RNase P Ribozymes Inhibit the Replication of Human Cytomegalovirus by Targeting Essential Viral Capsid Proteins
Abstract
:1. Introduction
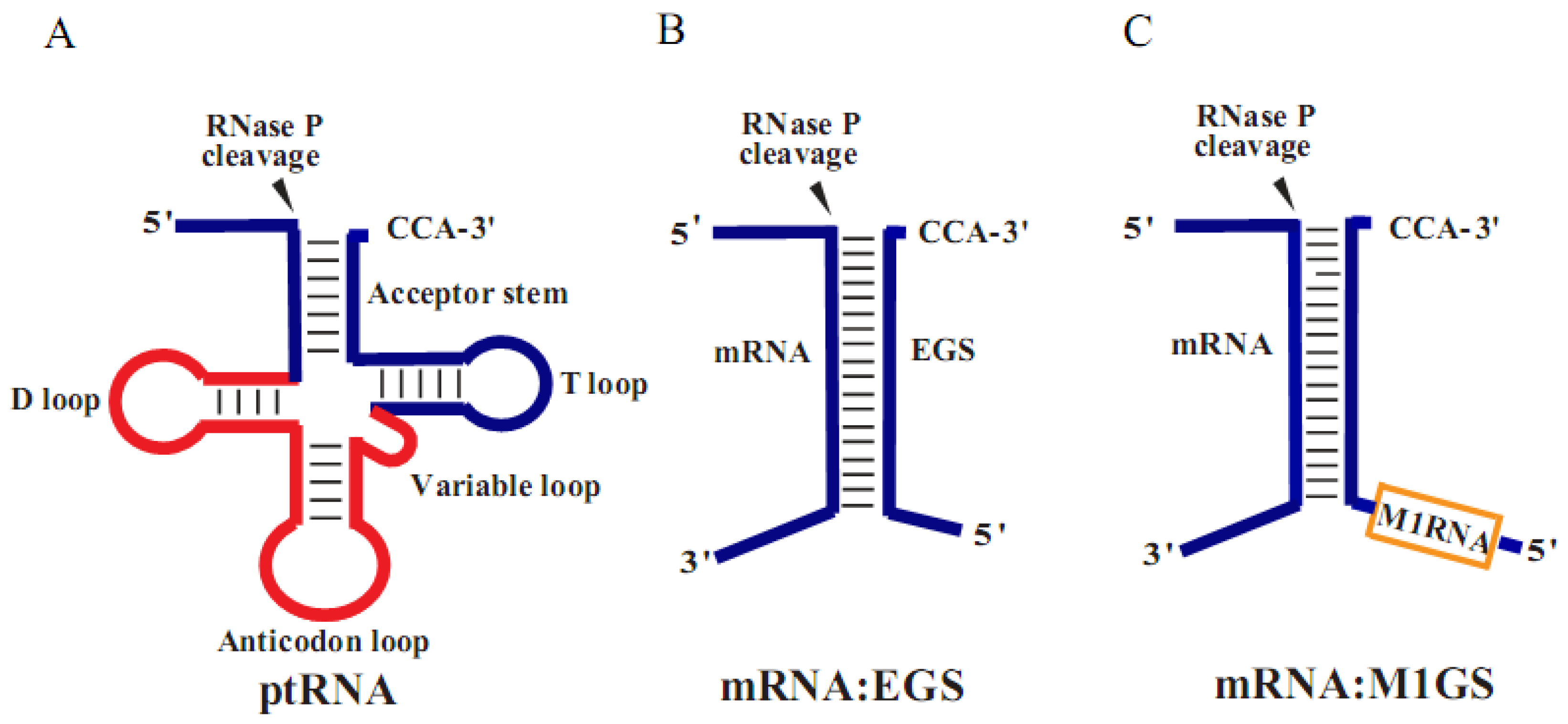
2. Materials and Methods
2.1. Viruses, Cells and Antibodies
2.2. Mapping of the AP mRNA Accessible Regions in Cells
2.3. In Vitro Ribozyme Studies
2.4. Construction of Ribozyme-Expressing Cell Lines
2.5. Studies of Viral Gene Expression and Growth in M1GS-Expressing Cells
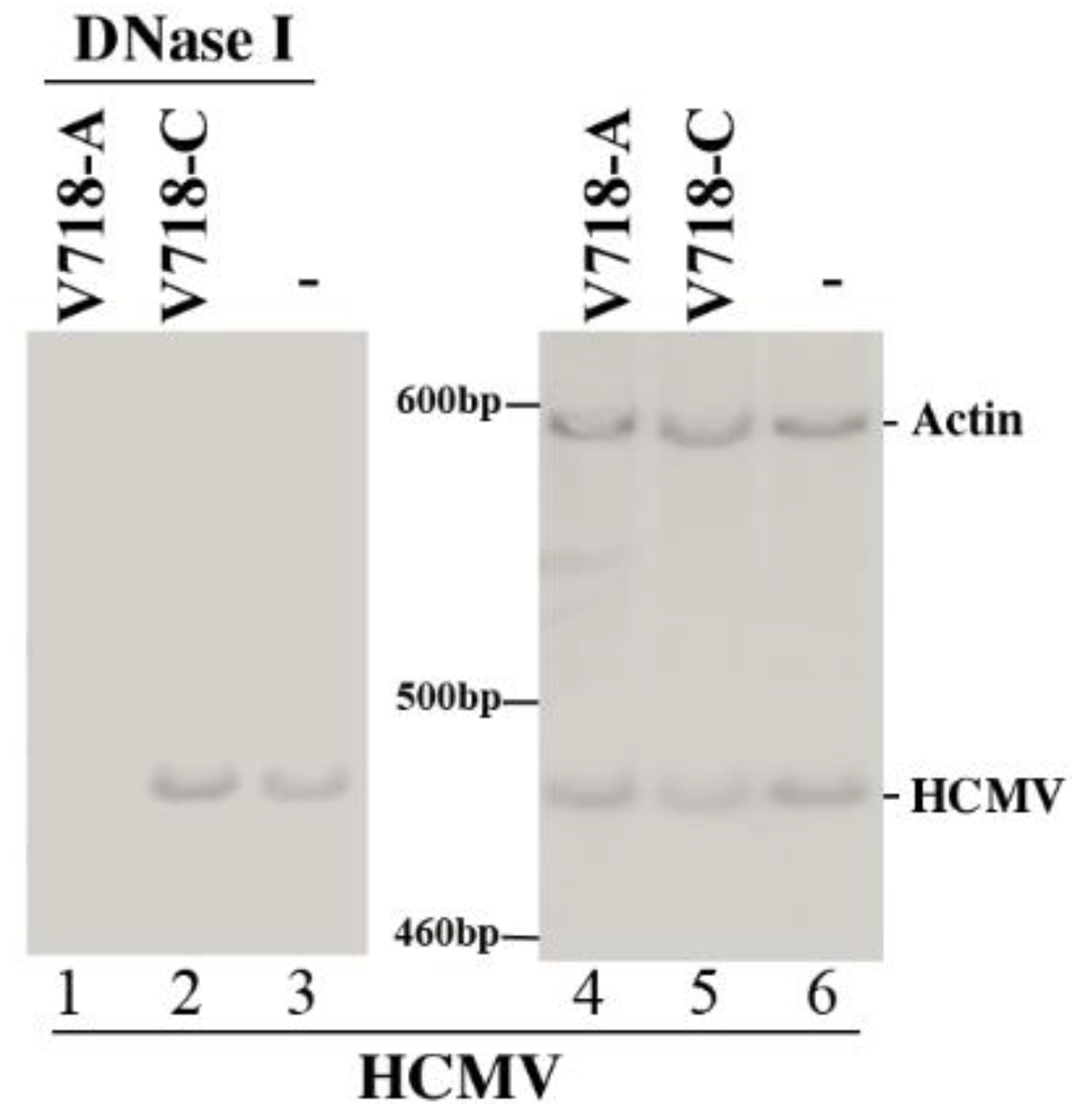
2.6. Determination of the HCMV DNA Level
2.7. Statistical Analysis
3. Results
3.1. Ribozyme-Mediated Cleavage of the AP mRNA Sequence in Vitro
| Enzyme | (kcat/Km)s (µM−1·min−1) | Kd (nM) |
|---|---|---|
| M1-A | 0.22 ± 0.05 | 0.31 ± 0.05 |
| V718-A | 13.5 ± 2.5 | 0.35 ± 0.06 |
| M1-C | <5 × 10−6 | 0.32 ± 0.06 |
| V718-C | <5 × 10−6 | 0.34 ± 0.05 |
| M1-TK | <5 × 10−6 | ND |
3.2. Ribozyme Expression in the Cultured Cells

3.3. Ribozyme-Mediated Reduction of HCMV AP and PR Expression

| Viral Gene Class | Ribozymes | ||||||
|---|---|---|---|---|---|---|---|
| U251 | M1-C | V718-C | M1-A | V718-A | M1-TK | ||
| IE2 mRNA | α | 0% | 0% | 0% | 0% | 0% | 0% |
| US2 mRNA | β | 0% | 0% | 0% | 0% | 0% | 0% |
| AP mRNA | γ | 0% | 3% | 6% | 78% ± 8% | 99% ± 7% | 0% |
| PR mRNA | γ | 0% | 3% | 7% | 75% ± 7% | 98% ± 7% | 0% |
| IE2 protein | α | 0% | 0% | 0% | 0% | 0% | 0% |
| UL44 protein | β, γ | 0% | 0% | 0% | 0% | 0% | 0% |
| UL83 protein | γ | 0% | 0% | 0% | 0% | 0% | 0% |
| AP protein | γ | 0% | 3% | 6% | 75% ± 7% | 99% ± 8% | 0% |
| PR protein | γ | 0% | 3% | 6% | 72% ± 7% | 98% ± 8% | 0% |

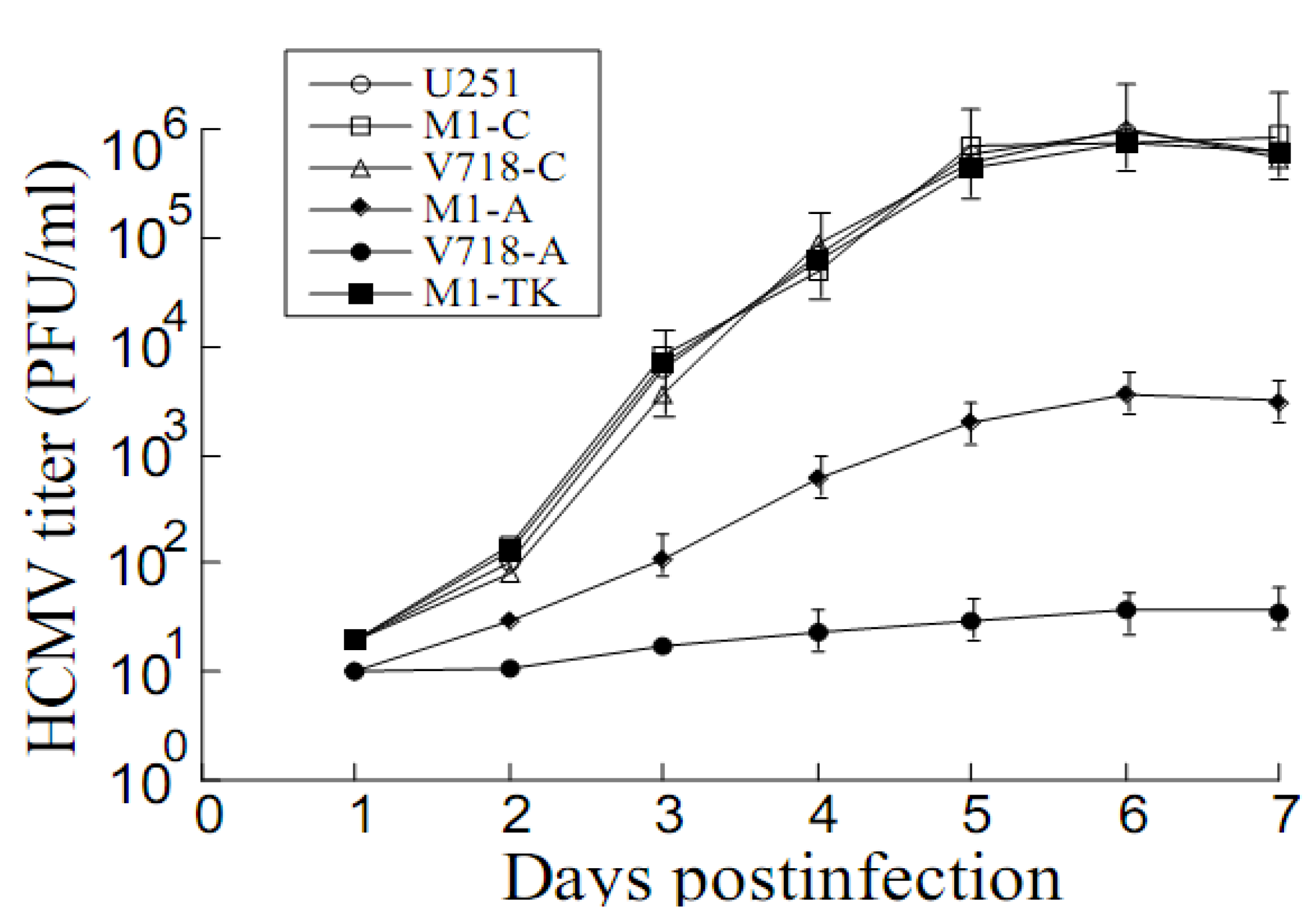
3.4. Inhibition of HCMV Replication Mediated by the Ribozymes
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mocarski, E.S.; Shenk, T.; Pass, R.F. Cytomegalovirus. In Fields Virology; Knipe, D.M., Howley, P.M., Griffin, D.E., Martin, M.A., Lamb, R.A., Roizman, B., Straus, S.E., Eds.; Lippincott-William & Wilkins: Philadelphia, PA, USA, 2007; pp. 2701–2772. [Google Scholar]
