Differential Susceptibility of Fetal Retinal Pigment Epithelial Cells, hiPSC- Retinal Stem Cells, and Retinal Organoids to Zika Virus Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells
2.2. Differentiation of hiPSCs and Development of Retinal Organoids
2.3. Zika Virus
2.4. Zika Viral Infection
2.5. ZIKV Infection of Retinal Organoids
2.6. Caspase 3/7 Assay to Measure Apoptosis
2.7. Cell Viability Assay
2.8. Proteomic Sample Preparations and Liquid Chromatography Mass Spectrometry (LC-MS/MS) Analysis
2.9. Proteomic Bioinformatics
2.10. RNA Library Preparation and Sequencing
2.11. Analysis of RNA Sequencing Data
2.12. Reverse Transcription-Quantitative PCR Analysis
2.13. Antiviral Compound Analysis
2.14. Western Blot Analysis
2.15. Immunocytochemistry
2.16. Statistical Analysis
3. Results
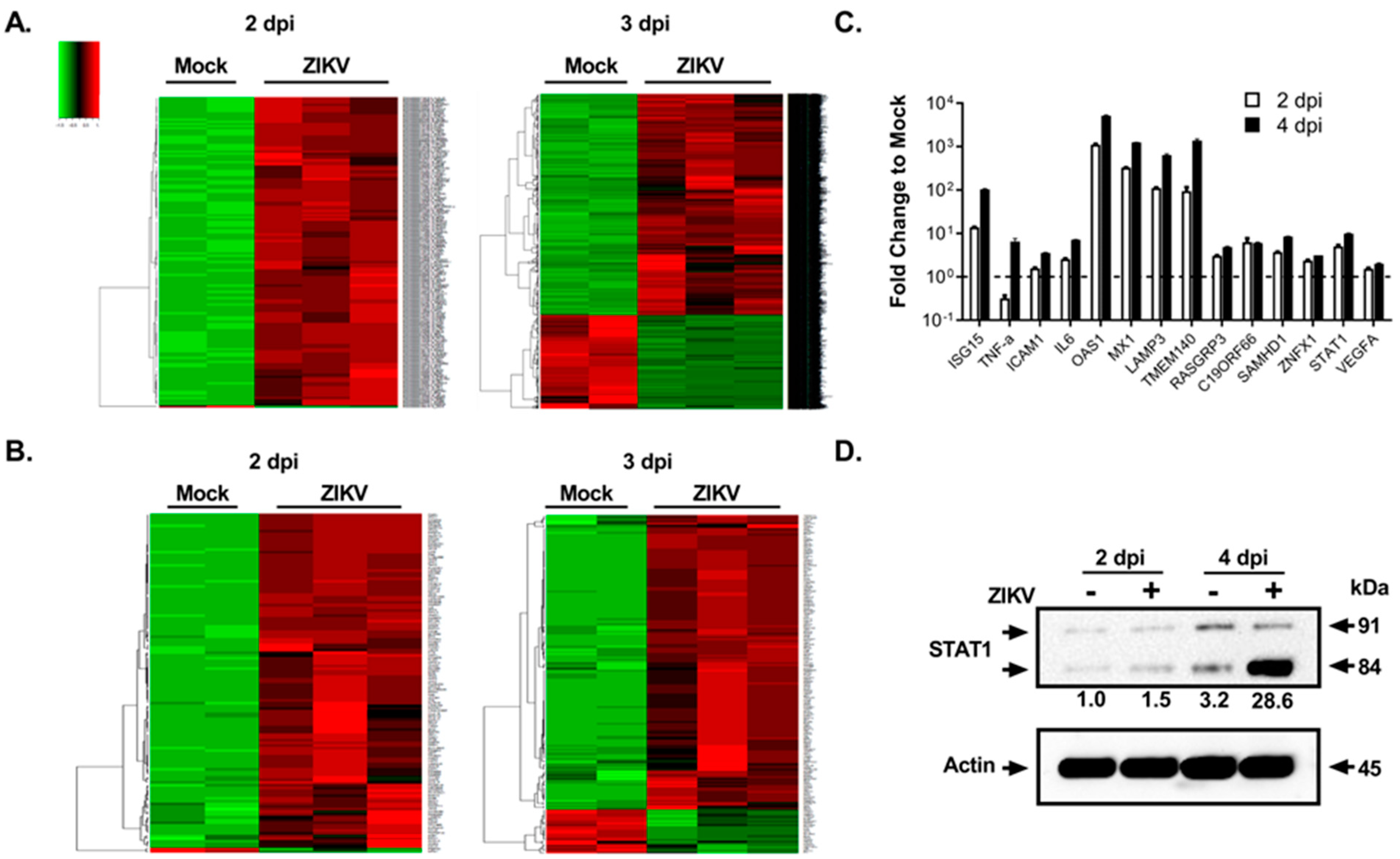
3.1. Fetal Retinal Pigment Epithelial Cells Are Susceptible to ZIKV Infection
3.2. Global Transcriptomics and Proteomics Analyses of ZIKV-Infected Fetal RPE Cells
3.3. Drug Treatment to Reduce ZIKV-Mediated Ocular Cell Death
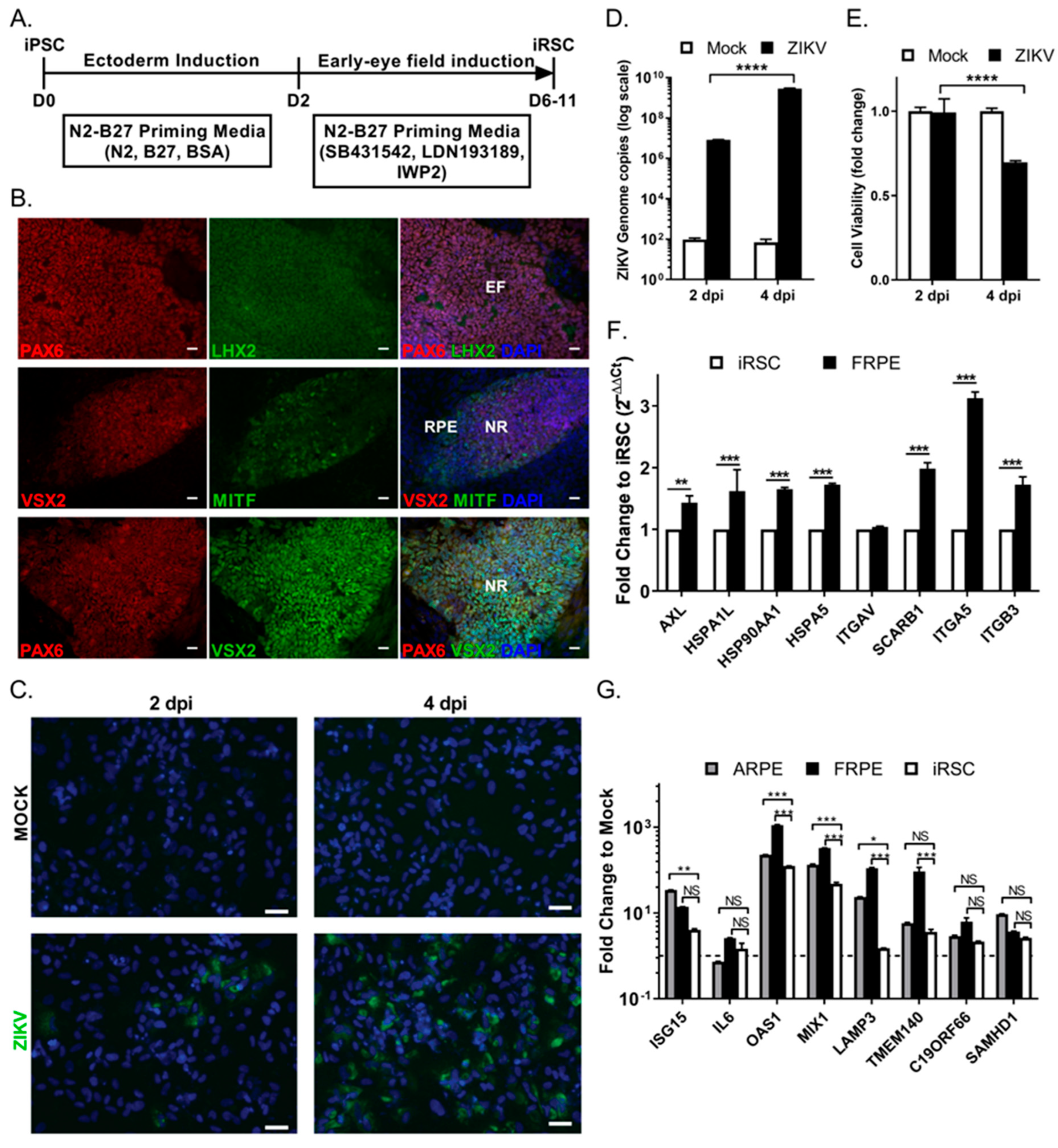
3.4. Human iPSC-Derived Retinal Stem Cells (iRSCs) Are Susceptible to ZIKV Infection
3.5. ZIKV Infects hiPSC-Derived 3-Dimensional Retinal Organoids and Dysregulates the Differentiation Program
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jampol, L.M.; Goldstein, D.A. Zika Virus Infection and the Eye. JAMA Ophthalmol. 2016, 134, 535–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brasil, P.; Pereira, J.P., Jr.; Moreira, M.E.; Ribeiro Nogueira, R.M.; Damasceno, L.; Wakimoto, M.; Rabello, R.S.; Valderramos, S.G.; Halai, U.A.; Salles, T.S.; et al. Zika Virus Infection in Pregnant Women in Rio de Janeiro. New Engl. J. Med. 2016, 375, 2321–2334. [Google Scholar] [CrossRef]
