Dissemination of the Flavivirus Subgenomic Replicon Genome and Viral Proteins by Extracellular Vesicles
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
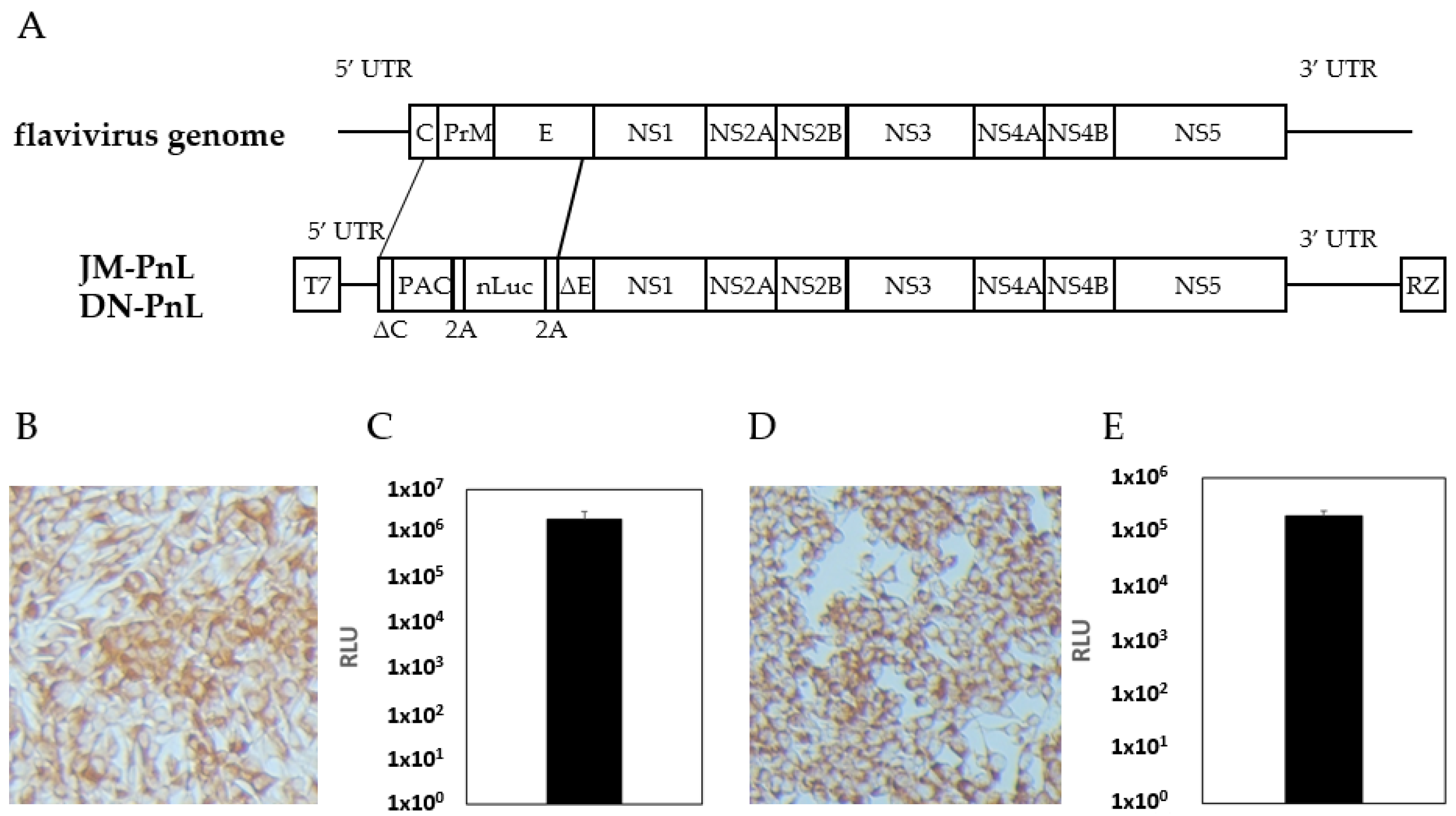
2.2. Establishment of Subgenomic Replicon Harboring Cell Lines
2.3. Immunostaining
2.4. EV Sample Preparation
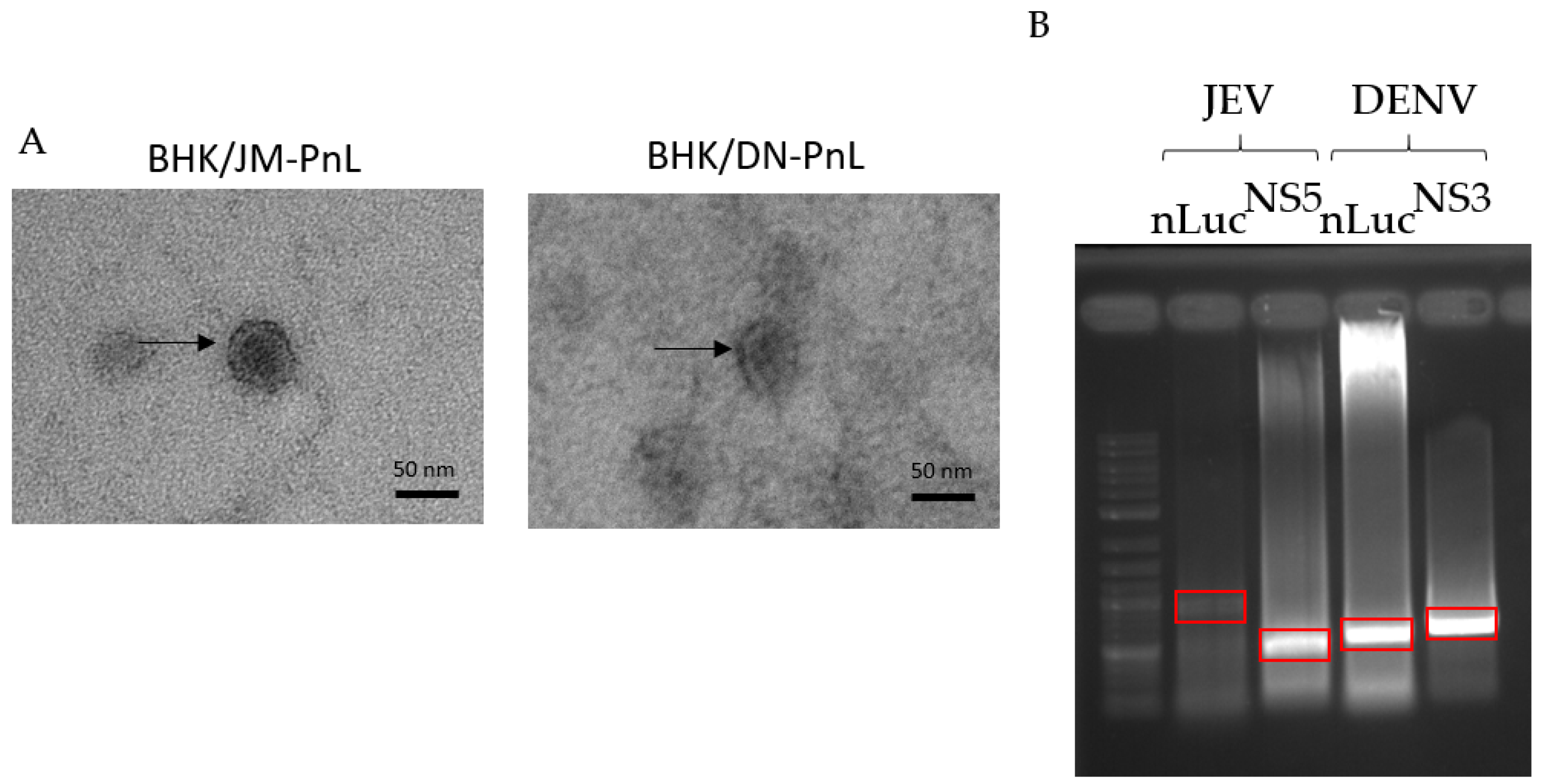
2.5. Electron Microscopy
2.6. EV Inoculation and Luciferase Assay
2.7. PCR
2.8. Real-Time PCR
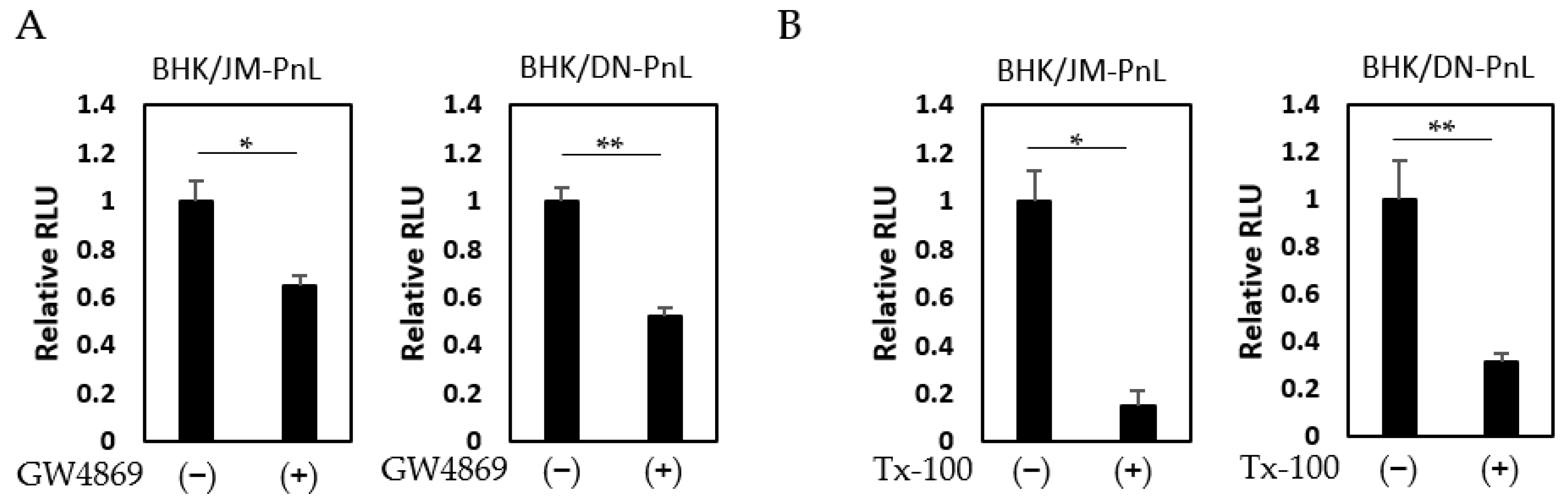
2.9. Treatment of the Cells with Antibodies and Inhibitors
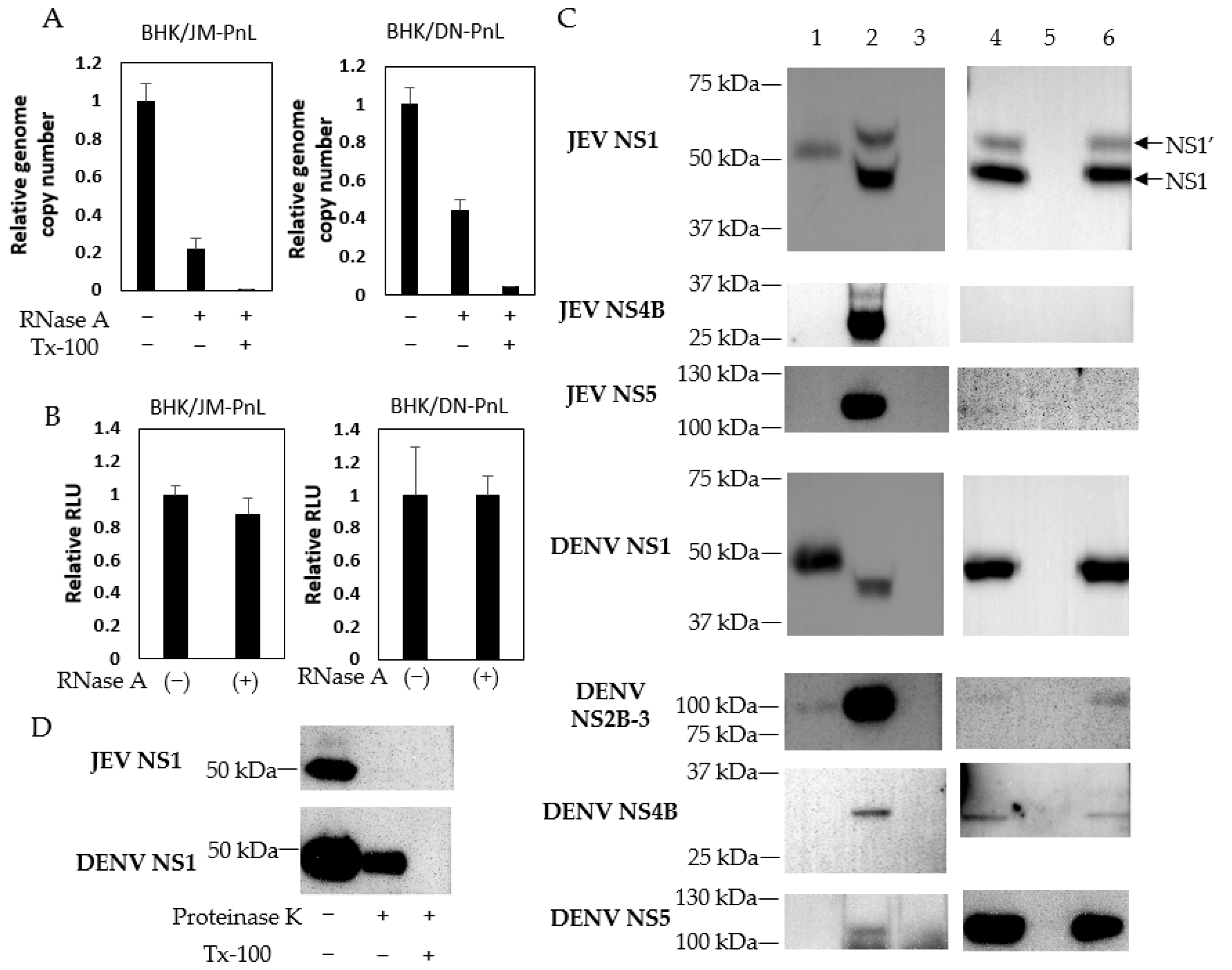
2.10. Western Blot
2.11. Proteinase K Protection Assay
2.12. Statistical Analysis
3. Results
3.1. Establishment of JEV and DENV Subgenomic Replicon Expressing Cells
3.2. Incorporation of the Replicon Genome and Viral Proteins in the EVs
3.3. Cell-to-Cell Transfer of the Replicon Genome Mediated by EVs
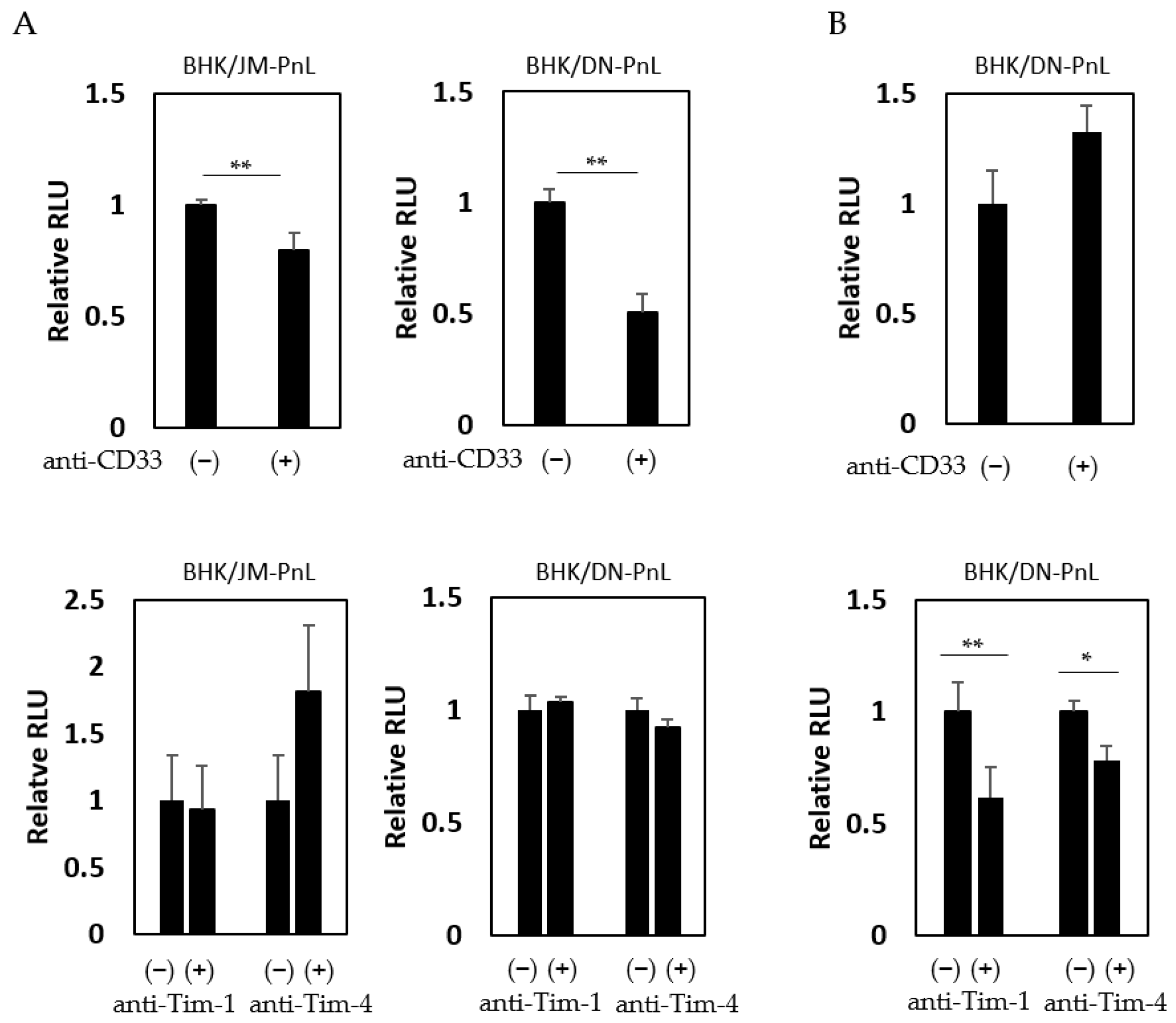
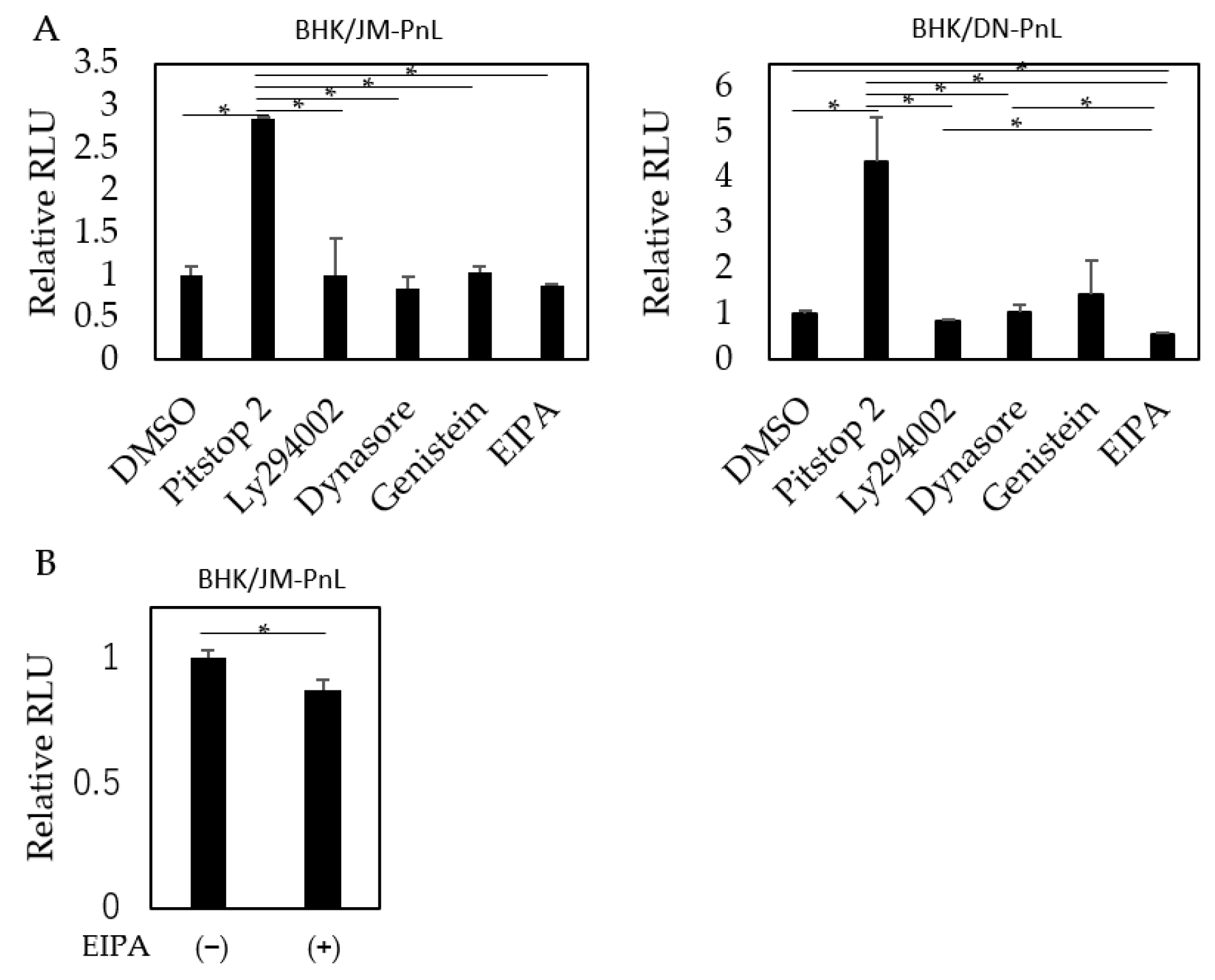
3.4. Cellular Factors Involved in the EV-Mediated Transfer of the Replicon Genome
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pierson, T.C.; Diamond, M.S. Flaviviruses. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; pp. 747–794. [Google Scholar]
- Pierson, T.C.; Diamond, M.S. The continued threat of emerging flaviviruses. Nat. Microbiol. 2020, 5, 796–812. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, T.; Yamanaka, A.; Konishi, E. A review of successful flavivirus vaccines and the problems with those flaviviruses for which vaccines are not yet available. Vaccine 2014, 32, 1326–1337. [Google Scholar] [CrossRef] [PubMed]
- Lindenbach, B.D.; Murray, C.; Thiel, H.J.; Rice, C.M. Flaviviridae. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; pp. 712–746. [Google Scholar]
- Latanova, A.; Starodubova, E.; Karpov, V. Flaviviridae Nonstructural Proteins: The Role in Molecular Mechanisms of Triggering Inflammation. Viruses 2022, 14, 1808. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; El Andaloussi, S.; Wood, M.J. Exosomes and microvesicles: Extracellular vesicles for genetic information transfer and gene therapy. Hum. Mol. Genet. 2012, 21, R125–R134. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Thakur, B.K.; Weiss, J.M.; Kim, H.S.; Peinado, H.; Lyden, D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell 2016, 30, 836–848. [Google Scholar] [CrossRef] [PubMed]
