New Insights into Hop Latent Viroid Detection, Infectivity, Host Range, and Transmission
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Infected Samples
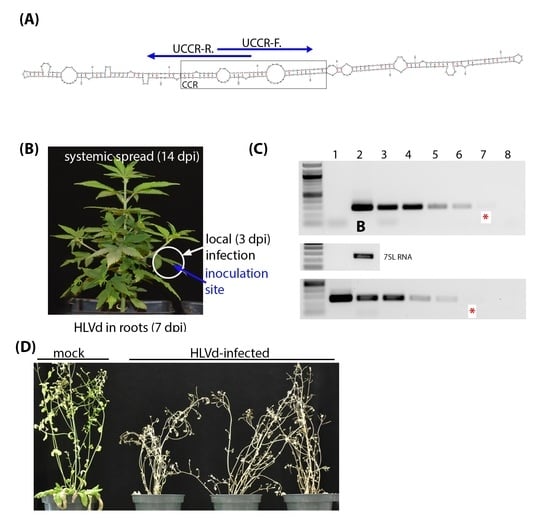
2.2. RNA Extraction, RT-PCR, and Primer Sensitivity
2.3. Viroid Inoculum and Plant Inoculation
2.4. Seed Transmission Assay
2.5. Sequence Analysis and Phylogeny
2.6. Imaging and Data Analysis
3. Results
3.1. Sensitive RT-PCR Diagnostic Detection of HLVd in Hemp Plants
3.2. Viroid Survey and Genetic Variation
3.3. Infectivity of HLVd Inoculum and Its Distribution in Mature Hemp Plants
3.4. HLVd Host Range and Disease Symptoms
3.5. Vertical Transmission of HLVd via Hemp Seeds
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ortola, B.; Daros, J.-A. Viroids: Non-Coding Circular RNAs Able to Autonomously Replicate and Infect Higher Plants. Biology 2023, 12, 172. [Google Scholar] [CrossRef] [PubMed]
- Tsagris, E.M.; de Alba, Á.E.M.; Gozmanova, M.; Kalantidis, K. Viroids. Cell. Microbiol. 2008, 10, 2168–2179. [Google Scholar] [CrossRef] [PubMed]
- Adkar-Purushothama, C.R.; Sano, T.; Perreault, J.P. Hop Latent Viroid: A Hidden Threat to the Cannabis Industry. Viruses 2023, 15, 681. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Wei, S.; Bian, R.; Luo, J.; Khan, H.A.; Tai, H.; Kondo, H.; Hadidi, A.; Andika, I.B.; Sun, L. Natural Cross-Kingdom Spread of Apple Scar Skin Viroid from Apple Trees to Fungi. Cells 2022, 11, 3686. [Google Scholar] [CrossRef] [PubMed]
- Leichtfried, T.; Reisenzein, H.; Steinkellner, S.; Gottsberger, R.A. Transmission Studies of the Newly Described Apple Chlorotic Fruit Spot Viroid Using a Combined RT-QPCR and Droplet Digital PCR Approach. Arch. Virol. 2020, 165, 2665–2671. [Google Scholar] [CrossRef] [PubMed]
- Hadidi, A.; Sun, L.; Randles, J.W. Modes of Viroid Transmission. Cells 2022, 11, 719. [Google Scholar] [CrossRef] [PubMed]
- Gandia, M.; Bernad, L.; Rubio, L.; Duran-Vila, N. Host Effect on the Molecular and Biological Properties of a Citrus Exocortis Viroid Isolate from Vicia Faba. Phytopathology 2007, 97, 1004–1010. [Google Scholar] [CrossRef] [PubMed]
- Diener, T.O. Circular RNAs: Relics of Precellular Evolution? Proc. Natl. Acad. Sci. USA 1989, 86, 9370–9374. [Google Scholar] [CrossRef]
- Diener, T.O. Subviral Pathogens of Plants: Viroids and Viroidlike Satellite RNAs. FASEB J. 1991, 5, 2808–2813. [Google Scholar] [CrossRef]
- Diener, T.O. Potato Spindle Tuber “Virus”. IV. A Replicating, Low Molecular Weight RNA. Virology 1971, 45, 411–428. [Google Scholar] [CrossRef]
- Flores, R.; Delgado, S.; Gas, M.-E.; Carbonell, A.; Molina, D.; Gago, S.; De la Peña, M. Viroids: The Minimal Non-Coding RNAs with Autonomous Replication. FEBS Lett. 2004, 567, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Puchta, H.; Ramm, K.; Sänger, H.L. The Molecular Structure of Hop Latent Viroid (HLV), a New Viroid Occurring Worldwide in Hops. Nucleic Acids Res. 1988, 16, 4197–4216. [Google Scholar] [CrossRef] [PubMed]
- Flores, R.; Serra, P.; Minoia, S.; Di Serio, F.; Navarro, B. Viroids: From Genotype to Phenotype Just Relying on RNA Sequence and Structural Motifs. Front. Microbiol. 2012, 3, 217. [Google Scholar] [CrossRef] [PubMed]
