Dynamics of the Presence of Israeli Acute Paralysis Virus in Honey Bee Colonies with Colony Collapse Disorder
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bee Samples
2.2. RNA Extraction
2.3. RT-PCR and cDNA
2.4. Quantitative Real-Time PCR
2.5. Infection with IAPV
2.6. Protein Extraction and Immunoblotting
2.7. Statistical Analysis
3. Results
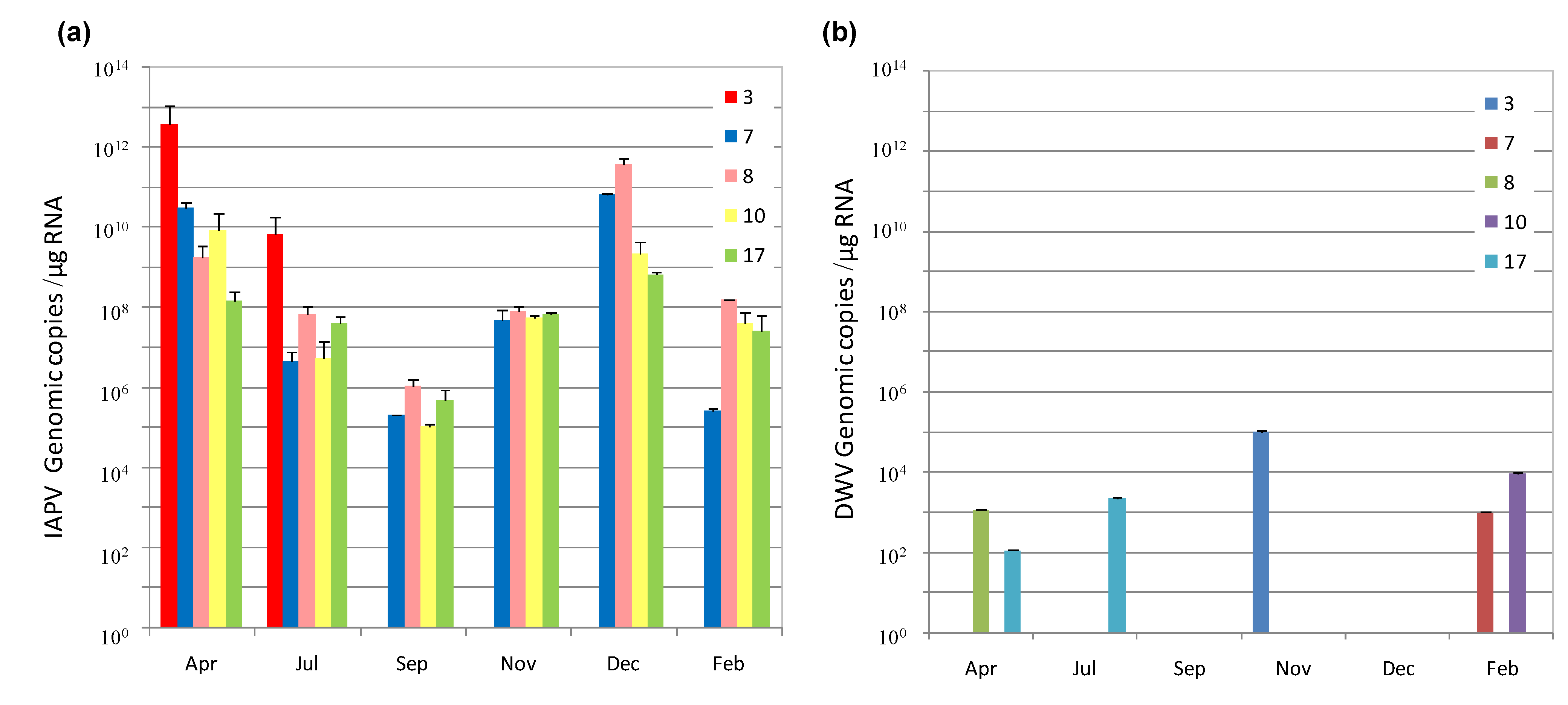
3.1. IAPV Fate in CCD Colonies
| Colony | Symptoms | Virus tested | |||||
|---|---|---|---|---|---|---|---|