- Wortzel, I.; Dror, S.; Kenific, C.M.; Lyden, D. Exosome-Mediated Metastasis: Communication from a Distance. Dev. Cell 2019, 49, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Saad, M.H.; Badierah, R.; Redwan, E.M.; El-Fakharany, E.M. A Comprehensive Insight into the Role of Exosomes in Viral Infection: Dual Faces Bearing Different Functions. Pharmaceutics 2021, 13, 1405. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Wu, N.; Gan, X.; Yan, W.; Morrell, J.C.; Gould, S.J. Higher-order oligomerization targets plasma membrane proteins and HIV gag to exosomes. PLoS Biol. 2007, 5, e158. [Google Scholar] [CrossRef]
- Jaworski, E.; Narayanan, A.; Van Duyne, R.; Shabbeer-Meyering, S.; Iordanskiy, S.; Saifuddin, M.; Das, R.; Afonso, P.V.; Sampey, G.C.; Chung, M.; et al. Human T-lymphotropic virus type 1-infected cells secrete exosomes that contain Tax protein. J. Biol. Chem. 2014, 289, 22284–22305. [Google Scholar] [CrossRef]
- Ramakrishnaiah, V.; Thumann, C.; Fofana, I.; Habersetzer, F.; Pan, Q.; de Ruiter, P.E.; Willemsen, R.; Demmers, J.A.; Stalin Raj, V.; Jenster, G.; et al. Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc. Natl. Acad. Sci. USA 2013, 110, 13109–13113. [Google Scholar] [CrossRef] [PubMed]
- Chivero, E.T.; Bhattarai, N.; Rydze, R.T.; Winters, M.A.; Holodniy, M.; Stapleton, J.T. Human pegivirus RNA is found in multiple blood mononuclear cells in vivo and serum-derived viral RNA-containing particles are infectious in vitro. J. Gen. Virol. 2014, 95, 1307–1319. [Google Scholar] [CrossRef] [PubMed]
- Kadiu, I.; Narayanasamy, P.; Dash, P.K.; Zhang, W.; Gendelman, H.E. Biochemical and biologic characterization of exosomes and microvesicles as facilitators of HIV-1 infection in macrophages. J. Immunol. 2012, 189, 744–754. [Google Scholar] [CrossRef]