- Di Serio, F.; Owens, R.A.; Li, S.F.; Matoušek, J.; Pallás, V.; Randles, J.W.; Sano, T.; Verhoeven, J.T.J.; Vidalakis, G.; Flores, R. ICTV Virus Taxonomy Profile: Pospiviroidae. J. Gen. Virol. 2021, 102, 001543. [Google Scholar] [CrossRef] [PubMed]
- Schindler, I.-M.; Mühlbach, H.-P. Involvement of Nuclear DNA-Dependent RNA Polymerases in Potato Spindle Tuber Viroid Replication: A Reevaluation. Plant Sci. 1992, 84, 221–229. [Google Scholar] [CrossRef]
- Owens, R.A.; Diener, T.O. RNA Intermediates in Potato Spindle Tuber Viroid Replication. Proc. Natl. Acad. Sci. USA 1982, 79, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Branch, A.D.; Robertson, H.D.; Dickson, E. Longer-than-Unit-Length Viroid Minus Strands Are Present in RNA from Infected Plants. Proc. Natl. Acad. Sci. USA 1981, 78, 6381–6385. [Google Scholar] [CrossRef]
- Branch, A.D.; Robertson, H.D. A Replication Cycle for Viroids and Other Small Infectious RNA’s. Science 1984, 223, 450–455. [Google Scholar] [CrossRef]
- Bruening, G.; Gould, A.R.; Murphy, P.J.; Symons, R.H. Oligomers of Avocado Sunblotch Viroid Are Found in Infected Avocado Leaves. FEBS Lett. 1982, 148, 71–78. [Google Scholar] [CrossRef]
- Muhlbach, H.-P.; Sanger, H.L. Viroid Replication Is Inhibited by α-Amanitin. Nature 1979, 278, 185–188. [Google Scholar] [CrossRef]
- Nohales, M.-Á.; Flores, R.; Daròs, J.-A. Viroid RNA Redirects Host DNA Ligase 1 to Act as an RNA Ligase. Proc. Natl. Acad. Sci. USA 2012, 109, 13805–13810. [Google Scholar] [CrossRef] [PubMed]
- Maria-Eugenia, G.; Diego, M.-S.; Carmen, H.; Ricardo, F.; José-Antonio, D. Monomeric Linear RNA of Citrus Exocortis Viroid Resulting from Processing In Vivo Has 5′-Phosphomonoester and 3′-Hydroxyl Termini: Implications for the RNase and RNA Ligase Involved in Replication. J. Virol. 2008, 82, 10321–10325. [Google Scholar] [CrossRef]
- Gas, M.-E.; Hernández, C.; Flores, R.; Daròs, J.-A. Processing of Nuclear Viroids In Vivo: An Interplay between RNA Conformations. PLoS Pathog. 2007, 3, e182. [Google Scholar] [CrossRef] [PubMed]
- Góra-Sochacka, A. Viroids: Unusual Small Pathogenic RNAs. Acta Biochim. Pol. 2004, 51, 587–607. [Google Scholar] [CrossRef]
- Cottilli, P.; Belda-Palazón, B.; Adkar-Purushothama, C.R.; Perreault, J.-P.; Schleiff, E.; Rodrigo, I.; Ferrando, A.; Lisón, P. Citrus Exocortis Viroid Causes Ribosomal Stress in Tomato Plants. Nucleic Acids Res. 2019, 47, 8649–8661. [Google Scholar] [CrossRef]
- Nath, V.S.; Shrestha, A.; Awasthi, P.; Mishra, A.K.; Kocábek, T.; Matoušek, J.; Sečnik, A.; Jakše, J.; Radišek, S.; Hallan, V. Mapping the Gene Expression Spectrum of Mediator Subunits in Response to Viroid Infection in Plants. Int. J. Mol. Sci. 2020, 21, 2498. [Google Scholar] [CrossRef]
- An, C.; Mou, Z. The Function of the Mediator Complex in Plant Immunity. Plant Signal. Behav. 2013, 8, e23182. [Google Scholar] [CrossRef]
- Wassenegger, M.; Heimes, S.; Riedel, L.; Sänger, H.L. RNA-Directed de Novo Methylation of Genomic Sequences in Plants. Cell 1994, 76, 567–576. [Google Scholar] [CrossRef]
