Unexpected Pathogen Diversity Detected in Australian Avifauna Highlights Potential Biosecurity Challenges
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Sample Collection
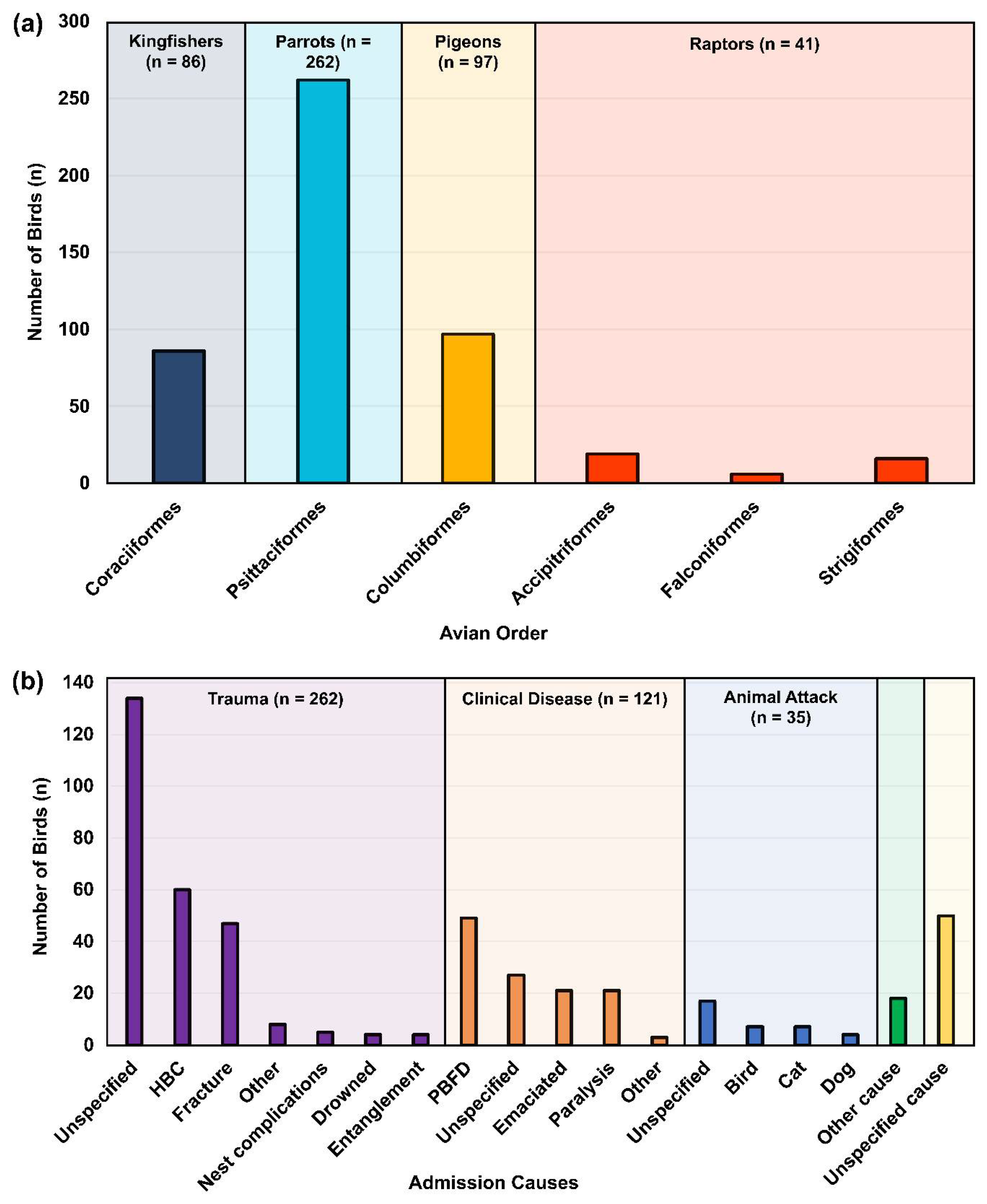
2.3. Sampled Avian Species and Hospital Admission Causes
2.4. New Sample Processing and DNA Extraction
2.5. Chlamydiaceace, BFDV, Avipoxvirus, CoAHV1 and PsAHV1 qPCR Detection
2.6. Molecular Characterisation of Avipoxvirus, BFDV and PsAHV1
2.7. Sequence and Phylogenetic Analysis
2.8. Statistical Analysis
3. Results
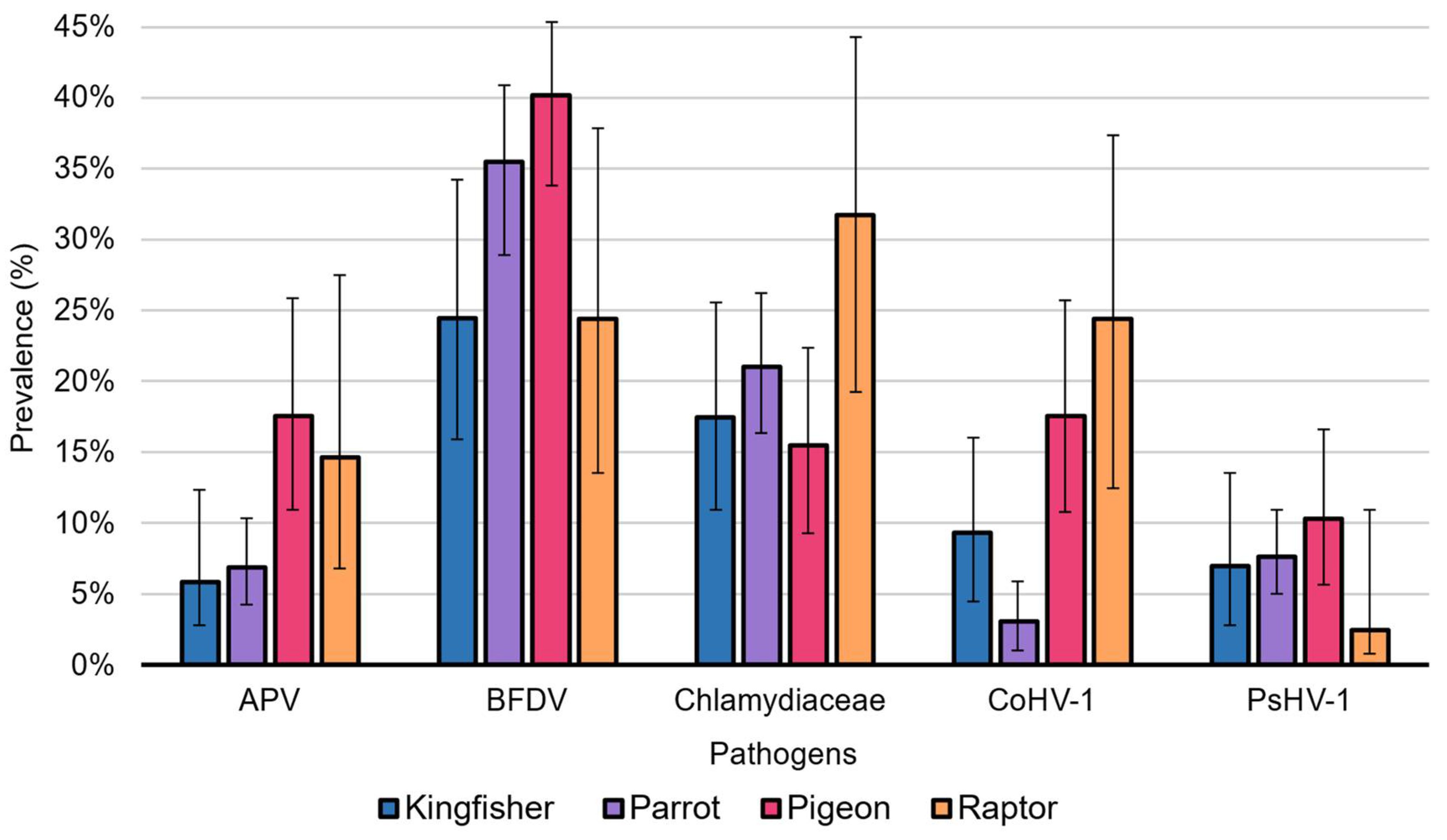
3.1. Beak and Feather Disease Virus Prevalence
| Total Pos. | Apparent Prevalence (%) a | True Prevalence (%) b | |
|---|---|---|---|
| Birds BFDV pos. | 163/486 | 33.54 (CI 29.49–37.85) | 33.58 (CI 28.81–38.65) |
| Swabs BFDV pos. | 212/688 | 30.81 (CI 27.48–34.36) | 30.37 (CI 26.45–34.54) |
| Tissue BFDV pos. | 59/139 | 42.45 (CI 34.54–50.76) | 44.05 (34.75–53.83) |
| Birds Chlamydiaceae pos. | 98/486 | 20.16 (CI 16.84–23.96) | 17.84 (CI 13.93–22.31) |
| Swabs Chlamydiaceae pos. | 138/688 | 20.06 (CI 17.24–23.21) | 17.72 (CI 14.40–21.43) |
| Tissue Chlamydiaceae pos. | 1/139 | 0.72 (CI 0.13–3.96) | NA |
