Identification and Characterization of a Novel Hypovirus from the Phytopathogenic Fungus Botryosphaeria dothidea
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Culture Conditions
2.2. RNA Extraction, High-Throughput Sequencing, and Data Analysis
2.3. Extraction and Purification of dsRNA
2.4. Validation of Virus-like Contigs via RT-PCR and Viral Sequencing Amplification
2.5. Sequences Alignment and Phylogenetic Analysis
2.6. Vertical and Horizontal Transmission of the Viruses
2.7. Biological Properties of Fungal Strains
3. Results
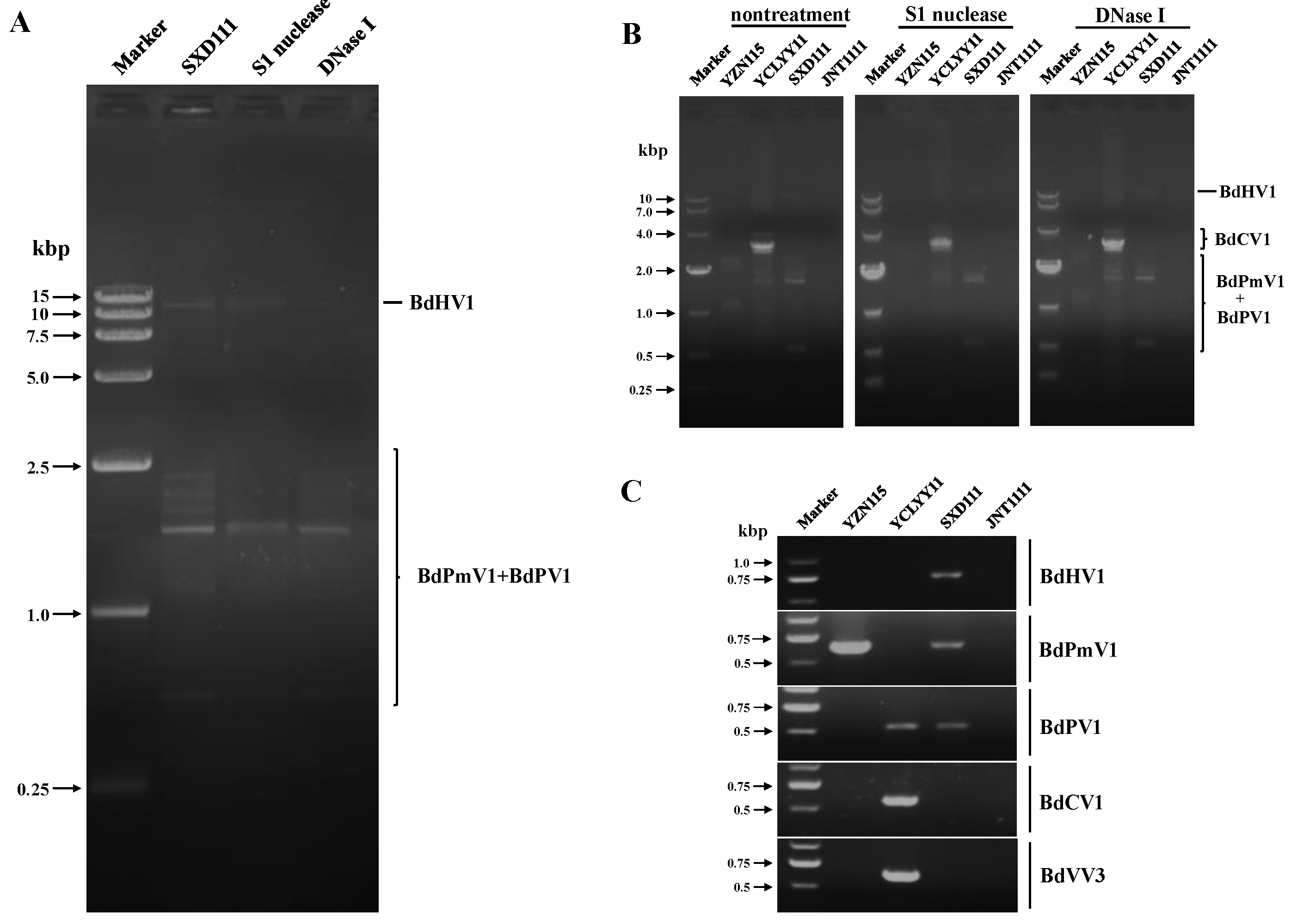
3.1. Diversity of Viruses in the Strains SXD111, YZN115, and YCLYY11
3.2. Detection and Validation of Mycoviruses in the Test Strains
3.3. Genetic Analysis of BdHV1/SXD111
3.4. Phylogenetic Analysis of BdHV1/SXD111
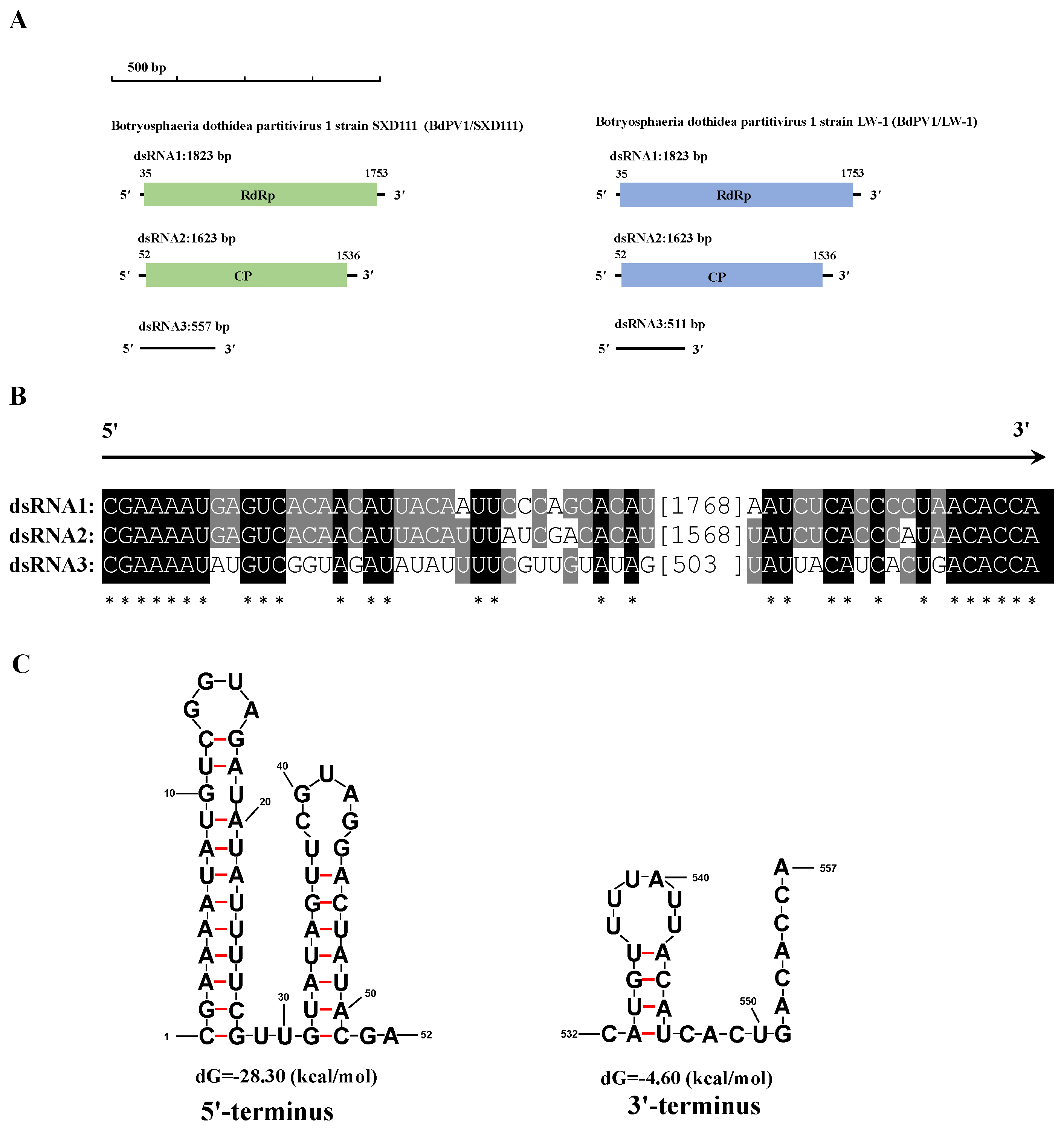
3.5. Genetic Analysis of BdPmV1/SXD111 and BdPV1/SXD111
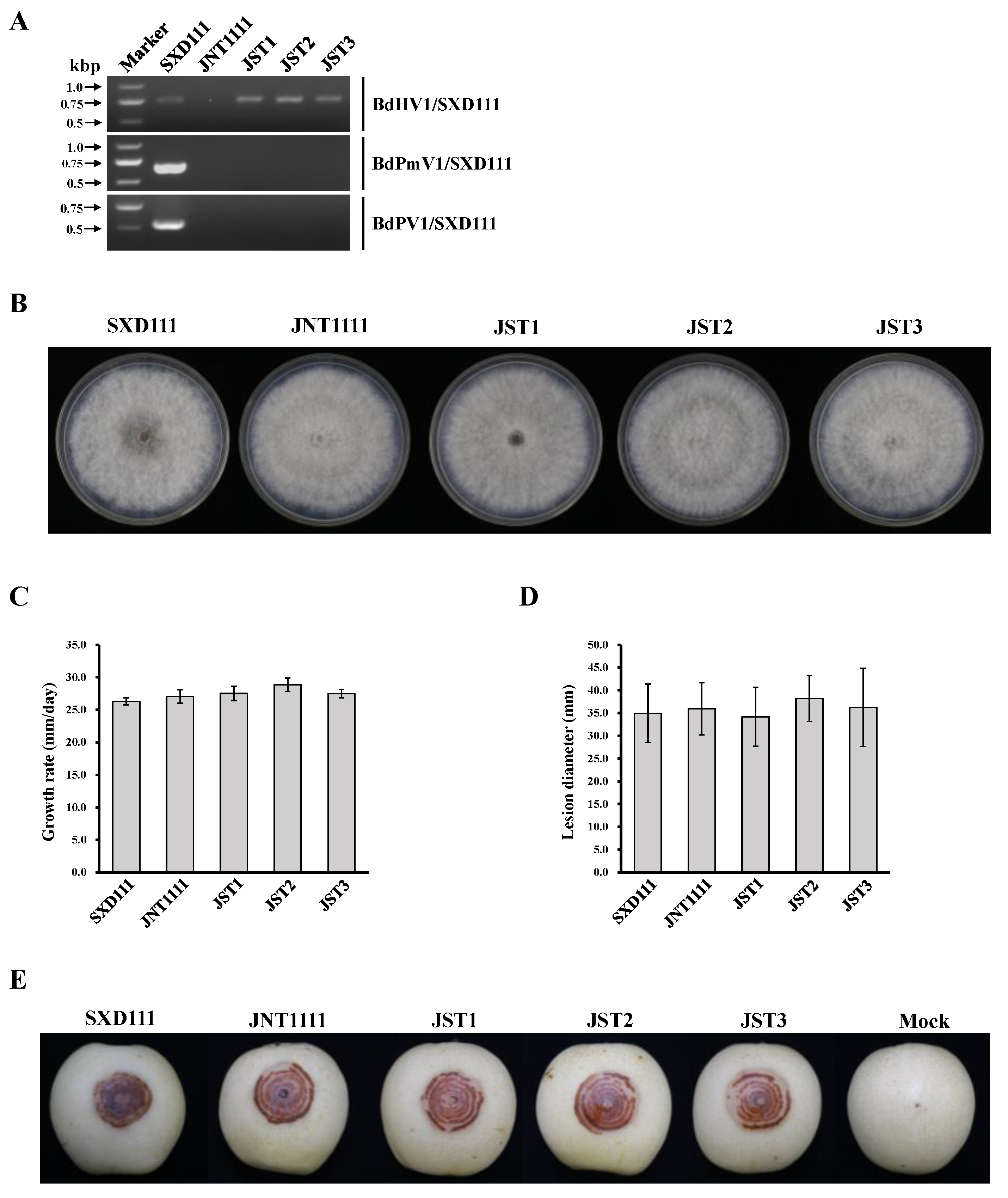
3.6. Vertical and Horizontal Transmission of BdHV1/SXD111
3.7. Effects of BdHV1/SXD111 on B. dothidea
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ayllón, M.A.; Vainio, E.J. Mycoviruses as a part of the global virome: Diversity, evolutionary links and lifestyle. Adv. Virus Res. 2023, 115, 1–86. [Google Scholar] [PubMed]
- Ghabrial, S.; Caston, J.; Jiang, D.H.; Nibert, M.; Suzuki, N. 50-plus years of fungal viruses. Virology 2015, 479–480, 356–368. [Google Scholar] [CrossRef]
- Kondo, H.; Botella, L.; Suzuki, N. Mycovirus diversity and evolution revealed/inferred from recent studies. Annu. Rev. Phytopathol. 2022, 60, 307–336. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.T.; Jiang, D.H. New insights into mycoviruses and exploration for the biological control of crop fungal diseases. Annu. Rev. Phytopathol. 2014, 52, 45–68. [Google Scholar] [CrossRef] [PubMed]
- Chiba, S.; Velasco, L.; Ayllón, M.A.; Suzuki, N.; Lee-Marzano, S.Y.; Sun, L.; Sabanadzovic, S.; Turina, M. ICTV virus taxonomy profile: Hypoviridae 2023. J. Gen. Virol. 2023, 104, 001848. [Google Scholar] [CrossRef] [PubMed]
- Deakin, G.; Dobbs, E.; Bennett, J.M.; Jones, I.M.; Grogan, H.M.; Burton, K.S. Multiple viral infections in Agaricus bisporus-characterisation of 18 unique RNA viruses and 8 ORFans identified by deep sequencing. Sci. Rep. 2017, 7, 2469. [Google Scholar] [CrossRef]
- Shi, M.; Lin, X.D.; Tian, J.H.; Chen, L.J.; Chen, X.; Li, C.X.; Qin, X.C.; Li, J.; Cao, J.P.; Eden, J.S.; et al. Redefining the invertebrate RNA virosphere. Nature 2016, 540, 539–543. [Google Scholar] [CrossRef]