- Boppana, S.B.; Ross, S.A.; Fowler, K.B. Congenital cytomegalovirus infection: Clinical outcome. Clin. Infect. Dis. 2013, 57 (Suppl. 4), S178–S181. [Google Scholar] [CrossRef] [PubMed]
- Gopalan, V.; Altman, S. RNase P: Structure and catalysis. In The RNA World; Gesteland, R., Cech, T., Atkins, J., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2006; Volume 277. [Google Scholar]
- Guerrier-Takada, C.; Gardiner, K.; Marsh, T.; Pace, N.; Altman, S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 1983, 35, 849–857. [Google Scholar] [CrossRef]
- Gopalan, V.; Vioque, A.; Altman, S. RNase P: Variations and uses. J. Biol. Chem. 2002, 277, 6759–6762. [Google Scholar] [CrossRef] [PubMed]
- Forster, A.C.; Altman, S. External guide sequences for an RNA enzyme. Science 1990, 249, 783–786. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.N.; Harris, M.; Pace, N.R. Rational design of self-cleaving pre-tRNA-ribonuclease P RNA conjugates. Biochemistry 1994, 33, 10800–10808. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Altman, S. Inhibition of viral gene expression by the catalytic RNA subunit of RNase P from Escherichia coli. Genes Dev. 1995, 9, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Weiss, B.; Davidkova, G.; Zhou, L.W. Antisense RNA gene therapy for studying and modulating biological processes. Cell. Mol. Life Sci. 1999, 55, 334–358. [Google Scholar] [CrossRef] [PubMed]
- Van der Ree, M.H.; van der Meer, A.J.; de Bruijne, J.; Maan, R.; van Vliet, A.; Welzel, T.M.; Zeuzem, S.; Lawitz, E.J.; Rodriguez-Torres, M.; Kupcova, V.; et al. Long-term safety and efficacy of microRNA-targeted therapy in chronic hepatitis C patients. Antiviral Res. 2014, 111, 53–59. [Google Scholar]
- Ditzler, M.A.; Bose, D.; Shkriabai, N.; Marchand, B.; Sarafianos, S.G.; Kvaratskhelia, M.; Burke, D.H. Broad-spectrum aptamer inhibitors of HIV reverse transcriptase closely mimic natural substrates. Nucleic Acids Res. 2011, 39, 8237–8247. [Google Scholar] [CrossRef] [PubMed]