| Birds PsAHV1 pos. | 37/486 | 7.61 (CI 5.57–10.32) | 3.07 (CI 0.67–6.26) |
| Swabs PsAHV1 pos. | 23/688 | 3.34 (CI 2.24–4.97) | NA |
| Tissue PsAHV1 pos. | 20/139 | 14.39 (CI 9.51–21.18) | 11.05 (CI 5.31–19.04) |
| Birds CoAHV1 pos. | 43/486 | 8.85 (CI 6.63–11.71) | 4.53 (CI 1.92–7.89) |
| Swabs CoAHV1 pos. | 40/688 | 5.81 (CI 4.30–7.82) | 0.96 (CI 0.00–3.32) |
| Tissue CoAHV1 pos. | 9/139 | 6.47 (CI 3.44–11.85) | 1.74 (CI 0.00–8.06) |
| Birds Avipoxvirus pos. | 46/486 | 9.47 (CI 7.17–12.39) | 5.25 (CI 2.55–8.70) |
| Swabs Avipoxvirus pos. | 36/688 | 5.23 (CI 3.80–7.16) | 0.27 (CI 0.00–2.54) |
| Tissue Avipoxvirus pos. | 17/139 | 12.23 (CI 7.78–18.71) | 8.51 (CI 3.27–16.13) |
3.2. Chlamydiaceae Prevalence
3.3. CoAHV1 Prevalence
3.4. PsAHV1 Prevalence
3.5. Avipoxvirus Prevalence
3.6. Pathogen Coinfection and the Manifestation of Clinical Disease
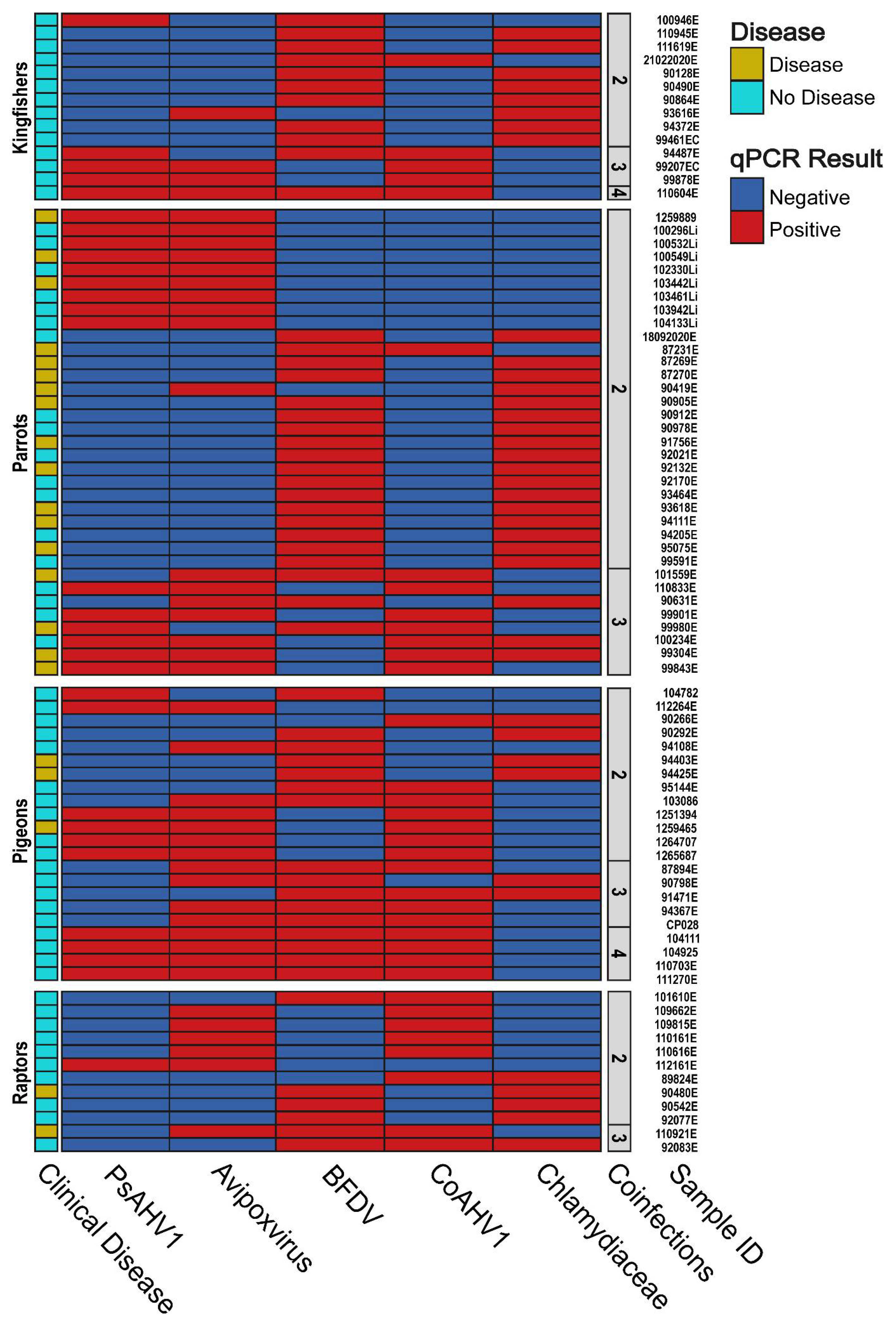
3.7. Sequence Analysis of Avipoxvirus, BFDV and PsAHV1
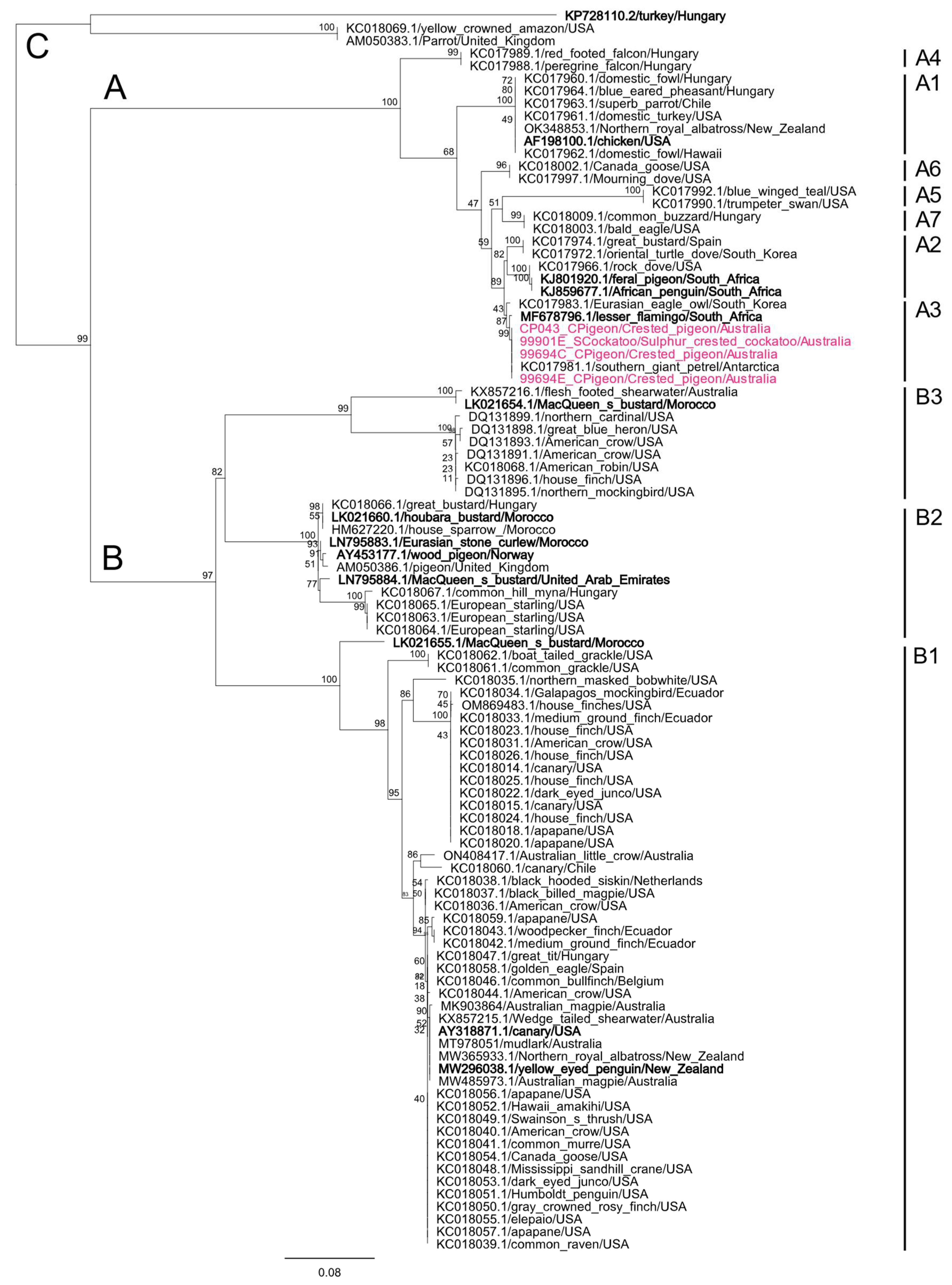
3.7.1. BFDV Phylogenetic Analyses

3.7.2. Avipoxvirus Phylogenetic Analyses
3.7.3. PsAHV1 Phylogenetic Analyses
4. Discussion
4.1. Avipoxvirus, BFDV, Chlamydiaceae, CoAHV1 and PsAHV1 Detection in Wild Avian Hosts from Southeast Queensland