| IAPV | SBV | ABPV | BQCV | CBPV | DWV | ||
| 1 | CCD | + | − | − | + | − | + |
| 2 | CCD | + | − | − | + | − | + |
| 3 | CCD | + | − | − | + | − | + |
| 4 | CCD | + | − | − | + | − | + |
| 5 | CCD | + | − | − | − | − | + |
| 6 | CCD | + | − | − | − | + | + |
| 7 | CCD | + | − | + | − | − | + |
| 8 | CCD | + | − | − | − | − | + |
| 9 | CCD | + | − | − | + | − | + |
| 10 | CCD | + | − | − | + | − | + |
| 11 | CCD | + | − | − | − | − | − |
| 12 | CCD | + | − | − | − | − | + |
| 13 | CCD | + | − | − | − | − | + |
| 16 | Control | − | − | − | + | − | + |
| 17 | Non-CCD | + | − | − | − | − | + |
| 18 | Control | − | − | − | − | − | − |
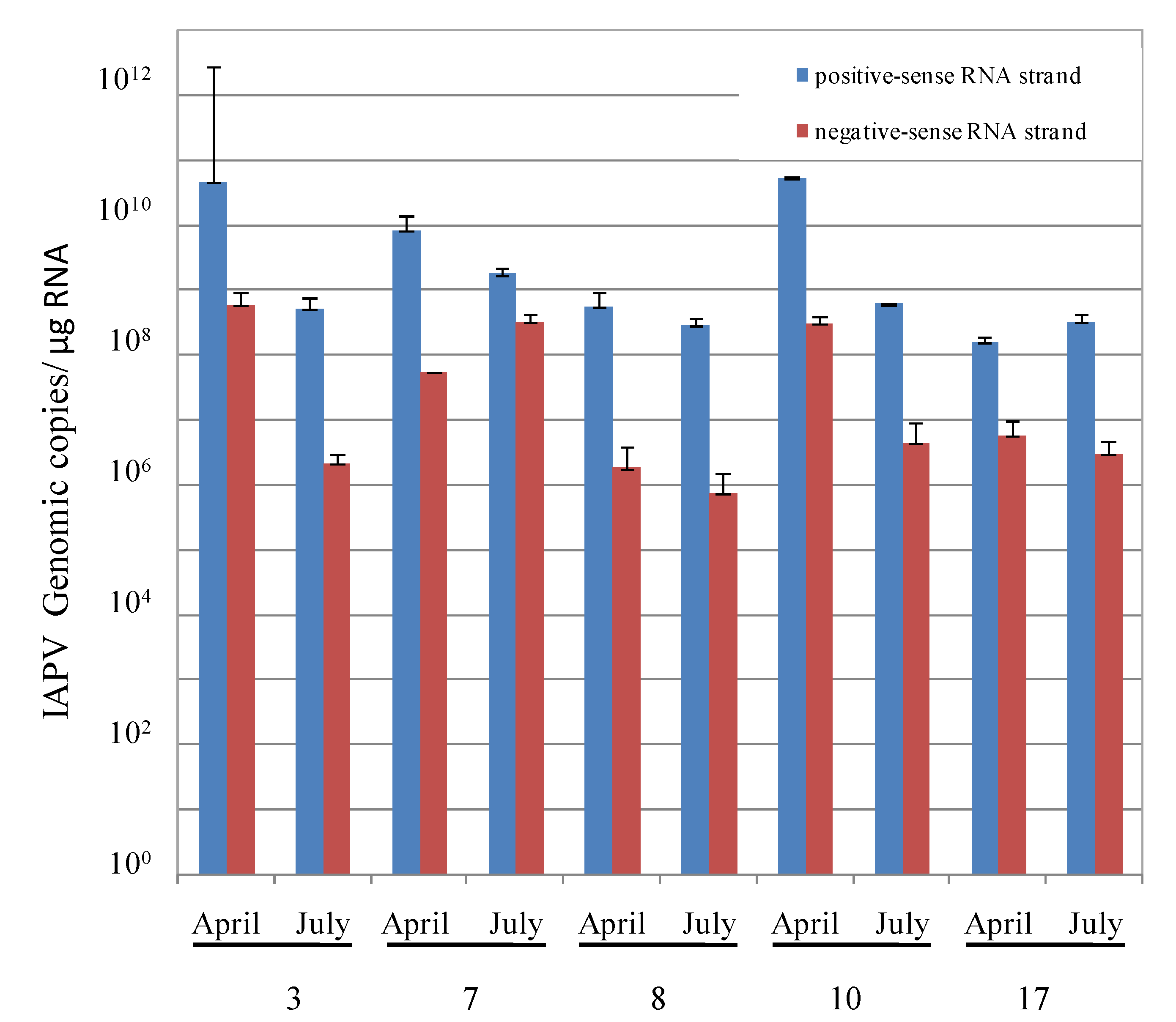
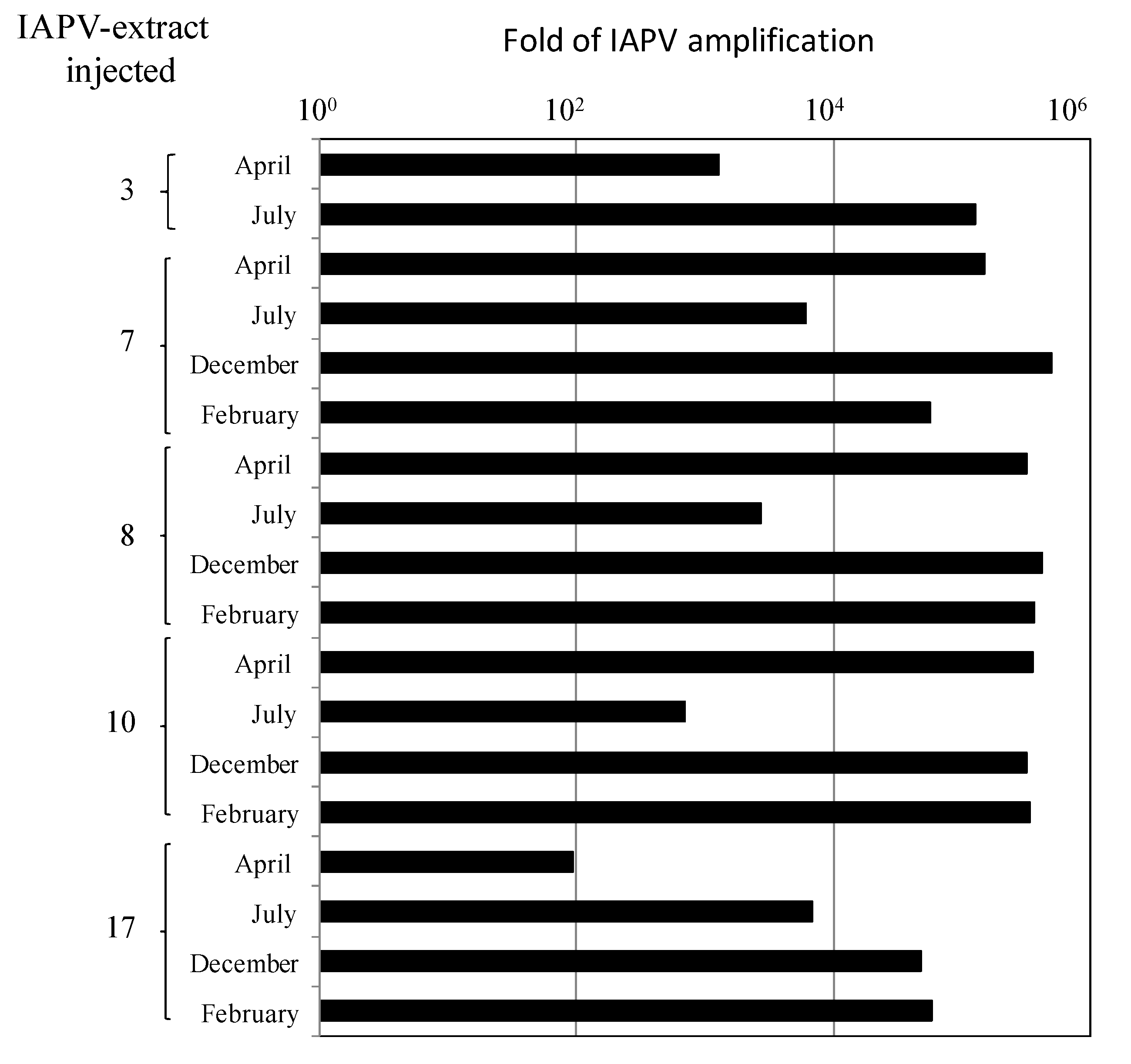
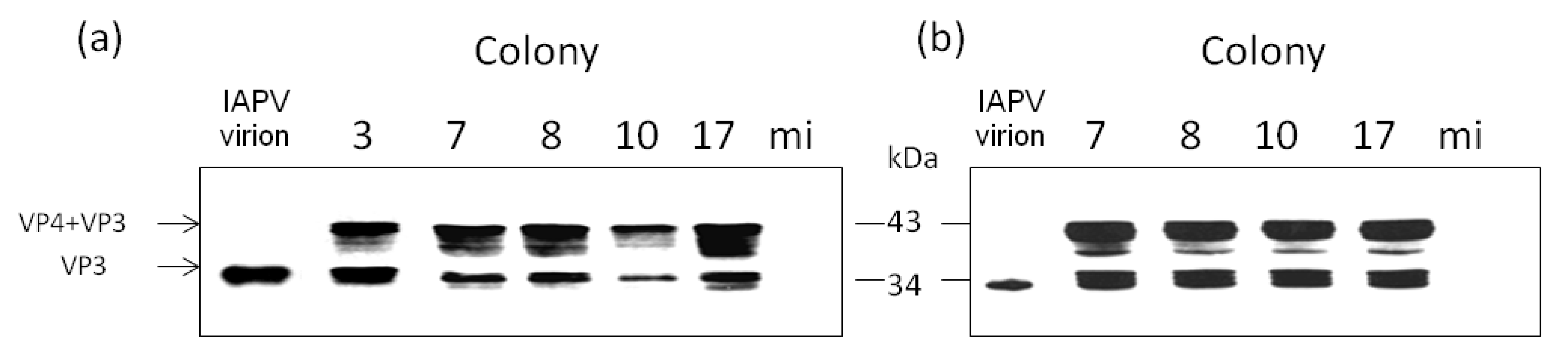
3.2. Replication and Infectivity of IAPV in CCD-Colonies
- By measuring the relative levels of positive- vs. negative-sense RNA strands of IAPV present in individual adult bees of these colonies, a common criterion used for evaluation of replicating positive-strand RNA viruses [37].
- By injecting virus-free pupae from healthy colonies with extracts of virus from infected bees from the above colonies and measuring if IAPV titers increased significantly following the infection.
- By injecting virus-free pupae from healthy colonies with extracts of virus from infected bees from the above colonies and following the expression of the viral capsid protein VP3 due to the infection.