- Sarver, N.; Cantin, E.M.; Chang, P.S.; Zaia, J.A.; Ladne, P.A.; Stephens, D.A.; Rossi, J.J. Ribozymes as potential anti-HIV-1 therapeutic agents. Science 1990, 247, 1222–1225. [Google Scholar] [CrossRef] [PubMed]
- Scherer, L.J.; Rossi, J.J. Approaches for the sequence-specific knockdown of mRNA. Nat. Biotechnol. 2003, 21, 1457–1465. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Rossi, J.J. Current progress in the development of RNAi-based therapeutics for HIV-1. Gene Ther. 2011, 18, 1134–1138. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.; Marquez, S.M.; Pace, N.R. RNase P: Interface of the RNA and protein worlds. Trends Biochem. Sci. 2006, 31, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Marvin, M.C.; Engelke, D.R. Broadening the mission of an RNA enzyme. J. Cell. Biochem. 2009, 108, 1244–1251. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Bai, Y.; Rider, P.; Kim, K.; Zhang, C.; Lu, S.; Liu, F. Engineered external guide sequences effectively block viral gene expression and replication in cultured cells. J. Biol. Chem. 2011, 286, 322–330. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Liu, F. Inhibition of gene expression in human cells using RNase P-derived ribozymes and external guide sequences. Biochim. Biophys. Acta 2007, 1769, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Kilani, A.F.; Trang, P.; Jo, S.; Hsu, A.; Kim, J.; Nepomuceno, E.; Liou, K.; Liu, F. RNase P ribozymes selected in vitro to cleave a viral mRNA effectively inhibit its expressionin cell culture. J. Biol. Chem. 2000, 275, 10611–10622. [Google Scholar] [CrossRef] [PubMed]
- Trang, P.; Lee, M.; Nepomuceno, E.; Kim, J.; Zhu, H.; Liu, F. Effective inhibition of human cytomegalovirus gene expression and replication by a ribozyme derived from the catalytic RNA subunit of RNase P from Escherichia coli. Proc. Natl. Acad. Sci. USA 2000, 97, 5812–5817. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.Y.; Roizman, B. The promoter, transcriptional unit, and coding sequence of herpes simplex virus 1 family 35 proteins are contained within and in frame with the UL26 open reading frame. J. Virol. 1991, 65, 206–212. [Google Scholar] [PubMed]