- Marsberg, A.; Kemler, M.; Jami, F.; Nagel, J.H.; Postma-Smidt, A.; Naidoo, S.; Wingfield, M.J.; Crous, P.W.; Spatafora, J.W.; Hesse, C.N.; et al. Botryosphaeria dothidea: A latent pathogen of global importance to woody plant health. Mol. Plant Pathol. 2017, 18, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Ding, Z.; Zhou, Z.Q.; Wang, Y.Z.; Guo, L.Y. Phylogenetic and pathogenic analyses show that the causal agent of apple ring rot in China is Botryosphaeria dothidea. Plant Dis. 2012, 96, 486–496. [Google Scholar] [CrossRef]
- Zhai, L.; Zhang, M.; Lv, G.; Chen, X.; Jia, N.; Hong, N.; Wang, G. Biological and molecular characterization of four Botryosphaeria species isolated from pear plants showing stem wart and stem canker in China. Plant Dis. 2014, 98, 716–726. [Google Scholar] [CrossRef]
- Tian, Y.; Zhao, Y.; Sun, T.; Wang, L.; Liu, J.; Ma, X.; Hu, B. Identification and characterization of Phomopsis amygdali and Botryosphaeria dothidea associated with peach shoot blight in Yangshan, China. Plant Dis. 2018, 102, 2511–2518. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhao, L.; Li, G.; Huang, J.; Hsiang, T. Identification and characterization of Botryosphaeria spp. causing gummosis of peach trees in Hubei Province, central China. Plant Dis. 2011, 95, 1378–1384. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Xie, Y.; Zhang, W.; Wang, Y.; Liu, J.; Hyde, K.D.; Seem, R.C.; Zhang, G.; Wang, Z.; Yao, S.; et al. Species of Botryosphaeriaceae involved in grapevine dieback in China. Fungal Divers. 2013, 61, 221–236. [Google Scholar] [CrossRef]
- Wang, L.; Hou, H.; Zhou, Z.; Tu, H.; Yuan, H. Identification and detection of Botryosphaeria dothidea from kiwifruit (Actinidia chinensis) in China. Plants 2021, 10, 401. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Gong, G.; Cui, Y.; Zhang, D.; Chang, X.; Hu, R.; Liu, N.; Sun, X. Identification of Botryosphaeriaceae species causing kiwifruit rot in Sichuan province, China. Plant Dis. 2015, 99, 699–708. [Google Scholar] [CrossRef]
- Ding, Z.; Zhou, T.; Guo, L.Y. Characterization of a novel strain of Botryosphaeria dothidea chrysovirus 1 from the apple white rot pathogen Botryosphaeria dothidea. Arch. Virol. 2017, 162, 2097–2102. [Google Scholar] [CrossRef]