3.3. Honey Bee Population in Recovered CCD-Colonies

4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References and Notes
- Gallai, N.; Salles, J.M.; Settele, J.; Vaissière, B.E. Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecol. Economics 2009, 68, 810–821. [Google Scholar] [CrossRef] [Green Version]
- vanEngelsdorp, D.; Meixner, M.D. A historical review of managed honey bee populations in Europe and the United States and the factors that may affect them. J. Invertebr. Pathol. 2010, 103, S80–S95. [Google Scholar] [CrossRef]
- Potts, S.G.; Biesmeijer, J.C.; Kremen, C.; Neumann, P.; Schweiger, O.; Kunin, W.E. Global pollinator declines: Trends, impacts and drivers. Trends Ecol. Evol. 2010, 25, 345–353. [Google Scholar] [CrossRef]
- Evans, J.D.; Schwarz, R.S. Bees brought to their knees: Microbes affecting honey bee health. Trends Microbiol. 2011, 19, 614–620. [Google Scholar] [CrossRef]
- vanEngelsdorp, D.; Evans, J.D.; Saegerman, C.; Mullin, C.; Haubruge, E.; Nguyen, B.K.; Frazier, M.; Frazier, J.; Cox-Foster, D.; Chen, Y.; et al. Colony Collapse Disorder: A Descriptive Study. PLoS One 2009, 4, e6481. [Google Scholar] [CrossRef]
- Cornman, R.S.; Tarpy, D.R.; Chen, Y.; Jeffreys, L.; Lopez, D.; Pettis, J.S.; vanEngelsdorp, D.; Evans, J.D. Pathogen webs in collapsing honey bee colonies. PLoS One 2012, 7, e43562. [Google Scholar]
- Ravoet, J.; Maharramov, J.; Meeus, I.; De Smet, L.; Wenseleers, T.; Smagghe, G. ; de Graaf, D.C. Comprehensive Bee Pathogen Screening in Belgium Reveals Crithidia mellificae as a New Contributory Factor to Winter Mortality. PLoS One 2013, 8, e72443. [Google Scholar]
- Dainat, B.; Evans, J.D.; Chen, Y.P.; Gauthier, L.; Neumann, P. Predictive markers of honey bee colony collapse. PLoS One 2012, 7, e32151. [Google Scholar]
- Genersch, E.; von der Ohe, W.; Kaatz, H.; Schroeder, A.; Otten, C.; Berg, S.; Ritter, W.; Gisder, S.; Meixner, M.; Liebig, G.; et al. The German bee monitoring project: a long term study to understand periodically high winter losses of honey bee colonies. Apidologie 2010, 41, 332–352. [Google Scholar] [CrossRef]
- Shen, M.; Yang, X.; Cox-Foster, D.; Cui, L. The role of varroa mites in infections of Kashmir bee virus (KBV) and deformed wing virus (DWV) in honey bees. Virology 2005, 342, 141–149. [Google Scholar]
- Runckel, C.; Flenniken, M.L.; Engel, J.C.; Ruby, J.G.; Ganem, D.; Andino, R.; DeRisi, J.L. Temporal Analysis of the Honey Bee Microbiome Reveals Four Novel Viruses and Seasonal Prevalence of Known Viruses, Nosema, and Crithidia. PLoS One 2011, 6, e20656. [Google Scholar]
- Chen, Y.P.; Siede, R. Honey bee viruses. Adv. Virus Res. 2007, 70, 33–80. [Google Scholar] [CrossRef]
- de Miranda, J.R.; Genersch, E. Deformed wing virus. J. Invertebr. Pathol 2010, 103, S48–S61. [Google Scholar] [CrossRef]
- Maori, E.; Lavi, S.; Mozes-Koch, R.; Gantman, Y.; Peretz, Y.; Edelbaum, O.; Tanne, E.; Sela, I. Isolation and characterization of Israeli acute paralysis virus, a dicistrovirus affecting honeybees in Israel: Evidence for diversity due to intra- and inter-species recombination. J. Gen. Virol. 2007, 88, 3428–3438. [Google Scholar] [CrossRef]