- Hamel, R.; Liegeois, F.; Wichit, S.; Pompon, J.; Diop, F.; Talignani, L.; Thomas, F.; Despres, P.; Yssel, H.; Misse, D. Zika virus: Epidemiology, clinical features and host-virus interactions. Microbes Infect. 2016, 18, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Shresta, S. Neuroteratogenic Viruses and Lessons for Zika Virus Models. Trends Microbiol. 2016, 24, 622–636. [Google Scholar] [CrossRef] [Green Version]
- De Paula Freitas, B.; de Oliveira Dias, J.R.; Prazeres, J.; Sacramento, G.A.; Ko, A.I.; Maia, M.; Belfort, R., Jr. Ocular Findings in Infants with Microcephaly Associated With Presumed Zika Virus Congenital Infection in Salvador, Brazil. JAMA Ophthalmol. 2016, 134, 529–535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miner, J.J.; Sene, A.; Richner, J.M.; Smith, A.M.; Santeford, A.; Ban, N.; Weger-Lucarelli, J.; Manzella, F.; Ruckert, C.; Govero, J.; et al. Zika Virus Infection in Mice Causes Panuveitis with Shedding of Virus in Tears. Cell Rep. 2016, 16, 3208–3218. [Google Scholar] [CrossRef] [Green Version]
- Dos Santos, T.; Rodriguez, A.; Almiron, M.; Sanhueza, A.; Ramon, P.; de Oliveira, W.K.; Coelho, G.E.; Badaro, R.; Cortez, J.; Ospina, M.; et al. Zika Virus and the Guillain-Barre Syndrome—Case Series from Seven Countries. New Engl. J. Med. 2016, 375, 1598–1601. [Google Scholar] [CrossRef]
- Lucchese, G.; Kanduc, D. Zika virus and autoimmunity: From microcephaly to Guillain-Barre syndrome, and beyond. Autoimmun. Rev. 2016, 15, 801–808. [Google Scholar] [CrossRef]
- Parra, B.; Lizarazo, J.; Jimenez-Arango, J.A.; Zea-Vera, A.F.; Gonzalez-Manrique, G.; Vargas, J.; Angarita, J.A.; Zuniga, G.; Lopez-Gonzalez, R.; Beltran, C.L.; et al. Guillain-Barre Syndrome Associated with Zika Virus Infection in Colombia. New Engl. J. Med. 2016, 375, 1513–1523. [Google Scholar] [CrossRef]
- Chen, B.Y.; Chang, H.H.; Chen, S.T.; Tsao, Z.J.; Yeh, S.M.; Wu, C.Y.; Lin, D.P. Congenital eye malformations associated with extensive periocular neural crest apoptosis after influenza B virus infection during early embryogenesis. Mol. Vis. 2009, 15, 2821–2828. [Google Scholar]