- Gómez, G.; Martínez, G.; Pallás, V. Interplay between Viroid-Induced Pathogenesis and RNA Silencing Pathways. Trends Plant Sci. 2009, 14, 264–269. [Google Scholar] [CrossRef]
- Di Serio, F.; Flores, R.; Verhoeven, J.T.J.; Li, S.F.; Pallás, V.; Randles, J.W.; Sano, T.; Vidalakis, G.; Owens, R.A. Current Status of Viroid Taxonomy. Arch. Virol. 2014, 159, 3467–3478. [Google Scholar] [CrossRef]
- Pallas, V.; Navarro, A.; Flores, R. Isolation of a Viroid-like RNA from Hop Different from Hop Stunt Viroid. J. Gen. Virol. 1987, 68, 3201–3205. [Google Scholar] [CrossRef]
- Stajner, N.; Radisek, S.; Mishra, A.K.; Nath, V.S.; Matoušek, J.; Jakše, J. Evaluation of Disease Severity and Global Transcriptome Response Induced by Citrus Bark Cracking Viroid, Hop Latent Viroid, and Their Co-Infection in Hop (Humulus Lupulus L.). Int. J. Mol. Sci. 2019, 20, 3154. [Google Scholar] [CrossRef] [PubMed]
- De Jonghe, K.; Van Bogaert, N.; Vandierendonck, S.; Smagghe, G.; Maes, M. First Report of Hop Latent Viroid in Belgian Hops. Plant Dis. 2016, 100, 1956. [Google Scholar] [CrossRef]
- Guler, P.G.; Önelge, N. First Report Of Hop Latent Viroid In Turkey. J. Plant Pathol. 2017, 99, 815. [Google Scholar]
- Solarska, E.; Grudzinska, M. The Field Distribution Of Hop Latent Viroid Within Polish Hop Cultivars. In Acta Horticulturae; International Society for Horticultural Science (ISHS): Leuven, Belgium, 2001; pp. 331–336. [Google Scholar]
- Liu, S.; Li, S.; Zhu, J.; Xiang, B.; Cao, L. First Report of Hop Latent Viroid (HLVd) in China. Plant Pathol. 2008, 57, 400. [Google Scholar] [CrossRef]
- Eiras, M.; de Oliveira, A.M.; de Fátima Ramos, A.; Harakava, R.; Daròs, J.-A. First Report of Citrus Bark Cracking Viroid and Hop Latent Viroid Infecting Hop in Commercial Yards in Brazil. J. Plant Pathol. 2023, 105, 603. [Google Scholar] [CrossRef]
- Warren, J.G.; Mercado, J.; Grace, D. Occurrence of Hop Latent Viroid Causing Disease in Cannabis sativa in California. Plant Dis. 2019, 103, 2699. [Google Scholar] [CrossRef]
- Barbara, D.J.; Morton, A.; Adams, A.N.; Green, C.P. Some Effects of Hop Latent Viroid on Two Cultivars of Hop (Humulus Lupulus) in the UK. Ann. Appl. Biol. 1990, 117, 359–366. [Google Scholar] [CrossRef]
- Bektas, A.; Hardwick, K.M.; Waterman, K.; Kristof, J. Occurrence of Hop Latent Viroid in Cannabis sativa with Symptoms of Cannabis Stunting Disease in California. Plant Dis. 2019, 103, 2699. [Google Scholar] [CrossRef]
- Jarugula, S.; Wagstaff, C.; Mitra, A.; Crowder, D.; Gang, D.; Rayapati, N. First Reports of Beet Curly Top Virus, Citrus Yellow Vein-Associated Virus, and Hop Latent Viroid in Industrial Hemp (Cannabis sativa) in Washington State. Plant Dis. 2023, 107, 2897. [Google Scholar] [CrossRef]
- Pethybridge, S.J.; Hay, F.S.; Barbara, D.J.; Eastwell, K.C.; Wilson, C.R. Viruses and Viroids Infecting Hop: Significance, Epidemiology, and Management. Plant Dis. 2008, 92, 324–338. [Google Scholar] [CrossRef] [PubMed]
- Matoušek, J.; Patzak, J. A Low Transmissibility of Hop Latent Viroid through a Generative Phase of Humulus Lupulus L. Biol. Plant. 2000, 43, 145–148. [Google Scholar] [CrossRef]