4.2. Pathogen Biosecurity Concerns and Precautions
4.3. Phylogenetic Analyses of BFDV Sequences Shows the Formation of Lorikeet and Host-Generalist Clades
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thompson, A.; Kutz, S. Introduction to the Special Issue on ‘Emerging Zoonoses and Wildlife’. Int. J. Parasitol. Parasites Wildl. 2019, 9, 322. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.T.; Sobur, M.A.; Islam, M.S.; Ievy, S.; Hossain, M.J.; El Zowalaty, M.E.; Rahman, A.T.; Ashour, H.M. Zoonotic diseases: Etiology, impact, and control. Microorganisms 2020, 8, 1405. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.-W.; To, K.K.-W.; Chen, H.; Yuen, K.-Y. Cross-species transmission and emergence of novel viruses from birds. Curr. Opin. Virol. 2015, 10, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Sarker, S. Special Issue: Emerging wildlife viral diseases. Viruses 2022, 14, 807. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, M.; Sarker, S.; Vaz, P.K.; Legione, A.R.; Devlin, J.M.; Macwhirter, P.L.; Whiteley, P.L.; Raidal, S.R. Disease surveillance in wild Victorian cacatuids reveals co-infection with multiple agents and detection of novel avian viruses. Vet. Microbiol. 2019, 235, 257–264. [Google Scholar] [CrossRef]