- Ribiere, M.; Olivier, V.; Blanchard, P. Chronic bee paralysis: A disease and a virus like no other? J. Invertebr. Pathol. 2010, 103, S120–S131. [Google Scholar] [CrossRef]
- de Miranda, J.R.; Cordoni, G.; Budge, G. The Acute bee paralysis virus-Kashmir bee virus-Israeli acute paralysis virus complex. J. Invertebr. Pathol. 2010, 103, S30–S47. [Google Scholar] [CrossRef]
- Yang, X.; Cox-Foster, D.L. Impact of an ectoparasite on the immunity and pathology of an invertebrate: Evidence for host immunosuppression and viral amplification. Proc. Natl. Acad. Sci. USA 2005, 102, 7470–7475. [Google Scholar] [CrossRef]
- Locke, B.; Forsgren, E.; Fries, I.; de Miranda, J.R. Acaricide treatment affects viral dynamics in Varroa destructor-infested honey bee colonies via both host physiology and mite control. App. Environ. Microbiol. 2012, 78, 227–235. [Google Scholar] [CrossRef]
- Di Prisco, G.; Cavaliere, V.; Annoscia, D.; Varricchio, P.; Caprio, E; Nazzi, F.; Gargiulo, G.; Pennacchio, F. Neonicotinoid clothianidin adversely affects insect immunity and promotes replication of a viral pathogen in honey bees. Proc. Natl. Acad. Sci. USA 2013, 110, 18466–18471. [Google Scholar] [CrossRef]
- Nazzi, F.; Brown, S.P.; Annoscia, D.; Del Piccolo, F.; Di Prisco, G.; Varricchio, P.; Della Vedova, G.; Cattonaro, F.; Caprio, E.; Pennacchio, F. Synergistic Parasite-Pathogen Interactions Mediated by Host Immunity Can Drive the Collapse of Honeybee Colonies. PLoS Pathog. 2012, 8, e1002735. [Google Scholar] [CrossRef]
- Berthoud, H.; Imdorf, A.; Haueter, M.; Radloff, S.; Neumann, P. Virus infections and winter losses of honey bee colonies (Apis mellifera). J. Apic. Res. 2010, 49, 60–65. [Google Scholar] [CrossRef]
- Cox-Foster, D.L.; Conlan, S.; Holmes, E.C.; Palacios, G.; Evans, J.D.; Moran, N.A.; Quan, P.L.; Briese, T.; Hornig, M.; Geiser, D.M.; et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science 2007, 318, 283–287. [Google Scholar] [CrossRef]
- Boncristiani, H.F.; Evans, J.D.; Chen, Y.; Pettis, J.; Murphy, C.; Lopez, D.L.; Simone-Finstrom, M.; Strand, M.; Tarpy, D.R.; Rueppell, O. In Vitro Infection of Pupae with Israeli Acute Paralysis Virus Suggests Disturbance of Transcriptional Homeostasis in Honey Bees (Apis mellifera). PLoS One 2013, 8, e73429. [Google Scholar] [CrossRef]
- Palacios, G.; Hui, J.; Quan, P.L.; Kalkstein, A.; Honkavuori, K.S; Bussetti, A.V.; Conlan, S.; Evans, J.; Chen, Y.P.; vanEngelsdorp, D.; et al. Genetic analysis of Israel acute paralysis virus: Distinct clusters are circulating in the United States. J. Virol. 2008, 82, 6209–6217. [Google Scholar] [CrossRef]
- Blanchard, P.; Schurr, F.; Celle, O.; Cougoule, N.; Drajnudel, P.; Thiéry, R.; Faucon, J.P.; Ribière, M. First detection of Israeli acute paralysis virus (IAPV) in France, a dicistrovirus affecting honeybees (Apis mellifera). J. Invertebr. Pathol. 2008, 99, 348–350. [Google Scholar] [CrossRef]
- Formato, G.; Giacomelli, A.; Olivia, M.; Aubin, L.; Glick, E.; Paldi, N.; Cardeti, G.; Cersini, A.; Ciabatti, I.M.; Palazzetti, M. First detection of Israeli acute paralysis virus (IAPV) in Italy. J. Apicult. Res. 2011, 50, 176–177. [Google Scholar] [CrossRef]