- Graw, J. The genetic and molecular basis of congenital eye defects. Nat. Rev. Genet. 2003, 4, 876–888. [Google Scholar] [CrossRef] [PubMed]
- Lupo, G.; Andreazzoli, M.; Gestri, G.; Liu, Y.; He, R.Q.; Barsacchi, G. Homeobox genes in the genetic control of eye development. Int. J. Dev. Biol. 2000, 44, 627–636. [Google Scholar] [PubMed]
- Ventura, C.V.; Maia, M.; Dias, N.; Ventura, L.O.; Belfort, R., Jr. Zika: Neurological and ocular findings in infant without microcephaly. Lancet 2016, 387, 2502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ventura, C.V.; Maia, M.; Travassos, S.B.; Martins, T.T.; Patriota, F.; Nunes, M.E.; Agra, C.; Torres, V.L.; van der Linden, V.; Ramos, R.C.; et al. Risk Factors Associated With the Ophthalmoscopic Findings Identified in Infants With Presumed Zika Virus Congenital Infection. JAMA Ophthalmol. 2016, 134, 912–918. [Google Scholar] [CrossRef]
- Vogel, G. Experts fear Zika’s effects may be even worse than thought. Science 2016, 352, 1375–1376. [Google Scholar] [CrossRef]
- Mohr, E.L.; Block, L.N.; Newman, C.M.; Stewart, L.M.; Koenig, M.; Semler, M.; Breitbach, M.E.; Teixeira, L.B.C.; Zeng, X.; Weiler, A.M.; et al. Ocular and uteroplacental pathology in a macaque pregnancy with congenital Zika virus infection. PLoS ONE 2018, 13, e0190617. [Google Scholar] [CrossRef] [Green Version]
- Rasmussen, S.A.; Jamieson, D.J.; Honein, M.A.; Petersen, L.R. Zika Virus and Birth Defects—Reviewing the Evidence for Causality. N. Engl. J. Med. 2016, 374, 1981–1987. [Google Scholar] [CrossRef]
- Ventura, C.V.; Ventura, L.O. Ophthalmologic Manifestations Associated With Zika Virus Infection. Pediatrics 2018, 141 (suppl. 2), S161–S166. [Google Scholar] [CrossRef] [Green Version]
- Cugola, F.R.; Fernandes, I.R.; Russo, F.B.; Freitas, B.C.; Dias, J.L.; Guimaraes, K.P.; Benazzato, C.; Almeida, N.; Pignatari, G.C.; Romero, S.; et al. The Brazilian Zika virus strain causes birth defects in experimental models. Nature 2016, 534, 267–271. [Google Scholar] [CrossRef] [Green Version]