- Li, J.; Zhai, L.; Zhang, M.; Luo, G.; Wen, Y.; Cao, T.; Xia, H.; Zhang, J.; Liu, M. Molecular characterization of a novel victorivirus isolated from Botryosphaeria dothidea, the causal agent of longan leaf spot disease. Arch. Virol. 2022, 167, 2417–2422. [Google Scholar] [CrossRef]
- Liu, H.; Wang, H.; Zhou, Q. A novel mycovirus isolated from the plant-pathogenic fungus Botryosphaeria dothidea. Arch. Virol. 2021, 166, 1267–1272. [Google Scholar] [CrossRef]
- Wang, L.; Jiang, J.; Wang, Y.; Hong, N.; Zhang, F.; Xu, W.; Wang, G. Hypovirulence of the phytopathogenic fungus Botryosphaeria dothidea: Association with a coinfecting chrysovirus and a partitivirus. J. Virol. 2014, 88, 7517–7527. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, H.; Cao, J.; Yin, X.; Guo, Y.; Guo, L.; Wu, H.; Zhang, M. Characterization of a novel mycovirus from the phytopathogenic fungus Botryosphaeria dothidea. Viruses 2022, 14, 331. [Google Scholar] [CrossRef]
- Yang, M.; Zhai, L.; Xiao, F.; Guo, Y.; Fu, M.; Hong, N.; Wang, G. Characterization of a novel victorivirus isolated from the phytopathogenic fungus Botryosphaeria dothidea. Arch. Virol. 2019, 164, 1609–1617. [Google Scholar] [CrossRef]
- Zhai, L.; Hong, N.; Zhang, M.; Wang, G. Complete dsRNA sequence of a novel victorivirus isolated from the pear stem wart fungus Botryosphaeria dothidea. Arch. Virol. 2015, 160, 613–616. [Google Scholar] [CrossRef]
- Zhai, L.; Xiang, J.; Zhang, M.; Fu, M.; Yang, Z.; Hong, N.; Wang, G. Characterization of a novel double-stranded RNA mycovirus conferring hypovirulence from the phytopathogenic fungus Botryosphaeria dothidea. Virology 2016, 493, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Zhai, L.; Yang, M.; Zhang, M.; Hong, N.; Wang, G. Characterization of a botybirnavirus conferring hypovirulence in the phytopathogenic fungus Botryosphaeria dothidea. Viruses 2019, 11, 266. [Google Scholar] [CrossRef]
- He, Y.; Zou, Q.; Li, S.; Zhu, H.; Hong, N.; Wang, G.; Wang, L. Molecular characterization of a new fusarivirus infecting Botryosphaeria dothidea, the causal agent of pear ring rot disease. Arch. Virol. 2022, 167, 1893–1897. [Google Scholar] [CrossRef]
- Liu, W.; Hai, D.; Mu, F.; Yu, X.; Zhao, Y.; He, B.; Xie, J.; Jiang, D.; Liu, H. Molecular characterization of a novel fusarivirus infecting the plant-pathogenic fungus Botryosphaeria dothidea. Arch. Virol. 2020, 165, 1033–1037. [Google Scholar] [CrossRef]