- Granberg, F.; Vicente-Rubiano, M.; Rubio-Guerri, C.; Karlsson, O.E.; Kukielka, D.; Belák, S.; Sánchez-Vizcaíno, J.M. Metagenomic detection of viral pathogens in spanish honeybees: Co-infection by aphid lethal paralysis, Israel acute paralysis and lake sinai viruses. PLoS One 2013, 8, e57459. [Google Scholar] [CrossRef]
- zkirim, A.; Schiesser, A. Israeli acute paralysis virus (IAPV) in Turkish bees. J. Apicult. Res. 2013, 52, 56–57. [Google Scholar] [CrossRef]
- Pohorecka, K.; Zdańska, D.; Bober, A.; Skubida, M. First detection of Israeli acute paralysis virus (IAPV) in Poland and phylogenetic analysis of the isolates. J. Apicult. Sci. 2011, 55, 149–159. [Google Scholar]
- Reynaldi, F.J.; Sguazza, G.H.; Tizzano, M.A.; Fuentealba, N.; Galosi, C.M.; Pecoraro, M.R. First report of Israeli acute paralysis virus in asymptomatic hives of Argentina. Revista Argentina de microbiología 2011, 43, 84–86. [Google Scholar]
- Al-Abbadi, A.A.; Hassawi, D.S.; Abu-Mallouh, S.A.; Al-Mazra'awi, M.S. Novel detection of Israel acute paralysis virus and Kashmir bee virus from honeybees Apis mellifera L. (Hymenoptera: Apidae) of Jordan using reverse transcriptase PCR technique. Applied. Entomol. Zool. 2010, 45, 183–190. [Google Scholar] [CrossRef]
- Dainat, B.; vanEngelsdorp, D.; Neumann, P. Colony collapse disorder in Europe. Environ. Microbiol. Reports 2012, 4, 123–125. [Google Scholar] [CrossRef]
- Di Prisco, G.; Pennacchio, F.; Caprio, E.; Boncristiani, H.F., Jr.; Evans, J.D.; Chen, Y.; et al. Varroa destructor is an effective vector of Israeli acute paralysis virus in the honeybee, Apis mellifera. J. Gen. Virol. 2010, 92, 151–155. [Google Scholar]
- Johnson, R.M.; Evans, J.D.; Robinson, G.E.; Berenbaum, M.R. Changes in transcript abundance relating to colony collapse disorder in honey bees (Apis mellifera). Proc. Natl. Acad. Sci. USA 2009, 106, 14790–14795. [Google Scholar]
- Higes, M.; Martin-Hernandez, R.; Botias, C.; Bailon, E.G.; Gonzalez-Porto, A.; VBarrios, L.; Del Nozal, M.J.; Bernal, J.L.; Jiménez, J.J.; Palencia, P.G.; et al. How natural infection by Nosema ceranae causes honeybee colony collapse. Environ. Microbiol. 2008, 10, 2659–2669. [Google Scholar] [CrossRef]
- Soroker, V.; Hetzroni, A.; Yakobson, B.; David, D.; David, A.; Voet, H.; Slabetzki, Y.; Efrat, H.; Levski, S.; Kamer, Y.; et al. Evaluation of colony losses in Israel in relation to the incidence of pathogens and pests. Apidologie 2011, 42, 192–199. [Google Scholar] [CrossRef]
- Chejanovsky, N.; Ophir, R.; Sharabi-Schwager, M.; Slabezki, Y.; Grossman, S.; Cox-Foster, D. Characterization of viral siRNA populations in honey bee colony collapse disorder. Virology 2014, 454–455, 176–183. [Google Scholar]
- Vicente-Rubiano, M.; Kukielka, D.; de las Heras, A.I.; Sanchez-Vizcaino, J.M. Short communication. Presence, quantification and phylogeny of Israeli acute paralysis virus of honeybees in Andalusia (Spain). Spanish J. Agricult. Res. 2013, 11, 708–713. [Google Scholar] [CrossRef]
- Yue, C.; Genersch, E. RT-PCR analysis of Deformed wing virus in honeybees (Apis mellifera) and mites (Varroa destructor). J. Gen. Virol. 2005, 8, 3419–3424. [Google Scholar]
- Evans, J.D. Beepath: an ordered quantitative-PCR array for exploring honey bee immunity and disease. J. Invertebr. Pathol. 2006, 93, 135–139. [Google Scholar] [CrossRef]