- Lazear, H.M.; Govero, J.; Smith, A.M.; Platt, D.J.; Fernandez, E.; Miner, J.J.; Diamond, M.S. A Mouse Model of Zika Virus Pathogenesis. Cell Host Microbe 2016, 19, 720–730. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Xu, D.; Ye, Q.; Hong, S.; Jiang, Y.; Liu, X.; Zhang, N.; Shi, L.; Qin, C.F.; Xu, Z. Zika Virus Disrupts Neural Progenitor Development and Leads to Microcephaly in Mice. Cell Stem Cell 2016, 19, 672. [Google Scholar] [CrossRef] [PubMed]
- Miner, J.J.; Cao, B.; Govero, J.; Smith, A.M.; Fernandez, E.; Cabrera, O.H.; Garber, C.; Noll, M.; Klein, R.S.; Noguchi, K.K.; et al. Zika Virus Infection during Pregnancy in Mice Causes Placental Damage and Fetal Demise. Cell 2016, 165, 1081–1091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Shi, S.; Xia, F.; Shan, C.; Ha, Y.; Zou, J.; Adam, A.; Zhang, M.; Wang, T.; Liu, H.; et al. Zika virus induces neuronal and vascular degeneration in developing mouse retina. Acta Neuropathol. Commun. 2021, 9, 97. [Google Scholar] [CrossRef] [PubMed]
- Garcia, G., Jr.; Paul, S.; Beshara, S.; Ramanujan, V.K.; Ramaiah, A.; Nielsen-Saines, K.; Li, M.M.H.; French, S.W.; Morizono, K.; Kumar, A.; et al. Hippo Signaling Pathway Has a Critical Role in Zika Virus Replication and in the Pathogenesis of Neuroinflammation. Am. J. Pathol. 2020, 190, 844–861. [Google Scholar] [CrossRef]
- Martinez, L.E.; Garcia, G., Jr.; Contreras, D.; Gong, D.; Sun, R.; Arumugaswami, V. Zika Virus Mucosal Infection Provides Protective Immunity. J. Virol. 2020, 94, e00067-20. [Google Scholar] [CrossRef]
- Miner, J.J.; Diamond, M.S. Zika Virus Pathogenesis and Tissue Tropism. Cell Host Microbe 2017, 21, 134–142. [Google Scholar] [CrossRef] [Green Version]
- Maleski, A.L.A.; Rosa, J.G.S.; Bernardo, J.T.G.; Astray, R.M.; Walker, C.I.B.; Lopes-Ferreira, M.; Lima, C. Recapitulation of Retinal Damage in Zebrafish Larvae Infected with Zika Virus. Cells 2022, 11, 1457. [Google Scholar] [CrossRef]