- Feng, Z.; Hensley, L.; McKnight, K.L.; Hu, F.; Madden, V.; Ping, L.; Jeong, S.H.; Walker, C.; Lanford, R.E.; Lemon, S.M. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 2013, 496, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.H.; Du, W.; Hagemeijer, M.C.; Takvorian, P.M.; Pau, C.; Cali, A.; Brantner, C.A.; Stempinski, E.S.; Connelly, P.S.; Ma, H.C.; et al. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell 2015, 160, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Moulin, C.; Crupi, M.J.F.; Ilkow, C.S.; Bell, J.C.; Boulton, S. Extracellular Vesicles and Viruses: Two Intertwined Entities. Int. J. Mol. Sci. 2023, 24, 1306. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Woodson, M.; Neupane, B.; Bai, F.; Sherman, M.B.; Choi, K.H.; Neelakanta, G.; Sultana, H. Exosomes serve as novel modes of tick-borne flavivirus transmission from arthropod to human cells and facilitates dissemination of viral RNA and proteins to the vertebrate neuronal cells. PLoS Pathog. 2018, 14, e1006764. [Google Scholar] [CrossRef] [PubMed]
- Vora, A.; Zhou, W.; Londono-Renteria, B.; Woodson, M.; Sherman, M.B.; Colpitts, T.M.; Neelakanta, G.; Sultana, H. Arthropod EVs mediate dengue virus transmission through interaction with a tetraspanin domain containing glycoprotein Tsp29Fb. Proc. Natl. Acad. Sci. USA 2018, 115, E6604–E6613. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Woodson, M.; Sherman, M.B.; Neelakanta, G.; Sultana, H. Exosomes mediate Zika virus transmission through SMPD3 neutral Sphingomyelinase in cortical neurons. Emerg. Microbes Infect. 2019, 8, 307–326. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, T.; Abe, M.; Masuda, M. Construction of an infectious molecular clone of Japanese encephalitis virus genotype V and its derivative subgenomic replicon capable of expressing a foreign gene. Virus Res. 2015, 195, 153–161. [Google Scholar] [CrossRef]
- Honjo, S.; Masuda, M.; Ishikawa, T. Effects of the Japanese Encephalitis Virus Genotype V-Derived Sub-Viral Particles on the Immunogenicity of the Vaccine Characterized by a Novel Virus-Like Particle-Based Assay. Vaccines 2019, 7, 81. [Google Scholar] [CrossRef] [PubMed]