- Lian, Z.; Das, S.; Luo, J.; Andika, I.B.; Sun, L. Complete genome sequence of a novel ourmia-like mycovirus infecting the phytopathogenic fungus Botryosphaeria dothidea. Arch. Virol. 2021, 166, 3461–3465. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Cao, J.; Xie, S.; Wang, Y.; Yin, X.; Guo, Y.; Xu, C.; Guo, L.; Wu, H.; Zhang, M. Molecular characterization of a novel ourmia-like virus from the phytopathogenic fungus Botryosphaeria dothidea. Arch. Virol. 2023, 168, 106. [Google Scholar] [CrossRef]
- Yang, M.; Zhou, X.; Zhai, L.; Xiao, F.; Hong, N.; Wang, G. Molecular characterization of a novel mycovirus infecting the phytopathogenic fungus Botryosphaeria dothidea. Arch. Virol. 2020, 165, 1667–1670. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Xu, W.; Zhou, X.; Yang, Z.; Wang, Y.; Xiao, F.; Guo, Y.; Hong, N.; Wang, G. Discovery and characterization of a novel bipartite botrexvirus from the phytopathogenic fungus Botryosphaeria dothidea. Front. Microbiol. 2021, 12, 696125. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Liu, M.; Zhu, H.; Zhong, J.; Liao, X.; Zhou, Q. Molecular characterization of a novel mitovirus from the plant-pathogenic fungus Botryosphaeria dothidea. Arch. Virol. 2021, 166, 633–637. [Google Scholar] [CrossRef]
- Wang, H.; Liu, H.; Lu, X.; Wang, Y.; Zhou, Q. A novel mitovirus isolated from the phytopathogenic fungus Botryosphaeria dothidea. Arch. Virol. 2021, 166, 1507–1511. [Google Scholar] [CrossRef]
- Zou, Q.; Gao, Y.; Wang, Q.; Yang, Y.; Wang, F.; Hong, N.; Wang, G.; Wang, L. The full-length genome sequence of a novel mitovirus from Botryosphaeria dothidea, the causal agent of pear ring rot disease. Arch. Virol. 2021, 166, 2881–2885. [Google Scholar] [CrossRef] [PubMed]
- Dong, K.; Xu, C.; Kotta-Loizou, I.; Jiang, J.; Lv, R.; Kong, L.; Li, S.; Hong, N.; Wang, G.; Coutts, R.H.A.; et al. Novel viroid-like RNAs naturally infect a filamentous fungus. Adv. Sci. 2023, 10, e2204308. [Google Scholar] [CrossRef]
- Wang, W.; Wang, X.; Tu, C.; Yang, M.; Xiang, J.; Wang, L.; Hong, N.; Zhai, L.; Wang, G. Novel mycoviruses discovered from a metatranscriptomics survey of the phytopathogenic Alternaria fungus. Viruses 2022, 14, 2552. [Google Scholar] [CrossRef] [PubMed]
- Valverde, R.A. Analysis of double-stranded RNA for plant virus diagnosis. Plant Dis. 1990, 74, 255–258. [Google Scholar]