- Amery-Gale, J.; Hartley, C.A.; Vaz, P.K.; Marenda, M.S.; Owens, J.; Eden, P.A.; Devlin, J.M. Avian viral surveillance in Victoria, Australia, and detection of two novel avian herpesviruses. PLoS ONE 2018, 13, e0194457. [Google Scholar] [CrossRef] [Green Version]
- Lupiani, B.; Reddy, S.M. The history of avian influenza. Comp. Immunol. Microbiol. Infect. Dis. 2009, 32, 311–323. [Google Scholar] [CrossRef]
- Rabiei, M.; Low, W.Y.; Ren, Y.; Cahyono, M.I.; Doan, P.T.K.; Dharmayanti, I.; Grande, E.D.; Hemmatzadeh, F. Indicators of the molecular pathogenesis of virulent Newcastle disease virus in chickens revealed by transcriptomic profiling of spleen. Sci. Rep. 2021, 11, 17570. [Google Scholar] [CrossRef]
- Walkden-Brown, S.W.; Islam, A.F.; Groves, P.J.; Rubite, A.; Sharpe, S.M.; Burgess, S.K. Development, application, and results of routine monitoring of Marek’s disease virus in broiler house dust using real-time quantitative PCR. Avian Dis. 2013, 57, 544–554. [Google Scholar] [CrossRef]