- Zioni, N.; Soroker, V.; Chejanovsky, N. Replication of Varroa destructor virus 1 (VDV-1) and a Varroa destructor virus 1-deformed wing virus recombinant (VDV-1-DWV) in the head of the honey bee. Virology 2011, 417, 106–112. [Google Scholar]
- Sokal, R.R.; Rohlf, F.J. Biometry; W. H. Freeman and Company: New York, NY, USA, 1981; p. 859. [Google Scholar]
- Williams, G.R.; Tarpy, D.R.; vanEngelsdorp, D.; Chauzat, M-P.; Cox-Foster, D.L.; Delaplane, K.S.; Neumann, P.; Pettis, J.S.; Rogers, R.E.L.; Shutler, D. Colony Collapse Disorder in context. Bioessays 2010, 32, 845–846. [Google Scholar] [CrossRef]
- Highfield, A.C.; El Nagar, A.; Mackinder, L.C.M.; Noel, L.; Hall, M.J.; Martin, S.J.; Schroeder, D.C. Deformed Wing Virus Implicated in Overwintering Honeybee Colony Losses. Appl. Environ. Microbiol. 2009, 75, 7212–7220. [Google Scholar] [CrossRef]
- Neumann, P.; Carreck, N.L. Honey bee colony losses. J. Apicult. Res. 2010, 49, 1–6. [Google Scholar] [CrossRef]
- Dainat, B.; vanEngelsdorp, D.; Neumann, P. Colony collapse disorder in Europe. (Thematic Issue: Taxonomy and biodiversity). Environ. Microbiol. Reports 2012, 4, 123–125. [Google Scholar] [CrossRef]
- Higes, M.; Martín-Hernández, R.; Botías, C.; Bailón, E.G.; González-Porto, A.V.; Barrios, L.; Del Nozal, M.J.; Bernal, J.L.; Jiménez, J.J.; Palencia, P.G.; et al. How natural infection by Nosema ceranae causes honeybee colony collapse. Environ. Microbiol. 2008, 10, 2659–2669. [Google Scholar] [CrossRef]
- Plaskon, N.E.; Adelman, Z.N.; Myles, K.M. Accurate Strand-Specific Quantification of Viral RNA. PLoS One 2009, 4, e7468. [Google Scholar] [CrossRef]
- Singh, R.; Levitt, A.L.; Rajotte, E.G.; Holmes, E.C.; Ostiguy, N.; vanEngelsdorp, D.; Lipkin, W.I; Depamphilis, C.W.; Toth, A.L.; Cox-Foster, D.L. RNA viruses in hymenopteran pollinators: Evidence of inter-Taxa virus transmission via pollen and potential impact on non-Apis hymenopteran species. PLoS One 2010, 5, e14357. [Google Scholar] [CrossRef]
- Chejanovsky, N.; Entomology Department, Institute of Plant Protection, The Volcani Center, Bet Dagan, Israel. Unpublished observations. 2010.
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Hou, C.; Rivkin, H.; Slabezki, Y.; Chejanovsky, N. Dynamics of the Presence of Israeli Acute Paralysis Virus in Honey Bee Colonies with Colony Collapse Disorder. Viruses 2014, 6, 2012-2027. https://doi.org/10.3390/v6052012
Hou C, Rivkin H, Slabezki Y, Chejanovsky N. Dynamics of the Presence of Israeli Acute Paralysis Virus in Honey Bee Colonies with Colony Collapse Disorder. Viruses. 2014; 6(5):2012-2027. https://doi.org/10.3390/v6052012
Chicago/Turabian StyleHou, Chunsheng, Hadassah Rivkin, Yossi Slabezki, and Nor Chejanovsky. 2014. "Dynamics of the Presence of Israeli Acute Paralysis Virus in Honey Bee Colonies with Colony Collapse Disorder" Viruses 6, no. 5: 2012-2027. https://doi.org/10.3390/v6052012
APA StyleHou, C., Rivkin, H., Slabezki, Y., & Chejanovsky, N. (2014). Dynamics of the Presence of Israeli Acute Paralysis Virus in Honey Bee Colonies with Colony Collapse Disorder. Viruses, 6(5), 2012-2027. https://doi.org/10.3390/v6052012