- Lambden, P.R.; Cooke, S.J.; Caul, E.O.; Clarke, I.N. Cloning of noncultivatable human rotavirus by single primer amplifcation. J. Virol. 1992, 66, 1817–1822. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment; interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Yaegashi, H.; Kanematsu, S.; Ito, T. Molecular characterization of a new hypovirus infecting a phytopathogenic fungus, Valsa ceratosperma. Virus Res. 2012, 165, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Smart, C.D.; Yuan, W.; Foglia, R.; Nuss, D.L.; Fulbright, D.W.; Hillman, B.I. Cryphonectria hypovirus 3, a virus species in the family Hypoviridae with a single open reading frame. Virology 1999, 265, 66–73. [Google Scholar] [CrossRef]
- Linder-Basso, D.; Dynek, J.N.; Hillman, B.I. Genome analysis of Cryphonectria hypovirus 4, the most common hypovirus species in North America. Virology 2005, 337, 192–203. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Xiao, X.; Fu, Y.; Liu, H.; Cheng, J.; Ghabrial, S.A.; Li, G.; Jiang, D. A novel mycovirus closely related to hypoviruses that infects the plant pathogenic fungus Sclerotinia sclerotiorum. Virology 2011, 418, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Fu, Y.; Jiang, D.; Mu, F.; Cheng, J.; Lin, Y.; Li, B.; Marzano, S.L.; Xie, J. Interannual dynamics, diversity and evolution of the virome in Sclerotinia sclerotiorum from a single crop field. Virus Evol. 2021, 7, veab032. [Google Scholar] [CrossRef]
- Hao, F.; Ding, T.; Wu, M.; Zhang, J.; Yang, L.; Chen, W.; Li, G. Two novel hypovirulence-associated mycoviruses in the phytopathogenic fungus Botrytis cinerea: Molecular characterization and suppression of infection cushion formation. Viruses 2018, 10, 254. [Google Scholar] [CrossRef]
- Ruiz-Padilla, A.; Rodríguez-Romero, J.; Gómez-Cid, I.; Pacifico, D.; Ayllón, M.A. Novel mycoviruses discovered in the mycovirome of a necrotrophic fungus. mBio 2021, 12, e03705-20. [Google Scholar] [CrossRef]
- Mizutani, Y.; Uesaka, K.; Ota, A.; Calassanzio, M.; Ratti, C.; Suzuki, T.; Fujimori, F.; Chiba, S. De novo sequencing of novel mycoviruses from Fusarium sambucinum: An attempt on direct RNA sequencing of viral dsRNAs. Front. Microbiol. 2021, 12, 641484. [Google Scholar] [CrossRef]
- Torres-Trenas, A.; Cañizares, M.C.; García-Pedrajas, M.D.; Pérez-Artés, E. Molecular and biological characterization of the first hypovirus identified in Fusarium oxysporum. Front. Microbiol. 2020, 10, 3131. [Google Scholar] [CrossRef]
- Koloniuk, I.; El-Habbak, M.H.; Petrzik, K.; Ghabrial, S.A. Complete genome sequence of a novel hypovirus infecting Phomopsis longicolla. Arch. Virol. 2014, 159, 1861–1863. [Google Scholar] [CrossRef]