- Patzak, J.; Henychová, A.; Krofta, K.; Svoboda, P.; Malířová, I. The Influence of Hop Latent Viroid (Hlvd) Infection on Gene Expression and Secondary Metabolite Contents in Hop (Humulus Lupulus L.) Glandular Trichomes. Plants 2021, 10, 2297. [Google Scholar] [CrossRef] [PubMed]
- Patzak, J.; Matoušek, J.; Krofta, K.; Svoboda, P. Hop Latent Viroid (HLVd)-Caused Pathogenesis: Effects of HLVd Infection on Lupulin Composition of Meristem Culture-Derived Humulus Lupulus. Biol. Plant. 2001, 44, 579–585. [Google Scholar] [CrossRef]
- Matoušek, J. Hop Latent Viroid (HLVd) Microevolution: An Experimental Transmission of HLVd “Thermomutants” to Solanaceous Species. Biol. Plant. 2003, 46, 607–610. [Google Scholar] [CrossRef]
- Schroeder, M.; Weidemann, H.-L. Simplified Application of Return Gel Electrophoresis for the Routine Detection of Potato Spindle Tuber Viroid. EPPO Bull. 1989, 19, 661–665. [Google Scholar] [CrossRef]
- Diener, T.O.; Hadidi, A.; Owens, R.A. Methods for Studying Viroids [Plant Pathogens]. In Methods in Virology; Academic Press: New York, NY, USA, 1977. [Google Scholar]
- Owens, R.A.; Sano, T.; Duran-Vila, N. Plant Viroids: Isolation, Characterization/Detection, and Analysis. Methods Mol. Biol. 2012, 894, 253–271. [Google Scholar] [CrossRef]
- Gucek, T.; Trdan, S.; Jakse, J.; Javornik, B.; Matousek, J.; Radisek, S. Diagnostic Techniques for Viroids. Plant Pathol. 2017, 66, 339–358. [Google Scholar] [CrossRef]
- Hadidi, A.; Yang, X. Detection of Pome Fruit Viroids by Enzymatic CDNA Amplification. J. Virol. Methods 1990, 30, 261–269. [Google Scholar] [CrossRef]
- Boonham, N.; Pérez, L.G.; Mendez, M.S.; Peralta, E.L.; Blockley, A.; Walsh, K.; Barker, I.; Mumford, R.A. Development of a Real-Time RT-PCR Assay for the Detection of Potato Spindle Tuber Viroid. J. Virol. Methods 2004, 116, 139–146. [Google Scholar] [CrossRef]
- Elleuch, A.; Hamdi, I.; Bessaies, N.; Fakhfakh, H. Single-Strand Conformation Polymorphism for Molecular Variability Studies of Six Viroid Species. Biosci. Biotechnol. Biochem. 2013, 77, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.X.; Hong, N.; Zhang, J.K.; Wang, G.P. Improving the Sensitivity of Single-Strand Conformation Polymorphism (SSCP) to Study the Variability of PLMVd. J. Virol. Methods 2006, 135, 276–280. [Google Scholar] [CrossRef] [PubMed]
- Marti, A.F.I.; Parungao, M.; Hollin, J.; Selimotic, B.; Farrar, G.; Seyler, T.; Anand, A.; Ahmad, R. A Novel, Precise and High-Throughput Technology for Viroid Detection in Cannabis (MFDetectTM). bioRxiv 2023. [Google Scholar] [CrossRef]
- Boubourakas, I.N.; Kyriakopoulou, P.E. Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP) at the Service of Viroid Detection. Methods Mol. Biol. 2022, 2316, 195–210. [Google Scholar] [CrossRef] [PubMed]
- Guček, T.; Jakše, J.; Radisek, S. Optimization and Validation of Singleplex and Multiplex RT-QPCR for Detection of Citrus Bark Cracking Viroid (CBCVd), Hop Latent Viroid (HLVd), and Hop Stunt Viroid (HSVd) in Hops (Humulus Lupulus L.). Plant Dis. 2023, 107, 3592–3601. [Google Scholar] [CrossRef] [PubMed]
- Bernad, L.; Duran-Vila, N. A Novel RT-PCR Approach for Detection and Characterization of Citrus Viroids. Mol. Cell. Probes 2006, 20, 105–113. [Google Scholar] [CrossRef]