- Welch, A.R.; Woods, A.S.; McNally, L.M.; Cotter, R.J.; Gibson, W. A herpesvirus maturational proteinase, assemblin: Identification of its gene, putative active site domain, and cleavage site. Proc. Natl. Acad. Sci. USA 1991, 88, 10792–10796. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Lee, J.; Umamoto, S.; Kilani, A.F.; Kim, J.; Trang, P.; Zhou, T.; Liu, F. Engineered RNase P ribozymes are efficient in cleaving a human cytomegalovirus mRNA in vitro and are effective in inhibiting viral gene expression and growth in human cells. J. Biol. Chem. 2003, 278, 37265–37274. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Trang, P.; Umamoto, S.; Hai, R.; Liu, F. RNase P ribozyme inhibits cytomegalovirus replication by blocking the expression of viral capsid proteins. Nucleic Acids Res. 2004, 32, 3427–3434. [Google Scholar] [CrossRef] [PubMed]
- Zaug, A.J.; Cech, T.R. Analysis of the structure of Tetrahymena nuclear RNAs in vivo: Telomerase RNA, the self-splicing rRNA intron, and U2 snRNA. RNA 1995, 1, 363–374. [Google Scholar] [PubMed]
- Trang, P.; Liu, F. Mapping the regions of RNase P catalytic RNA that are potentially in close contact with its protein cofactor. Methods Mol. Biol. 2008, 488, 267–277. [Google Scholar] [PubMed]
- Kim, J.J.; Kilani, A.F.; Zhan, X.; Altman, S.; Liu, F. The protein cofactor allows the sequence of an RNase P ribozyme to diversify by maintaining the catalytically active structure of the enzyme. RNA 1997, 3, 613–623. [Google Scholar] [PubMed]
- Bai, Y.; Trang, P.; Li, H.; Kim, K.; Zhou, T.; Liu, F. Effective inhibition in animals of viral pathogenesis by a ribozyme derived from RNase P catalytic RNA. Proc. Natl. Acad. Sci. USA 2008, 105, 10919–10924. [Google Scholar] [CrossRef] [PubMed]
- Trang, P.; Kim, K.; Zhu, J.; Liu, F. Expression of an RNase P ribozyme against the mRNA encoding human cytomegalovirus protease inhibits viral capsid protein processing and growth. J. Mol. Biol. 2003, 328, 1123–1135. [Google Scholar] [CrossRef]