- Gilbert, K.B.; Holcomb, E.E.; Allscheid, R.L.; Carrington, J.C. Hiding in plain sight: New virus genomes discovered via a systematic analysis of fungal public transcriptomes. PLoS ONE 2019, 14, e0219207. [Google Scholar] [CrossRef]
- De Miccolis Angelini, R.M.; Raguseo, C.; Rotolo, C.; Gerin, D.; Faretra, F.; Pollastro, S. The mycovirome in a worldwide collection of the brown rot fungus Monilinia fructicola. J. Fungi 2022, 8, 481. [Google Scholar] [CrossRef]
- Zhong, J.; Li, P.; Gao, B.D.; Zhong, S.Y.; Li, X.G.; Hu, Z.; Zhu, J.Z. Novel and diverse mycoviruses co-infecting a single strain of the phytopathogenic fungus Alternaria dianthicola. Front. Cell. Infect. Microbiol. 2022, 12, 980970. [Google Scholar] [CrossRef]
- Nibert, M.L.; Ghabrial, S.A.; Maiss, E.; Lesker, T.; Vainio, E.J.; Jiang, D.; Suzuki, N. Taxonomic reorganization of family Partitiviridae and other recent progress in partitivirus research. Virus Res. 2014, 188, 128–141. [Google Scholar] [CrossRef]
- Thapa, V.; Roossinck, M.J. Determinants of coinfection in the mycoviruses. Front. Cell. Infect. Microbiol. 2019, 9, 169. [Google Scholar] [CrossRef] [PubMed]
- Palukaitis, P. Satellite RNAs and satellite viruses. Mol. Plant Microbe Interact. 2016, 29, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Vainio, E.J.; Chiba, S.; Ghabrial, S.A.; Maiss, E.; Roossinck, M.; Sabanadzovic, S.; Suzuki, N.; Xie, J.; Nibert, M.; ICTV Report Consortium. ICTV virus taxonomy profile: Partitiviridae. J. Gen. Virol. 2018, 99, 17–18. [Google Scholar] [CrossRef]
- Oh, C.S.; Hillman, B.I. Genome organization of a partitivirus from the filamentous ascomycete Atkinsonella hypoxylon. J. Gen. Virol. 1995, 76, 1461–1470. [Google Scholar] [CrossRef]
- Liu, W.; Duns, G.; Chen, J. Genomic characterization of a novel partitivirus infecting. Aspergillus ochraceus. Virus Genes 2008, 37, 322–327. [Google Scholar] [CrossRef] [PubMed]
- Filippou, C.; Coutts, R.H.A.; Stevens, D.A.; Sabino, R.; Kotta-Loizou, I. Completion of the sequence of the Aspergillus fumigatus partitivirus 1 genome. Arch. Virol. 2020, 165, 1891–1894. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Wang, J.; Yang, B.; Wang, Q.; Zhou, J.; Yu, W. Molecular characterization of a debilitation-associated partitivirus infecting the pathogenic fungus Aspergillus flavus. Front. Microbiol. 2019, 10, 626. [Google Scholar] [CrossRef]