- Radisek, S.; Majer, A.; Jakse, J.; Javornik, B.; Matoušek, J. First Report of Hop stunt viroid Infecting Hop in Slovenia. Plant Dis. 2012, 96, 592. [Google Scholar] [CrossRef]
- Matousek, J.; Junker, V.; Vrba, L.; Schubert, J.; Patzak, J.; Steger, G. Molecular Characterization and Genome Organization of 7SL RNA Genes from Hop (Humulus Lupulus L.). Gene 1999, 239, 173–183. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Truszkowski, J.; Goldman, N. Maximum Likelihood Phylogenetic Inference Is Consistent on Multiple Sequence Alignments, with or without Gaps. Syst. Biol. 2016, 65, 328–333. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Urrutia, C.D.; Romay, G.; Shaw, B.D.; Verchot, J. Advancing the Rose Rosette Virus Minireplicon and Encapsidation System by Incorporating GFP, Mutations, and the CMV 2b Silencing Suppressor. Viruses 2022, 14, 836. [Google Scholar] [CrossRef] [PubMed]
- Tuiskunen, A.; Leparc-Goffart, I.; Boubis, L.; Monteil, V.; Klingström, J.; Tolou, H.J.; Lundkvist, A.; Plumet, S. Self-Priming of Reverse Transcriptase Impairs Strand-Specific Detection of Dengue Virus RNA. J. Gen. Virol. 2010, 91, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Warncke, S.R.; Knudsen, C.R. Detection Methods Targeting the Positive- and Negative-Sense RNA Transcripts from plus-Stranded RNA Viruses. APMIS 2022, 130, 284–292. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, S.; Sano, T. Threats to Japanese Agriculture from Newly Emerged Plant Viruses and Viroids. J. Gen. Plant Pathol. 2014, 80, 2–14. [Google Scholar] [CrossRef]
- Matsushita, Y.; Yanagisawa, H.; Sano, T. Vertical and Horizontal Transmission of Pospiviroids. Viruses 2018, 10, 706. [Google Scholar] [CrossRef]
- Eastwell, K.C.; Nelson, M.E. Occurrence of Viroids in Commercial Hop (Humulus Lupulus L.) Production Areas of Washington State. Plant Health Prog. 2007, 8, 1. [Google Scholar] [CrossRef]
- Rivedal, H.M.; Funke, C.N.; Frost, K.E. An Overview of Pathogens Associated with Biotic Stresses in Hemp Crops in Oregon, 2019 to 2020. Plant Dis. 2021, 106, 1334–1340. [Google Scholar] [CrossRef]
- Steinbachová, L.; Matoušek, J.; Steger, G.; Matoušková, H.; Radišek, S.; Honys, D. Transformation of Seed Non-Transmissible Hop Viroids in Nicotiana Benthamiana Causes Distortions in Male Gametophyte Development. Plants 2021, 10, 2398. [Google Scholar] [CrossRef]
- Ziegler, A.; Kawka, M.; Przybys, M.; Doroszewska, T.; Skomra, U.; Kastirr, U.; Matoušek, J.; Schubert, J. Detection and Molecular Analysis of Hop Latent Virus and Hop Latent Viroid in Hop Samples from Poland. J. Für Kult. 2014, 66, 248–254. [Google Scholar] [CrossRef]
- Punja, Z.K. Emerging Diseases of Cannabis sativa and Sustainable Management. Pest Manag. Sci. 2021, 77, 3857–3870. [Google Scholar] [CrossRef] [PubMed]
- Chiginsky, J.; Langemeier, K.; MacWilliams, J.; Albrecht, T.; Cranshaw, W.; Fulladolsa, A.C.; Kapuscinski, M.; Stenglein, M.; Nachappa, P. First Insights Into the Virus and Viroid Communities in Hemp (Cannabis sativa). Front. Agron. 2021, 3, 96. [Google Scholar] [CrossRef]