- Kitai, Y.; Shoda, M.; Kondo, T.; Konishi, E. Epitope-blocking enzyme-linked immunosorbent assay to differentiate west nile virus from Japanese encephalitis virus infections in equine sera. Clin. Vaccine Immunol. 2007, 14, 1024–1031. [Google Scholar] [CrossRef] [PubMed]
- Malenica, M.; Vukomanovic, M.; Kurtjak, M.; Masciotti, V.; Dal Zilio, S.; Greco, S.; Lazzarino, M.; Krusic, V.; Percic, M.; Jelovica Badovinac, I.; et al. Perspectives of Microscopy Methods for Morphology Characterisation of Extracellular Vesicles from Human Biofluids. Biomedicines 2021, 9, 603. [Google Scholar] [CrossRef] [PubMed]
- Gonda, A.; Kabagwira, J.; Senthil, G.N.; Wall, N.R. Internalization of Exosomes through Receptor-Mediated Endocytosis. Mol. Cancer Res. 2019, 17, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Zitvogel, L.; Regnault, A.; Lozier, A.; Wolfers, J.; Flament, C.; Tenza, D.; Ricciardi-Castagnoli, P.; Raposo, G.; Amigorena, S. Eradication of established murine tumors using a novel cell-free vaccine: Dendritic cell-derived exosomes. Nat. Med. 1998, 4, 594–600. [Google Scholar] [CrossRef] [PubMed]
- Kitai, Y.; Kawasaki, T.; Sueyoshi, T.; Kobiyama, K.; Ishii, K.J.; Zou, J.; Akira, S.; Matsuda, T.; Kawai, T. DNA-Containing Exosomes Derived from Cancer Cells Treated with Topotecan Activate a STING-Dependent Pathway and Reinforce Antitumor Immunity. J. Immunol. 2017, 198, 1649–1659. [Google Scholar] [CrossRef] [PubMed]
- De Wever, O.; Demetter, P.; Mareel, M.; Bracke, M. Stromal myofibroblasts are drivers of invasive cancer growth. Int. J. Cancer 2008, 123, 2229–2238. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Deng, T.; Liu, R.; Bai, M.; Zhou, L.; Wang, X.; Li, S.; Wang, X.; Yang, H.; Li, J.; et al. Exosome-delivered EGFR regulates liver microenvironment to promote gastric cancer liver metastasis. Nat. Commun. 2017, 8, 15016. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, S.; Yao, J.; Lowery, F.J.; Zhang, Q.; Huang, W.C.; Li, P.; Li, M.; Wang, X.; Zhang, C.; et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature 2015, 527, 100–104. [Google Scholar] [CrossRef]