- Miller, A.D.; Rosman, G.J. Improved retroviral vectors for gene transfer and expression. Biotechniques 1989, 7, 980–982, 984–986, 989–990. [Google Scholar] [PubMed]
- Yang, Y.H.; Li, H.; Zhou, T.; Kim, K.; Liu, F. Engineered external guide sequences are highly effective in inducing RNase P for inhibition of gene expression and replication of human cytomegalovirus. Nucleic Acids Res. 2006, 34, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Sunkara, N.; Lu, S.; Liu, F. Directing RNase P-mediated cleavage of target mRNAs by engineered external guide sequences in cultured cells. Methods Mol. Biol. 2014, 1103, 45–56. [Google Scholar] [PubMed]
- Yang, Z.; Vu, G.P.; Qian, H.; Chen, Y.C.; Wang, Y.; Reeves, M.; Zen, K.; Liu, F. Engineered RNase P ribozymes effectively inhibit human cytomegalovirus gene expression and replication. Viruses 2014, 6, 2376–2391. [Google Scholar] [CrossRef] [PubMed]
- Matusick-Kumar, L.; Hurlburt, W.; Weinheimer, S.P.; Newcomb, W.W.; Brown, J.C.; Gao, M. Phenotype of the herpes simplex virus type 1 protease substrate ICP35 mutant virus. J. Virol. 1994, 68, 5384–5394. [Google Scholar] [PubMed]
- Jiang, X.; Chen, Y.C.; Gong, H.; Trang, P.; Lu, S.; Liu, F. Ribonuclease P-mediated inhibition of human cytomegalovirus gene expression and replication induced by engineered external guide sequences. RNA Biol. 2012, 9, 1186–1195. [Google Scholar] [CrossRef] [PubMed]
- Chee, M.S.; Bankier, A.T.; Beck, S.; Bohni, R.; Brown, C.M.; Cerny, R.; Horsnell, T.; Hutchison, C.A., 3rd; Kouzarides, T.; Martignetti, J.A.; et al. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 1990, 154, 125–169. [Google Scholar] [PubMed]
- Davison, A.J.; Dolan, A.; Akter, P.; Addison, C.; Dargan, D.J.; Alcendor, D.J.; McGeoch, D.J.; Hayward, G.S. The human cytomegalovirus genome revisited: Comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 2003, 84, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Altman, S. Requirements for cleavage by a modified RNase P of a small model substrate. Nucleic Acids Res. 1996, 24, 2690–2696. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Kim, J.; Kilani, A.F.; Kim, K.; Dunn, W.; Jo, S.; Nepomuceno, E.; Liu, F. In vitro selection of external guide sequences for directing RNase P-mediated inhibition of viral gene expression. J. Biol. Chem. 2002, 277, 30112–30120. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Chen, Y.C.; Bai, Y.; Trang, P.; Vu, G.P.; Lu, S.; Wu, J.; Liu, F. Effective inhibition of human immunodeficiency virus 1 replication by engineered RNase P ribozyme. PLoS ONE 2012, 7, e51855. [Google Scholar] [CrossRef] [PubMed]
- Altman, S.; Kirsebom, L.A. Ribonuclease P. In The RNA World; Gesteland, R.F., Cech, T.R., Atkins, J.F., Eds.; Cold Spring Harbor Press: Cold Spring Harbor, NY, USA, 1999; pp. 351–380. [Google Scholar]
- Liu, F. Ribonuclease P as A Tool; Springer: New York, NY, USA, 2010; pp. 257–276. [Google Scholar]
- Dunn, W.; Chou, C.; Li, H.; Hai, R.; Patterson, D.; Stolc, V.; Zhu, H.; Liu, F. Functional profiling of human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA 2003, 100, 14223–14228. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Z.; Reeves, M.; Ye, J.; Trang, P.; Zhu, L.; Sheng, J.; Wang, Y.; Zen, K.; Wu, J.; Liu, F. RNase P Ribozymes Inhibit the Replication of Human Cytomegalovirus by Targeting Essential Viral Capsid Proteins. Viruses 2015, 7, 3345-3360. https://doi.org/10.3390/v7072775
Yang Z, Reeves M, Ye J, Trang P, Zhu L, Sheng J, Wang Y, Zen K, Wu J, Liu F. RNase P Ribozymes Inhibit the Replication of Human Cytomegalovirus by Targeting Essential Viral Capsid Proteins. Viruses. 2015; 7(7):3345-3360. https://doi.org/10.3390/v7072775
Chicago/Turabian StyleYang, Zhu, Michael Reeves, Jun Ye, Phong Trang, Li Zhu, Jingxue Sheng, Yu Wang, Ke Zen, Jianguo Wu, and Fenyong Liu. 2015. "RNase P Ribozymes Inhibit the Replication of Human Cytomegalovirus by Targeting Essential Viral Capsid Proteins" Viruses 7, no. 7: 3345-3360. https://doi.org/10.3390/v7072775