- Zhong, J.; Lei, X.H.; Zhu, J.Z.; Song, G.; Zhang, Y.D.; Chen, Y.; Gao, B.D. Detection and sequence analysis of two novel co-infecting double-strand RNA mycoviruses in Ustilaginoidea virens. Arch. Virol. 2014, 159, 3063–3070. [Google Scholar] [CrossRef]
- Kim, J.W.; Choi, E.Y.; Lee, J.I. Genome organization and expression of the Penicillium stoloniferum virus F. Virus Genes 2005, 31, 175–183. [Google Scholar] [CrossRef]
- Rong, R.; Rao, S.; Scott, S.W.; Carner, G.R.; Tainter, F.H. Complete sequence of the genome of two dsRNA viruses from Discula destructiva. Virus Res. 2002, 90, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Yang, B.; Liu, X.; Tian, X.; Wang, Q.; Wang, B.; Zhang, Q.; Yu, W.; Qi, X.; Jiang, Y.; et al. A satellite dsRNA attenuates the induction of helper virus-mediated symptoms in Aspergillus flavus. Front. Microbiol. 2022, 13, 895844. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Liu, J.; Kong, L.; He, Y.; Wu, H.; Xu, W. A special satellite-like RNA of a novel hypovirus from Pestalotiopsis fici broadens the definition of fungal satellite. PLoS Pathog. 2023, 19, e1010889. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Kondo, H.; Liu, L.; Guo, L.; Qiu, D. A novel virus in the family Hypoviridae from the plant pathogenic fungus Fusarium graminearum. Virus Res. 2013, 174, 69–77. [Google Scholar] [CrossRef]
- Fulbright, D.W. Effect of eliminating dsRNA in hypovirulent Endothia parasitica. Phytopathology 1984, 74, 722–724. [Google Scholar] [CrossRef]
- Hillman, B.I.; Shapira, R.; Nuss, D.L. Hypovirulence-associated suppression of host functions in Cryphonectria parasitica can be partially relieved by high light intensity. Phytopathology 1990, 80, 950–956. [Google Scholar] [CrossRef]
- Hillman, B.I.; Tian, Y.; Bedker, P.J.; Brown, M.P. A North American hypovirulent isolate of the chestnut blight fungus with European isolated-related dsRNA. J. Gen. Virol. 1992, 73, 681–686. [Google Scholar] [CrossRef]
- Li, P.; Zhang, H.; Chen, X.; Qiu, D.; Guo, L. Molecular characterization of a novel hypovirus from the plant pathogenic fungus Fusarium graminearum. Virology 2015, 481, 151–160. [Google Scholar] [CrossRef] [PubMed]


| Strain | Origin | Mycovirus |
|---|---|---|
| SXD111 | Pyrus bretschneideri cv. ‘Hongxiangsu’, Shaanxi province, China | BdHV1 *, BdPmV1, BdPV1 |
| YZN115 | P. bretschneideri cv. ‘Suli’, Henan province, China | BdPmV1 |
| YCLYY11 | Dimocarpus longan cv. ‘Shuguan’, Chongqing City, China | BdVV3, BdCV1, BdPV1 |
| JNT1111 | P. bretschneideri cv. ‘Suli’, Shanxi, China | Virus free |