- Cooks, T.; Pateras, I.S.; Jenkins, L.M.; Patel, K.M.; Robles, A.I.; Morris, J.; Forshew, T.; Appella, E.; Gorgoulis, V.G.; Harris, C.C. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat. Commun. 2018, 9, 771. [Google Scholar] [CrossRef]
- Temme, S.; Eis-Hubinger, A.M.; McLellan, A.D.; Koch, N. The herpes simplex virus-1 encoded glycoprotein B diverts HLA-DR into the exosome pathway. J. Immunol. 2010, 184, 236–243. [Google Scholar] [CrossRef]
- Vallhov, H.; Gutzeit, C.; Johansson, S.M.; Nagy, N.; Paul, M.; Li, Q.; Friend, S.; George, T.C.; Klein, E.; Scheynius, A.; et al. Exosomes containing glycoprotein 350 released by EBV-transformed B cells selectively target B cells through CD21 and block EBV infection in vitro. J. Immunol. 2011, 186, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Campbell, T.D.; Khan, M.; Huang, M.B.; Bond, V.C.; Powell, M.D. HIV-1 Nef protein is secreted into vesicles that can fuse with target cells and virions. Ethn. Dis. 2008, 18, S2-14-9. [Google Scholar] [PubMed]
- Pinto, D.O.; DeMarino, C.; Pleet, M.L.; Cowen, M.; Branscome, H.; Al Sharif, S.; Jones, J.; Dutartre, H.; Lepene, B.; Liotta, L.A.; et al. HTLV-1 Extracellular Vesicles Promote Cell-to-Cell Contact. Front. Microbiol. 2019, 10, 2147. [Google Scholar] [CrossRef] [PubMed]
- Kalamvoki, M.; Du, T.; Roizman, B. Cells infected with herpes simplex virus 1 export to uninfected cells exosomes containing STING, viral mRNAs, and microRNAs. Proc. Natl. Acad. Sci. USA 2014, 111, E4991–E4996. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Philip, P.S.; Tariq, S.; Khan, G. Epstein-Barr virus-encoded small RNAs (EBERs) are present in fractions related to exosomes released by EBV-transformed cells. PLoS ONE 2014, 9, e99163. [Google Scholar] [CrossRef]
- Bukong, T.N.; Momen-Heravi, F.; Kodys, K.; Bala, S.; Szabo, G. Exosomes from hepatitis C infected patients transmit HCV infection and contain replication competent viral RNA in complex with Ago2-miR122-HSP90. PLoS Pathog. 2014, 10, e1004424. [Google Scholar] [CrossRef] [PubMed]