- Salinas, S.; Erkilic, N.; Damodar, K.; Moles, J.P.; Fournier-Wirth, C.; Van de Perre, P.; Kalatzis, V.; Simonin, Y. Zika Virus Efficiently Replicates in Human Retinal Epithelium and Disturbs Its Permeability. J. Virol. 2017, 91, e02144-16. [Google Scholar] [CrossRef] [Green Version]
- Singh, P.K.; Khatri, I.; Jha, A.; Pretto, C.D.; Spindler, K.R.; Arumugaswami, V.; Giri, S.; Kumar, A.; Bhasin, M.K. Determination of system level alterations in host transcriptome due to Zika virus (ZIKV) Infection in retinal pigment epithelium. Sci. Rep. 2018, 8, 11209. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.J.; Afshari, N.A. Generation of Human Corneal Endothelial Cells via In Vitro Ocular Lineage Restriction of Pluripotent Stem Cells. Investig. Ophthalmol. Vis. Sci. 2016, 57, 6878–6884. [Google Scholar] [CrossRef] [Green Version]
- Zhong, X.; Gutierrez, C.; Xue, T.; Hampton, C.; Vergara, M.N.; Cao, L.H.; Peters, A.; Park, T.S.; Zambidis, E.T.; Meyer, J.S.; et al. Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat. Commun. 2014, 5, 4047. [Google Scholar] [CrossRef] [PubMed]
- Contreras, D.; Arumugaswami, V. Zika Virus Infectious Cell Culture System and the In Vitro Prophylactic Effect of Interferons. J. Vis. Exp. 2016, 114, e54767. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, E.W.; Mendoza, L.; Shteynberg, D.; Farrah, T.; Lam, H.; Tasman, N.; Sun, Z.; Nilsson, E.; Pratt, B.; Prazen, B.; et al. A guided tour of the Trans-Proteomic Pipeline. Proteomics 2010, 10, 1150–1159. [Google Scholar] [CrossRef] [Green Version]
- Keller, A.; Eng, J.; Zhang, N.; Li, X.J.; Aebersold, R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol. 2005, 1, 2005-0017. [Google Scholar] [CrossRef]
- MacLean, B.; Tomazela, D.M.; Shulman, N.; Chambers, M.; Finney, G.L.; Frewen, B.; Kern, R.; Tabb, D.L.; Liebler, D.C.; MacCoss, M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [Green Version]
- Gencode. Available online: http://www.gencodegenes.org (accessed on 14 November 2022).