- Das, S.; Smith, K.; Sarker, S.; Peters, A.; Adriaanse, K.; Eden, P.; Ghorashi, S.A.; Forwood, J.K.; Raidal, S.R. Repeat spillover of beak and feather disease virus into an endangered parrot highlights the risk associated with endemic pathogen loss in endangered species. J. Wildl. Dis. 2020, 56, 896–906. [Google Scholar] [CrossRef]
- Kasimov, V.; Dong, Y.; Shao, R.; Brunton, A.; Anstey, S.I.; Hall, C.; Chalmers, G.; Conroy, G.; Booth, R.; Timms, P.; et al. Emerging and well-characterized chlamydial infections detected in a wide range of wild Australian birds. Transbound. Emerg. Dis. 2022, 69, e3154–e3170. [Google Scholar] [CrossRef]
- Amery-Gale, J.; Marenda, M.S.; Owens, J.; Eden, P.A.; Browning, G.F.; Devlin, J.M. A high prevalence of beak and feather disease virus in non-psittacine Australian birds. J. Med. Microbiol. 2017, 66, 1005–1013. [Google Scholar] [CrossRef]
- Sarker, S.; Bowden, T.R.; Boyle, D.B. Genomic characterisation of a novel avipoxvirus, magpiepox virus 2, from an Australian magpie (Gymnorhina tibicen terraereginae). Virology 2021, 562, 121–127. [Google Scholar] [CrossRef]
- Birdlife International. Country Profile: Australia. Available online: http://datazone.birdlife.org/country/australia/species (accessed on 29 October 2022).