- Honegger, A.; Schilling, D.; Bastian, S.; Sponagel, J.; Kuryshev, V.; Sultmann, H.; Scheffner, M.; Hoppe-Seyler, K.; Hoppe-Seyler, F. Dependence of intracellular and exosomal microRNAs on viral E6/E7 oncogene expression in HPV-positive tumor cells. PLoS Pathog. 2015, 11, e1004712. [Google Scholar] [CrossRef]
- Sampey, G.C.; Saifuddin, M.; Schwab, A.; Barclay, R.; Punya, S.; Chung, M.C.; Hakami, R.M.; Zadeh, M.A.; Lepene, B.; Klase, Z.A.; et al. Exosomes from HIV-1-infected Cells Stimulate Production of Pro-inflammatory Cytokines through Trans-activating Response (TAR) RNA. J. Biol. Chem. 2016, 291, 1251–1266. [Google Scholar] [CrossRef]
- Hoshina, S.; Sekizuka, T.; Kataoka, M.; Hasegawa, H.; Hamada, H.; Kuroda, M.; Katano, H. Profile of Exosomal and Intracellular microRNA in Gamma-Herpesvirus-Infected Lymphoma Cell Lines. PLoS ONE 2016, 11, e0162574. [Google Scholar] [CrossRef]
- Fabbiano, F.; Corsi, J.; Gurrieri, E.; Trevisan, C.; Notarangelo, M.; D’Agostino, V.G. RNA packaging into extracellular vesicles: An orchestra of RNA-binding proteins? J. Extracell. Vesicles 2020, 10, e12043. [Google Scholar] [CrossRef]
- Katoh, H.; Mori, Y.; Kambara, H.; Abe, T.; Fukuhara, T.; Morita, E.; Moriishi, K.; Kamitani, W.; Matsuura, Y. Heterogeneous nuclear ribonucleoprotein A2 participates in the replication of Japanese encephalitis virus through an interaction with viral proteins and RNA. J. Virol. 2011, 85, 10976–10988. [Google Scholar] [CrossRef] [PubMed]
- Phillips, S.L.; Soderblom, E.J.; Bradrick, S.S.; Garcia-Blanco, M.A. Identification of Proteins Bound to Dengue Viral RNA In Vivo Reveals New Host Proteins Important for Virus Replication. mBio 2016, 7, e01865-15. [Google Scholar] [CrossRef] [PubMed]
- Viktorovskaya, O.V.; Greco, T.M.; Cristea, I.M.; Thompson, S.R. Identification of RNA Binding Proteins Associated with Dengue Virus RNA in Infected Cells Reveals Temporally Distinct Host Factor Requirements. PLoS Negl. Trop. Dis. 2016, 10, e0004921. [Google Scholar] [CrossRef] [PubMed]