- Atallah, O.O.; Kang, S.-H.; El-Mohtar, C.A.; Shilts, T.; Bergua, M.; Folimonova, S.Y. A 5′-Proximal Region of the Citrus Tristeza Virus Genome Encoding Two Leader Proteases Is Involved in Virus Superinfection Exclusion. Virology 2016, 489, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Syller, J. Facilitative and Antagonistic Interactions between Plant Viruses in Mixed Infections. Mol. Plant Pathol. 2012, 13, 204–216. [Google Scholar] [CrossRef] [PubMed]
- Dietzgen, R.G.; Mann, K.S.; Johnson, K.N. Plant Virus-Insect Vector Interactions: Current and Potential Future Research Directions. Viruses 2016, 8, 303. [Google Scholar] [CrossRef]
- Syller, J.; Grupa, A. Antagonistic Within-Host Interactions between Plant Viruses: Molecular Basis and Impact on Viral and Host Fitness. Mol. Plant Pathol. 2016, 17, 769–782. [Google Scholar] [CrossRef]
- Matousek, J.; Trnena, L.; Svoboda, P.; Oriniakova, P.; Lichtenstein, C.P. The Gradual Reduction of Viroid Levels in Hop Mericlones Following Heat Therapy: A Possible Role for a Nuclease Degrading DsRNA. Biol. Chem. Hoppe Seyler 1995, 376, 715–722. [Google Scholar] [CrossRef]
- Matoušek, J.; Patzak, J.; Orctová, L.; Schubert, J.; Vrba, L.; Steger, G.; Riesner, D. The Variability of Hop Latent Viroid as Induced upon Heat Treatment. Virology 2001, 287, 349–358. [Google Scholar] [CrossRef]
- Więsyk, A.; Candresse, T.; Zagórski-Ostoja, W.; Góra-Sochacka, A. Viability and Genetic Stability of Potato Spindle Tuber Viroid Mutants with Indels in Specific Loops of the Rod-like Secondary Structure. Virus Res. 2017, 240, 94–100. [Google Scholar] [CrossRef]
- Daròs, J.A.; Flores, R. Arabidopsis Thaliana Has the Enzymatic Machinery for Replicating Representative Viroid Species of the Family Pospiviroidae. Proc. Natl. Acad. Sci. USA 2004, 101, 6792–6797. [Google Scholar] [CrossRef]
- Juškytė, A.D.; Mažeikienė, I.; Stanys, V. An Effective Method of Ribes Spp. Inoculation with Blackcurrant Reversion Virus under In Vitro Conditions. Plants 2022, 11, 1635. [Google Scholar] [CrossRef] [PubMed]
- Tran, P.-T.; Zhang, C.F.; Citovsky, V. Rapid Generation of Inoculum of a Plant RNA Virus Using Overlap PCR. Virology 2021, 553, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Harper, S.J.; Cowell, S.J.; Robertson, C.J.; Dawson, W.O. Differential Tropism in Roots and Shoots Infected by Citrus Tristeza Virus. Virology 2014, 460–461, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Mehle, N.; Gutiérrez-Aguirre, I.; Prezelj, N.; Delic, D.; Vidic, U.; Ravnikar, M. Survival and Transmission of Potato Virus Y, Pepino Mosaic Virus, and Potato Spindle Tuber Viroid in Water. Appl. Environ. Microbiol. 2014, 80, 1455–1462. [Google Scholar] [CrossRef] [PubMed]
- Osman, F.; Dang, T.; Bodaghi, S.; Vidalakis, G. One-Step Multiplex RT-QPCR Detects Three Citrus Viroids from Different Genera in a Wide Range of Hosts. J. Virol. Methods 2017, 245, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Owens, R.A.; Verhoeven, J.T.J. Chapter 14—Potato Spindle Tuber Viroid. In Viroids and Satellites; Hadidi, A., Flores, R., Randles, J.W., Palukaitis, P., Eds.; Academic Press: Boston, MA, USA, 2017; pp. 149–158. ISBN 978-0-12-801498-1. [Google Scholar]