- Boyd, B.M.; Nguyen, N.-P.; Allen, J.M.; Waterhouse, R.M.; Vo, K.B.; Sweet, A.D.; Clayton, D.H.; Bush, S.E.; Shapiro, M.D.; Johnson, K.P. Long-distance dispersal of pigeons and doves generated new ecological opportunities for host-switching and adaptive radiation by their parasites. Proc. Biol. Sci. 2022, 289, 20220042. [Google Scholar] [CrossRef]
- Wright, T.F.; Schirtzinger, E.E.; Matsumoto, T.; Eberhard, J.R.; Graves, G.R.; Sanchez, J.J.; Capelli, S.; Müller, H.; Scharpegge, J.; Chambers, G.K.; et al. A Multilocus molecular phylogeny of the parrots (Psittaciformes): Support for a Gondwanan origin during the cretaceous. Mol. Biol. Evol. 2008, 25, 2141–2156. [Google Scholar] [CrossRef]
- Debus, S.J.S.; Davies, J.N.; BirdLife Australia. Birds of Prey of Australia: A Field Guide; CSIRO Publishing: Clayton, Australia, 2012. [Google Scholar]
- Marchant, S.; Higgins, P.J.; Ambrose, S.J.; Davies, S.J.J.F.; Steele, W.K. Handbook of Australian, New Zealand & Antarctic Birds: Parrots to Dollarbird; Higgins, P.J., Ed.; Oxford University Press: New York, NY, USA, 1990. [Google Scholar]
- Ehricht, R.; Slickers, P.; Goellner, S.; Hotzel, H.; Sachse, K. Optimized DNA microarray assay allows detection and genotyping of single PCR-amplifiable target copies. Mol. Cell Probes. 2006, 20, 60–63. [Google Scholar] [CrossRef]
- Tomaszewski, E.; Wilson, V.G.; Wigle, W.L.; Phalen, D.N. Detection and heterogeneity of herpesviruses causing Pacheco’s disease in parrots. J. Clin. Microbiol. 2001, 39, 533–538. [Google Scholar] [CrossRef] [Green Version]
- Woźniakowski, G.J.; Samorek-Salamonowicz, E.; Szymański, P.; Wencel, P.; Houszka, M. Phylogenetic analysis of Columbid herpesvirus-1 in rock pigeons, birds of prey and non-raptorial birds in Poland. BMC Vet. Res. 2013, 9, 52. [Google Scholar] [CrossRef] [Green Version]
- Sarker, S.; Das, S.; Lavers, J.L.; Hutton, I.; Helbig, K.; Imbery, J.; Upton, C.; Raidal, S.R. Genomic characterization of two novel pathogenic avipoxviruses isolated from pacific shearwaters (Ardenna spp.). BMC Genomics 2017, 18, 298. [Google Scholar] [CrossRef] [Green Version]
- Bassami, M.R.; Berryman, D.; Wilcox, G.E.; Raidal, S.R. Psittacine beak and feather disease virus nucleotide sequence analysis and its relationship to porcine circovirus, plant circoviruses, and chicken anaemia virus. Virology 1998, 249, 453–459. [Google Scholar] [CrossRef]
- Lee, L.H.; Lee, K.H. Application of the polymerase chain reaction for the diagnosis of fowl poxvirus infection. J. Virol. Methods 1997, 63, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Phalen, D.N.; Holz, P.; Rasmussen, L.; Bayley, C. Fatal columbid herpesvirus-1 infections in three species of Australian birds of prey. Aust. Vet. J. 2011, 89, 193–196. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sergeant, E.S.G. Epitools Epidemiological Calculators. Ausvet. 2018. Available online: http://epitools.ausvet.com.au (accessed on 15 October 2022).
- Gyuranecz, M.; Foster, J.T.; Dán, Á.; Ip, H.S.; Egstad, K.F.; Parker, P.G.; Higashiguchi, J.M.; Skinner, M.A.; Höfle, U.; Kreizinger, Z.; et al. Worldwide phylogenetic relationship of avian poxviruses. J. Virol. 2013, 87, 4938–4951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phalen, D.N.; Woods, R. Psittacid Herpesviruses and Mucosal Papillomas of Birds in Australia: Fact Sheet; Wildlife Health Australia: Mosman, Australia, 2017. [Google Scholar]
- Tomaszewski, E.K.; Gravendyck, M.; Kaleta, E.F.; Phalen, D.N. Genetic characterization of a herpesvirus isolate from a superb starling (Lamprotornis superbus) as a psittacid herpesvirus genotype 1. Avian Dis. 2004, 48, 212–214. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, M.; Sarker, S.; Raidal, S.R. Molecular and microscopic characterisation of a novel pathogenic herpesvirus from Indian ringneck parrots (Psittacula krameri). Vet. Microbiol. 2019, 239, 108428. [Google Scholar] [CrossRef]
- Gabor, M.; Gabor, L.J.; Peacock, L.; Srivastava, M.; Rosenwax, A.; Phalen, D. Psittacid herpesvirus 3 infection in the eclectus parrot (Eclectus roratus) in Australia. Vet. Pathol. 2013, 50, 1053–1057. [Google Scholar] [CrossRef]
- Phalen, D.N.; Alvarado, C.; Grillo, V.; Mason, P.; Dobson, E.; Holz, P. Prevalence of columbid herpesvirus infection in feral pigeons from New South Wales and Victoria, Australia, with spillover into a wild powerful owl (Ninox Struena). J. Wildl. Dis. 2017, 53, 543–551. [Google Scholar] [CrossRef]