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controling the false discovery rate: A practical and powerful approach to rapid testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar]
- DAVID Bioinformatics Resources. Available online: https://david.ncifcrf.gov (accessed on 14 November 2022).
- QIAGEN Ingenuity Pathway Analysis (IPA). Available online: http://www.ingenuity.com/products/ipa (accessed on 14 November 2022).
- Grant, A.; Ponia, S.S.; Tripathi, S.; Balasubramaniam, V.; Miorin, L.; Sourisseau, M.; Schwarz, M.C.; Sanchez-Seco, M.P.; Evans, M.J.; Best, S.M.; et al. Zika Virus Targets Human STAT2 to Inhibit Type I Interferon Signaling. Cell Host Microbe 2016, 19, 882–890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morrey, J.D.; Smee, D.F.; Sidwell, R.W.; Tseng, C. Identification of active antiviral compounds against a New York isolate of West Nile virus. Antivir. Res. 2002, 55, 107–116. [Google Scholar] [CrossRef]
- Shrestha, R.; Wen, Y.T.; Ding, D.C.; Tsai, R.K. Aberrant hiPSCs-Derived from Human Keratinocytes Differentiates into 3D Retinal Organoids that Acquire Mature Photoreceptors. Cells 2019, 8, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowe, A.; Harris, R.; Bhansali, P.; Cvekl, A.; Liu, W. Intercellular Adhesion-Dependent Cell Survival and ROCK-Regulated Actomyosin-Driven Forces Mediate Self-Formation of a Retinal Organoid. Stem Cell Rep. 2016, 6, 743–756. [Google Scholar] [CrossRef] [PubMed]
- Reichman, S.; Terray, A.; Slembrouck, A.; Nanteau, C.; Orieux, G.; Habeler, W.; Nandrot, E.F.; Sahel, J.A.; Monville, C.; Goureau, O. From confluent human iPS cells to self-forming neural retina and retinal pigmented epithelium. Proc. Natl. Acad. Sci. USA 2014, 111, 8518–8523. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Jin, Z.B. Directed Induction of Retinal Organoids from Human Pluripotent Stem Cells. J. Vis. Exp. 2021, 170, e62298. [Google Scholar] [CrossRef]
- Reh, T.A.; Fischer, A.J. Retinal stem cells. Methods Enzymol. 2006, 419, 52–73. [Google Scholar] [CrossRef]
- Volkner, M.; Zschatzsch, M.; Rostovskaya, M.; Overall, R.W.; Busskamp, V.; Anastassiadis, K.; Karl, M.O. Retinal Organoids from Pluripotent Stem Cells Efficiently Recapitulate Retinogenesis. Stem Cell Rep. 2016, 6, 525–538. [Google Scholar] [CrossRef] [Green Version]
- Parameswaran, S.; Balasubramanian, S.; Babai, N.; Qiu, F.; Eudy, J.D.; Thoreson, W.B.; Ahmad, I. Induced pluripotent stem cells generate both retinal ganglion cells and photoreceptors: Therapeutic implications in degenerative changes in glaucoma and age-related macular degeneration. Stem Cells 2010, 28, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Osakada, F.; Ikeda, H.; Mandai, M.; Wataya, T.; Watanabe, K.; Yoshimura, N.; Akaike, A.; Sasai, Y.; Takahashi, M. Toward the generation of rod and cone photoreceptors from mouse, monkey and human embryonic stem cells. Nat. Biotechnol. 2008, 26, 215–224. [Google Scholar] [CrossRef]
- Nakano, T.; Ando, S.; Takata, N.; Kawada, M.; Muguruma, K.; Sekiguchi, K.; Saito, K.; Yonemura, S.; Eiraku, M.; Sasai, Y. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 2012, 10, 771–785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGrath, E.L.; Rossi, S.L.; Gao, J.; Widen, S.G.; Grant, A.C.; Dunn, T.J.; Azar, S.R.; Roundy, C.M.; Xiong, Y.; Prusak, D.J.; et al. Differential Responses of Human Fetal Brain Neural Stem Cells to Zika Virus Infection. Stem Cell Rep. 2017, 8, 715–727. [Google Scholar] [CrossRef]