| JST1 | JNT1111 in a pairing culture of JNT1111 and SXD111 | BdHV1 |
| JST2 | JNT1111 in a pairing culture of JNT1111 and SXD111 | BdHV1 |
| JST3 | JNT1111 in a pairing culture of JNT1111 and SXD111 | BdHV1 |
| No. | Contig Number | Contig Length (nt/bp) | Best Match | Protein | Cover % | Aa Ident % | Taxon |
|---|---|---|---|---|---|---|---|
| 1 | contig 1 | 11,113 | Cryphonectria hypovirus 4 (CHV4) | polyprotein | 68 | 49.77 | Hypoviridae Betahypovirus |
| 2 | contig 135 | 3592 | Botryosphaeria dothidea chrysovirus 1 (BdCV1) | RdRp | 93 | 98.66 | Chrysoviridae Betachrysovirus |
| 3 | contig 2025 | 1401 | hypothetical protein P3 | 99 | 99.14 | ||
| 4 | Contig 2640 | 1216 | coat protein | 85 | 98.90 | ||
| 6 | contig 5902 | 703 | hypothetical protein | 91 | 98.13 | ||
| 7 | contig 744 | 2171 | Botryosphaeria dothidea polymycovirus 1 (BdPmV1) | hypothetical protein P2 | 95 | 100.00 | Polymycoviridae Polymycovirus |
| 8 | contig 1102 | 1855 | methyltransferase | 97 | 99.17 | ||
| 9 | contig 3852 | 960 | RdRp | 99 | 99.37 | ||
| 10 | contig 5123 | 784 | hypothetical protein P5 | 89 | 98.29 | ||
| 11 | Contig 5986 | 694 | PAS-rich protein | 97 | 98.22 | ||
| 12 | contig 1458 | 1635 | Botryosphaeria dothidea partitivirus 1 (BdPV1) | coat protein | 90 | 99.80 | Partitiviridae |
| 13 | contig 5134 | 783 | RdRp | 97 | 100 | ||
| 14 | contig 406 | 2641 | Botryosphaeria dothidea victorivirus 3 (BdVV3) | RdRp | 93 | 99.80 | Totiviridae Victorivirus |
| 15 | contig 421 | 2624 | coat protein | 81 | 99.86 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, Y.; Qu, J.; Zhang, H.; Yang, Y.; Huang, R.; Deng, J.; Zhang, J.; Xiao, Y.; Li, J.; Zhang, M.; et al. Identification and Characterization of a Novel Hypovirus from the Phytopathogenic Fungus Botryosphaeria dothidea. Viruses 2023, 15, 2059. https://doi.org/10.3390/v15102059
Wen Y, Qu J, Zhang H, Yang Y, Huang R, Deng J, Zhang J, Xiao Y, Li J, Zhang M, et al. Identification and Characterization of a Novel Hypovirus from the Phytopathogenic Fungus Botryosphaeria dothidea. Viruses. 2023; 15(10):2059. https://doi.org/10.3390/v15102059
Chicago/Turabian StyleWen, Yongqi, Jinyue Qu, Honglin Zhang, Yi Yang, Rui Huang, Jili Deng, Jiayu Zhang, Yanping Xiao, Jiali Li, Meixin Zhang, and et al. 2023. "Identification and Characterization of a Novel Hypovirus from the Phytopathogenic Fungus Botryosphaeria dothidea" Viruses 15, no. 10: 2059. https://doi.org/10.3390/v15102059