- Rose, N.; Warren, A.L.; Whiteside, D.; Bidulka, J.; Robinson, J.H.; Illanes, O.; Brookfield, C. Columbid herpesvirus-1 mortality in great horned owls (Bubo virginianus) from Calgary, Alberta. Can. Vet. J. 2012, 53, 265–268. [Google Scholar]
- Sarker, S.; Athukorala, A.; Bowden, T.R.; Boyle, D.B. Characterisation of an Australian fowlpox virus carrying a near-full-length provirus of reticuloendotheliosis virus. Arch. Virol. 2021, 166, 1485–1488. [Google Scholar] [CrossRef]
- Sarker, S.; Athukorala, A.; Raidal, S.R. Molecular characterisation of a novel pathogenic avipoxvirus from an Australian passerine bird, mudlark (Grallina cyanoleuca). Virology 2021, 554, 66–74. [Google Scholar] [CrossRef]
- Sarker, S.; Batinovic, S.; Talukder, S.; Das, S.; Park, F.; Petrovski, S.; Forwood, J.K.; Helbig, K.J.; Raidal, S.R. Molecular characterisation of a novel pathogenic avipoxvirus from the Australian magpie (Gymnorhina tibicen). Virology 2020, 540, 15053. [Google Scholar] [CrossRef]
- Slocombe, R.F.; McCowan, C.; Wang, J.; Holz, P. Avian pox in crimson rosellas (Platycercus elegans) in southern Australia. Avian Pathol. 2013, 42, 147–150. [Google Scholar] [CrossRef] [Green Version]
- Williams, R.A.J.; Truchado, D.A.; Benitez, L. A Review on the prevalence of poxvirus disease in free-living and captive wild birds. Microbiol. Res. 2021, 12, 403–418. [Google Scholar] [CrossRef]
- Hulbert, C.L.; Chamings, A.; Hewson, K.A.; Steer, P.A.; Gosbell, M.; Noormohammadi, A.H. Survey of captive parrot populations around Port Phillip Bay, Victoria, Australia, for psittacine beak and feather disease virus, avian polyomavirus and psittacine adenovirus. Aust. Vet. J. 2015, 93, 287–292. [Google Scholar] [CrossRef]
- Martens, J.M.; Stokes, H.S.; Berg, M.L.; Walder, K.; Bennett, A.T.D. Seasonal fluctuation of beak and feather disease virus (BFDV) infection in wild Crimson Rosellas (Platycercus elegans). Sci. Rep. 2020, 10, 7894. [Google Scholar] [CrossRef]
- Mattmann, P.; Marti, H.; Borel, N.; Jelocnik, M.; Albini, S.; Vogler, B.R. Chlamydiaceae in wild, feral and domestic pigeons in Switzerland and insight into population dynamics by Chlamydia psittaci multilocus sequence typing. PLoS ONE 2019, 14, e0226088. [Google Scholar] [CrossRef] [Green Version]
- Tiyawattanaroj, W.; Lindenwald, R.; Mohr, L.; Günther, E.; Legler, M. Monitoring of the infectious agent Chlamydia psittaci in common swifts (Apus apus) in the area of Hannover, Lower Saxony, Germany. Berl. Munich Vet. Wkly. J. 2021, 134, 1–5. [Google Scholar]
- Gass, J.D., Jr.; Dusek, R.J.; Hall, J.S.; Hallgrimsson, G.T.; Halldórsson, H.P.; Vignisson, S.R.; Ragnarsdottir, S.B.; Jónsson, J.E.; Krauss, S.; Wong, S.S.; et al. Global dissemination of Influenza A virus is driven by wild bird migration through arctic and subarctic zones. Mol. Ecol. 2023, 32, 198–213. [Google Scholar] [CrossRef]
- Jaeger, A.; Lebarbenchon, C.; Bourret, V.; Bastien, M.; Lagadec, E.; Thiebot, J.-B.; Boulinier, T.; Delord, K.; Barbraud, C.; Marteau, C.; et al. Avian cholera outbreaks threaten seabird species on Amsterdam Island. PLoS ONE 2018, 13, e0197291. [Google Scholar] [CrossRef]
- Loeb, J. Scottish seabirds hit by avian influenza. Vet. Rec. 2022, 190, 488. [Google Scholar] [CrossRef] [PubMed]
- Katoh, H.; Ohya, K.; Fukushi, H. Development of novel real-time PCR assays for detecting DNA virus infections in psittaciform birds. J. Virol. Methods 2008, 154, 92–98. [Google Scholar] [CrossRef] [PubMed]
- IUCN. The IUCN Red List of Threatened Species. Version 2022-1. 2022. Available online: https://www.iucnredlist.org/ (accessed on 29 October 2022).
- Mercier, A.; Hamel, J.; Toral-Granda, T.; Alvarado, J.; Paola Ortiz, E.; Benavides, M. Isostichopus fuscus. The IUCN Red List of Threatened Species 2013: E. T180373A1621878; International Union for Conservation of Nature and Natural Resource: Gland, Switzerland, 2013. [Google Scholar]
- White, N.E.; Phillips, M.J.; Gilbert, M.T.P.; Alfaro-Núñez, A.; Willerslev, E.; Mawson, P.R.; Spencer, P.B.S.; Bunce, M. The evolutionary history of cockatoos (Aves: Psittaciformes: Cacatuidae). Mol. Phylogenet. Evol. 2011, 59, 615–622. [Google Scholar] [CrossRef] [Green Version]
- Coroian, M.; Silaghi, C.; Tews, B.A.; Baltag, E.; Marinov, M.; Alexe, V.; Kalmár, Z.; Cintia, H.; Lupșe, M.S.; Mihalca, A.D. Serological survey of mosquito-borne arboviruses in wild birds from important migratory hotspots in Romania. Pathogens 2022, 11, 1270. [Google Scholar] [CrossRef] [PubMed]
- Department of Agriculture Fisheries and Forestry. Environment Protection and Biodiversity Conservation Act; Australian Government: Canberra, ACT, Australia, 2022.