- Ageta, H.; Ageta-Ishihara, N.; Hitachi, K.; Karayel, O.; Onouchi, T.; Yamaguchi, H.; Kahyo, T.; Hatanaka, K.; Ikegami, K.; Yoshioka, Y.; et al. UBL3 modification influences protein sorting to small extracellular vesicles. Nat. Commun. 2018, 9, 3936. [Google Scholar] [CrossRef] [PubMed]
- Richard, P.; Feng, S.; Manley, J.L. A SUMO-dependent interaction between Senataxin and the exosome, disrupted in the neurodegenerative disease AOA2, targets the exosome to sites of transcription-induced DNA damage. Genes Dev. 2013, 27, 2227–2232. [Google Scholar] [CrossRef] [PubMed]
- Villarroya-Beltri, C.; Gutierrez-Vazquez, C.; Sanchez-Cabo, F.; Perez-Hernandez, D.; Vazquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sanchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [PubMed]
- Buschow, S.I.; Liefhebber, J.M.; Wubbolts, R.; Stoorvogel, W. Exosomes contain ubiquitinated proteins. Blood Cells Mol. Dis. 2005, 35, 398–403. [Google Scholar] [CrossRef]
- Shimoda, A.; Tahara, Y.; Sawada, S.I.; Sasaki, Y.; Akiyoshi, K. Glycan profiling analysis using evanescent-field fluorescence-assisted lectin array: Importance of sugar recognition for cellular uptake of exosomes from mesenchymal stem cells. Biochem. Biophys. Res. Commun. 2017, 491, 701–707. [Google Scholar] [CrossRef]
- Kobayashi, N.; Karisola, P.; Pena-Cruz, V.; Dorfman, D.M.; Jinushi, M.; Umetsu, S.E.; Butte, M.J.; Nagumo, H.; Chernova, I.; Zhu, B.; et al. TIM-1 and TIM-4 glycoproteins bind phosphatidylserine and mediate uptake of apoptotic cells. Immunity 2007, 27, 927–940. [Google Scholar] [CrossRef]
- Kondratowicz, A.S.; Lennemann, N.J.; Sinn, P.L.; Davey, R.A.; Hunt, C.L.; Moller-Tank, S.; Meyerholz, D.K.; Rennert, P.; Mullins, R.F.; Brindley, M.; et al. T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for Zaire Ebolavirus and Lake Victoria Marburgvirus. Proc. Natl. Acad. Sci. USA 2011, 108, 8426–8431. [Google Scholar] [CrossRef]
