysis children of India: Vital assessment before polio eradication. J. Paediatr. Child Health. 2009, 45, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Cabrerizo, M.; Echevarria, J.E.; González, I.; de Miguel, T.; Trallero, G. Molecular epidemiological study of HEV-B enteroviruses involved in the increase in meningitis cases occurred in Spain during 2006. J. Med. Virol. 2008, 80, 1018–1024. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.J.; Wang, J.R.; Liu, C.C.; Yang, H.B.; Su, I.J. An outbreak of enterovirus 71 infection in Taiwan 1998: A comprehensive pathological, virological, and molecular study on a case of fulminant encephalitis. J. Clin. Virol. 2000, 17, 13–22. [Google Scholar] [CrossRef]
- Ooi, M.H.; Wong, S.C.; Lewthwaite, P.; Cardosa, M.J.; Solomon, T. Clinical features, diagnosis, and management of enterovirus 71. Lancet Neurol. 2010, 9, 1097–1105. [Google Scholar] [CrossRef]
- Oberste, M.S.; Maher, K.; Kilpatrick, D.R.; Flemister, M.R.; Brown, B.A.; Pallansch, M.A. Typing of human enteroviruses by partial sequencing of VP1. J. Clin. Microbiol. 1999, 37, 1288–1293. [Google Scholar] [PubMed]
- Caro, V.; Guillot, S.; Delpeyroux, F.; Crainic, R. Molecular strategy for “serotyping” of human enteroviruses. J. Gen. Virol. 2001, 82, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Itturiza-Gomara, M.; Megson, B.; Gray, J. Molecular detection and characterization of human enteroviruses directly from clinical samples using RT-PCR and DNA sequencing. J. Med. Virol. 2006, 75, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Leitch, E.C.; Harvala, H.; Robertson, I.; Ubillos, I.; Templeton, K.; Simmonds, P. Direct identification of human enterovirus serotypes in cerebrospinal fluid by amplification and sequencing of the VP1 region. J. Clin. Virol. 2009, 44, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Papadakis, G.; Chibo, D.; Druce, J.; Catton, M.; Birch, C. Detection and genotyping of enteroviruses in celebrospinal fluid in patients in Victoria, Australia, 2007–2013. J. Med. Virol. 2014, 86, 1609–1613. [Google Scholar] [CrossRef] [PubMed]
- Chapman, N.M.; Kim, K.S.; Drescher, K.M.; Oka, K.; Tracy, S. 5′ terminal deletions in the genome of the coxsackievirus B2 strain occurred naturally in human heart. Virology 2008, 375, 480–491. [Google Scholar] [CrossRef] [PubMed]
- Kühl, U.; Pauschinger, M.; Seeberg, B.; Lassner, D.; Noutsias, M.; Poller, W.; Schultheiss, H.P. Viral persistence in the myocardium is associated with progressive cardiac dysfunction. Circulation 2005, 112, 1965–1970. [Google Scholar] [CrossRef] [PubMed]
- Feuer, R.; Ruller, C.M.; An, N.; Tabor-Godwin, J.M.; Rhoades, R.E.; Maciejewski, S.; Pagarigan, R.R.; Cornell, C.T.; Crocker, S.; Kiosses, W.; et al. Viral persistence and chronic immunopathology in the adult central nervous system following Coxsackievirus infection during the neonatal period. J. Virol. 2009, 83, 9356–9369. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krasota, A.; Loginovskih, N.; Ivanova, O.; Lipskaya, G. Direct Identification of Enteroviruses in Cerebrospinal Fluid of Patients with Suspected Meningitis by Nested PCR Amplification. Viruses 2016, 8, 10. https://doi.org/10.3390/v8010010
Krasota A, Loginovskih N, Ivanova O, Lipskaya G. Direct Identification of Enteroviruses in Cerebrospinal Fluid of Patients with Suspected Meningitis by Nested PCR Amplification. Viruses. 2016; 8(1):10. https://doi.org/10.3390/v8010010
Chicago/Turabian StyleKrasota, Alexandr, Natalia Loginovskih, Olga Ivanova, and Galina Lipskaya. 2016. "Direct Identification of Enteroviruses in Cerebrospinal Fluid of Patients with Suspected Meningitis by Nested PCR Amplification" Viruses 8, no. 1: 10. https://doi.org/10.3390/v8010010