- Raidal, S.R.; Sabine, M.; Cross, G.M. Laboratory diagnosis of psittacine beak and feather disease by haemagglutination and haemagglutination inhibition. Aust. Vet. J. 1993, 70, 133–137. [Google Scholar] [CrossRef]
- Raidal, S.R.; Johnsen Bonne, N.; Stewart, M. Development of Recombinant Proteins as a Candidate Vaccine for Psittacine Beak and Feather Disease; Murdoch University: Perth, WA, Australia, 2005. [Google Scholar]
- Cross, G. Hygiene Protocols for the Prevention and Control of Diseases (Particularly Beak and Feather Disease) in Australian Birds; Department of the Environment and Heritage (now Department of the Environment, Water, Heritage and the Arts), The Australian Government, Australia; 2006. Available online: https://www.dcceew.gov.au/sites/default/files/documents/hygiene-protocols-all.pdf (accessed on 29 October 2022).
- Tomaszewski, E.K.; Wigle, W.; Phalen, D.N. Tissue Distribution of psittacid herpesviruses in latently infected parrots, repeated sampling of latently infected parrots and prevalence of latency in parrots submitted for necropsy. Vet. Diagn. Investig. 2006, 18, 536–544. [Google Scholar] [CrossRef] [Green Version]
- Katoh, H.; Ogawa, H.; Ohya, K.; Fukushi, H. A review of DNA viral infections in psittacine birds. J. Vet. Med. Sci. 2010, 72, 1099–1106. [Google Scholar] [CrossRef] [Green Version]
- Sarker, S.; Ghorashi, S.A.; Forwood, J.K.; Bent, S.J.; Peters, A.; Raidal, S.R. Phylogeny of beak and feather disease virus in cockatoos demonstrates host generalism and multiple-variant infections within Psittaciformes. Virology 2014, 460–461, 72–82. [Google Scholar] [CrossRef]
- Das, S.; Sarker, S.; Peters, A.; Ghorashi, S.A.; Phalen, D.; Forwood, J.K.; Raidal, S.R. Evolution of circoviruses in lorikeets lags behind its hosts. Mol. Phylogenet. Evol. 2016, 100, 281–291. [Google Scholar] [CrossRef]
- Goldingay, R.L. Characteristics of tree hollows used by Australian birds and bats. Wildl. Res. 2009, 36, 394–409. [Google Scholar] [CrossRef]
- Sarker, S.; Moylan, K.G.; Ghorashi, S.A.; Forwood, J.K.; Peters, A.; Raidal, S.R. Evidence of a deep viral host switch event with beak and feather disease virus infection in rainbow bee-eaters (Merops ornatus). Sci Rep. 2015, 5, 14511. [Google Scholar] [CrossRef] [Green Version]
- Sarker, S.; Lloyd, C.; Forwood, J.; Raidal, S.R. Forensic genetic evidence of beak and feather disease virus infection in a Powerful Owl, Ninox strenua. Emu 2015, 116, 71–74. [Google Scholar] [CrossRef]
- Higgins, P.J. (Ed.) Laughing Kookaburra. Handbook of Australian, New Zealand, and Antarctic Birds; Oxford University Press: Melbourne, Australia, 1999; Volume 4. [Google Scholar]
- König, C.; Weick, F.; Becking, J.H. Owls of the World; Bloomsbury Publishing: London, UK, 2009. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kasimov, V.; Wille, M.; Sarker, S.; Dong, Y.; Shao, R.; Hall, C.; Potvin, D.; Conroy, G.; Valenza, L.; Gillett, A.; et al. Unexpected Pathogen Diversity Detected in Australian Avifauna Highlights Potential Biosecurity Challenges. Viruses 2023, 15, 143. https://doi.org/10.3390/v15010143
Kasimov V, Wille M, Sarker S, Dong Y, Shao R, Hall C, Potvin D, Conroy G, Valenza L, Gillett A, et al. Unexpected Pathogen Diversity Detected in Australian Avifauna Highlights Potential Biosecurity Challenges. Viruses. 2023; 15(1):143. https://doi.org/10.3390/v15010143
Chicago/Turabian StyleKasimov, Vasilli, Michelle Wille, Subir Sarker, Yalun Dong, Renfu Shao, Clancy Hall, Dominique Potvin, Gabriel Conroy, Ludovica Valenza, Amber Gillett, and et al. 2023. "Unexpected Pathogen Diversity Detected in Australian Avifauna Highlights Potential Biosecurity Challenges" Viruses 15, no. 1: 143. https://doi.org/10.3390/v15010143