- McKelvey, K.J.; Powell, K.L.; Ashton, A.W.; Morris, J.M.; McCracken, S.A. Exosomes: Mechanisms of Uptake. J. Circ. Biomark. 2015, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal 2021, 19, 47. [Google Scholar] [CrossRef]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3, 24641. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.P.; Gleeson, P.A. Macropinocytosis: An endocytic pathway for internalising large gulps. Immunol. Cell Biol. 2011, 89, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Ruiz, J.M.; Osuna-Ramos, J.F.; De Jesus-Gonzalez, L.A.; Hurtado-Monzon, A.M.; Farfan-Morales, C.N.; Cervantes-Salazar, M.; Bolanos, J.; Cigarroa-Mayorga, O.E.; Martin-Martinez, E.S.; Medina, F.; et al. Isolation and characterization of exosomes released from mosquito cells infected with dengue virus. Virus Res. 2019, 266, 1–14. [Google Scholar] [CrossRef]
- Martinez-Rojas, P.P.; Quiroz-Garcia, E.; Monroy-Martinez, V.; Agredano-Moreno, L.T.; Jimenez-Garcia, L.F.; Ruiz-Ordaz, B.H. Participation of Extracellular Vesicles from Zika-Virus-Infected Mosquito Cells in the Modification of Naive Cells’ Behavior by Mediating Cell-to-Cell Transmission of Viral Elements. Cells 2020, 9, 123. [Google Scholar] [CrossRef]
- Mayor, S.; Pagano, R.E. Pathways of clathrin-independent endocytosis. Nat. Rev. Mol. Cell Biol. 2007, 8, 603–612. [Google Scholar] [CrossRef]
- Verdera, C.H.; Gitz-Francois, J.J.; Schiffelers, R.M.; Vader, P. Cellular uptake of extracellular vesicles is mediated by clathrin-independent endocytosis and macropinocytosis. J. Control. Release 2017, 266, 100–108. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishikawa, T.; Narita, K.; Matsuyama, K.; Masuda, M. Dissemination of the Flavivirus Subgenomic Replicon Genome and Viral Proteins by Extracellular Vesicles. Viruses 2024, 16, 524. https://doi.org/10.3390/v16040524
Ishikawa T, Narita K, Matsuyama K, Masuda M. Dissemination of the Flavivirus Subgenomic Replicon Genome and Viral Proteins by Extracellular Vesicles. Viruses. 2024; 16(4):524. https://doi.org/10.3390/v16040524
Chicago/Turabian StyleIshikawa, Tomohiro, Kentaro Narita, Kinichi Matsuyama, and Michiaki Masuda. 2024. "Dissemination of the Flavivirus Subgenomic Replicon Genome and Viral Proteins by Extracellular Vesicles" Viruses 16, no. 4: 524. https://doi.org/10.3390/v16040524