- Matoušek, J.; Orctová, L.; Škopek, J.; Pešina, K.; Steger, G. Elimination of Hop Latent Viroid upon Developmental Activation of Pollen Nucleases. Biol. Chem. 2008, 389, 905–918. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, H.; Matsushita, Y. Effect of Potato Spindle Tuber Viroid Variants and Infection Stage on Seed Transmission through Pollen. Lett. Appl. Microbiol. 2022, 75, 836–843. [Google Scholar] [CrossRef]
- Kryczyński, S.; Paduch-Cichal, E.; Skrzeczkowski, L.J. Transmission of Three Viroids Through Seed and Pollen of Tomato Plants. J. Phytopathol. 1988, 121, 51–57. [Google Scholar] [CrossRef]
- Yanagisawa, H.; Matsushita, Y. Differences in Dynamics of Horizontal Transmission of Tomato Planta Macho Viroid and Potato Spindle Tuber Viroid after Pollination with Viroid-Infected Pollen. Virology 2018, 516, 258–264. [Google Scholar] [CrossRef]
- Yanagisawa, H.; Sano, T.; Hase, S.; Matsushita, Y. Influence of the Terminal Left Domain on Horizontal and Vertical Transmissions of Tomato Planta Macho Viroid and Potato Spindle Tuber Viroid through Pollen. Virology 2019, 526, 22–31. [Google Scholar] [CrossRef]
- Hammond, R.W. Chapter 48—Seed, Pollen, and Insect Transmission of Viroids. In Viroids and Satellites; Hadidi, A., Flores, R., Randles, J.W., Palukaitis, P., Eds.; Academic Press: Boston, MA, USA, 2017; pp. 521–530. ISBN 978-0-12-801498-1. [Google Scholar]
- Bhuvitarkorn, S.; Reanwarakorn, K. Pollen and Seed Transmission of Columnea Latent Viroid in Eggplants. Eur. J. Plant Pathol. 2019, 154, 1067–1075. [Google Scholar] [CrossRef]








| Target | Primer | Primer Sequence 5′-3′ | Amplicon Size (bp) | Reference |
|---|---|---|---|---|
| HLVd | UCCR-F | GGGATCCCCGGGGAAACCTACTCG | 256 | This study |
| UCCR-R | GGGATCCCTCTTCGAGCCCTTGCC | |||
| HSVd | F1 | GGGGCAACTCTTCTCAGAATCC | 302 | [58] |
| R1 | GGGGCTCCTTTCTCAGGTAAGTC | |||
| 7SL RNA | alpha | TGTAACCCAAGTGGGGG | 231 | [60] |
| anti-beta | GCACCGGCCCGTTATCC |
| Plant Family: | RT-PCR Detection in: | Symptoms | |||
|---|---|---|---|---|---|
| Inoculated Leaves | Roots | Upper Leaves | |||
| Negative control | - | 0 | 0 | 0 | Asymptomatic |
| Arabidopsis | (Brassicaceae) | 100 | 100 | 70 | Die-back, gradual decline |
| N. benthamiana | (Solanaceae) | 100 | 60 | 0 | Asymptomatic |
| Tomato | (Solanaceae) | 100 | 100 | 80 | Asymptomatic |
| Cucumber | (Cucurbitaceae) | 100 | 100 | 100 | Asymptomatic |
| Chrysanthemum | (Asteraceae) | 60 | 40 | 40 | Asymptomatic |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Atallah, O.O.; Yassin, S.M.; Verchot, J. New Insights into Hop Latent Viroid Detection, Infectivity, Host Range, and Transmission. Viruses 2024, 16, 30. https://doi.org/10.3390/v16010030
Atallah OO, Yassin SM, Verchot J. New Insights into Hop Latent Viroid Detection, Infectivity, Host Range, and Transmission. Viruses. 2024; 16(1):30. https://doi.org/10.3390/v16010030
Chicago/Turabian StyleAtallah, Osama O., Sherin M. Yassin, and Jeanmarie Verchot. 2024. "New Insights into Hop Latent Viroid Detection, Infectivity, Host Range, and Transmission" Viruses 16, no. 1: 30. https://doi.org/10.3390/v16010030