- Tang, H.; Hammack, C.; Ogden, S.C.; Wen, Z.; Qian, X.; Li, Y.; Yao, B.; Shin, J.; Zhang, F.; Lee, E.M.; et al. Zika Virus Infects Human Cortical Neural Progenitors and Attenuates Their Growth. Cell Stem Cell 2016, 18, 587–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Souza, B.S.; Sampaio, G.L.; Pereira, C.S.; Campos, G.S.; Sardi, S.I.; Freitas, L.A.; Figueira, C.P.; Paredes, B.D.; Nonaka, C.K.; Azevedo, C.M.; et al. Zika virus infection induces mitosis abnormalities and apoptotic cell death of human neural progenitor cells. Sci. Rep. 2016, 6, 39775. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Buth, J.E.; Vishlaghi, N.; de la Torre-Ubieta, L.; Taxidis, J.; Khakh, B.S.; Coppola, G.; Pearson, C.A.; Yamauchi, K.; Gong, D.; et al. Self-Organized Cerebral Organoids with Human-Specific Features Predict Effective Drugs to Combat Zika Virus Infection. Cell Rep. 2017, 21, 517–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roulston, A.; Marcellus, R.C.; Branton, P.E. Viruses and apoptosis. Annu. Rev. Microbiol. 1999, 53, 577–628. [Google Scholar] [CrossRef]
- Jurado, K.A.; Simoni, M.K.; Tang, Z.; Uraki, R.; Hwang, J.; Householder, S.; Wu, M.; Lindenbach, B.D.; Abrahams, V.M.; Guller, S.; et al. Zika virus productively infects primary human placenta-specific macrophages. JCI Insight 2016, 1, e88461. [Google Scholar] [CrossRef] [Green Version]
- Hottz, E.D.; Oliveira, M.F.; Nunes, P.C.; Nogueira, R.M.; Valls-de-Souza, R.; Da Poian, A.T.; Weyrich, A.S.; Zimmerman, G.A.; Bozza, P.T.; Bozza, F.A. Dengue induces platelet activation, mitochondrial dysfunction and cell death through mechanisms that involve DC-SIGN and caspases. J. Thromb. Haemost. 2013, 11, 951–962. [Google Scholar] [CrossRef] [Green Version]
- Ornelas, A.M.M.; Paula, P.; Silveira, P.P.; Melo, F.O.; Ferreira, T.A.; Oliveira-Szejnfeld, P.S.; Leal, J.I.; Amorim, M.M.R.; Hamilton, S.; Rawlinson, W.D.; et al. Immune activation in amniotic fluid from Zika virus associated microcephaly. Ann. Neurol. 2016, 81, 152–156. [Google Scholar] [CrossRef]
- Ford, K.M.; Saint-Geniez, M.; Walshe, T.; Zahr, A.; D’Amore, P.A. Expression and role of VEGF in the adult retinal pigment epithelium. Investig. Ophthalmol. Vis. Sci. 2011, 52, 9478–9487. [Google Scholar] [CrossRef] [Green Version]
- Nagineni, C.N.; Kommineni, V.K.; William, A.; Detrick, B.; Hooks, J.J. Regulation of VEGF expression in human retinal cells by cytokines: Implications for the role of inflammation in age-related macular degeneration. J. Cell. Physiol. 2012, 227, 116–126. [Google Scholar] [CrossRef]
- Eichmann, A.; Simons, M. VEGF signaling inside vascular endothelial cells and beyond. Curr. Opin. Cell Biol. 2012, 24, 188–193. [Google Scholar] [CrossRef] [Green Version]
- Kovach, J.L.; Schwartz, S.G.; Flynn, H.W., Jr.; Scott, I.U. Anti-VEGF Treatment Strategies for Wet AMD. J. Ophthalmol. 2012, 2012, 786870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crance, J.M.; Scaramozzino, N.; Jouan, A.; Garin, D. Interferon, ribavirin, 6-azauridine and glycyrrhizin: Antiviral compounds active against pathogenic flaviviruses. Antivir. Res. 2003, 58, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Pascoalino, B.S.; Courtemanche, G.; Cordeiro, M.T.; Gil, L.H.; Freitas-Junior, L. Zika antiviral chemotherapy: Identification of drugs and promising starting points for drug discovery from an FDA-approved library. F1000Research 2016, 5, 2523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adcock, R.S.; Chu, Y.K.; Golden, J.E.; Chung, D.H. Evaluation of anti-Zika virus activities of broad-spectrum antivirals and NIH clinical collection compounds using a cell-based, high-throughput screen assay. Antivir. Res. 2017, 138, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Lee, E.M.; Wen, Z.; Cheng, Y.; Huang, W.K.; Qian, X.; Tcw, J.; Kouznetsova, J.; Ogden, S.C.; Hammack, C.; et al. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat. Med. 2016, 22, 1101–1107. [Google Scholar] [CrossRef]
- Micewicz, E.D.; Khachatoorian, R.; French, S.W.; Ruchala, P. Identification of novel small-molecule inhibitors of Zika virus infection. Bioorg. Med. Chem. Lett. 2018, 28, 452–458. [Google Scholar] [CrossRef]
- Baz, M.; Boivin, G. Antiviral Agents in Development for Zika Virus Infections. Pharmaceuticals 2019, 12, 101. [Google Scholar] [CrossRef] [Green Version]
- Shrestha, R.; Wen, Y.T.; Tsai, R.K. Effective Differentiation and Biological Characterization of Retinal Pigment Epithelium Derived from Human Induced Pluripotent Stem Cells. Curr. Eye Res. 2020, 45, 1155–1167. [Google Scholar] [CrossRef]
- Osakada, F.; Jin, Z.B.; Hirami, Y.; Ikeda, H.; Danjyo, T.; Watanabe, K.; Sasai, Y.; Takahashi, M. In vitro differentiation of retinal cells from human pluripotent stem cells by small-molecule induction. J. Cell Sci. 2009, 122, 3169–3179. [Google Scholar] [CrossRef] [Green Version]
- Reichman, S.; Slembrouck, A.; Gagliardi, G.; Chaffiol, A.; Terray, A.; Nanteau, C.; Potey, A.; Belle, M.; Rabesandratana, O.; Duebel, J.; et al. Generation of Storable Retinal Organoids and Retinal Pigmented Epithelium from Adherent Human iPS Cells in Xeno-Free and Feeder-Free Conditions. Stem Cells 2017, 35, 1176–1188. [Google Scholar] [CrossRef] [Green Version]
- Kamao, H.; Mandai, M.; Okamoto, S.; Sakai, N.; Suga, A.; Sugita, S.; Kiryu, J.; Takahashi, M. Characterization of human induced pluripotent stem cell-derived retinal pigment epithelium cell sheets aiming for clinical application. Stem Cell Rep. 2014, 2, 205–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maeda, T.; Lee, M.J.; Palczewska, G.; Marsili, S.; Tesar, P.J.; Palczewski, K.; Takahashi, M.; Maeda, A. Retinal pigmented epithelial cells obtained from human induced pluripotent stem cells possess functional visual cycle enzymes in vitro and in vivo. J. Biol. Chem. 2013, 288, 34484–34493. [Google Scholar] [CrossRef] [PubMed]
- Simonin, Y.; Erkilic, N.; Damodar, K.; Clé, M.; Desmetz, C.; Bolloré, K.; Taleb, M.; Torriano, S.; Barthelemy, J.; Dubois, G.; et al. Zika virus induces strong inflammatory responses and impairs homeostasis and function of the human retinal pigment epithelium. EBioMedicine 2019, 39, 315–331. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Contreras, D.; Garcia, G., Jr.; Jones, M.K.; Martinez, L.E.; Jayakarunakaran, A.; Gangalapudi, V.; Tang, J.; Wu, Y.; Zhao, J.J.; Chen, Z.; et al. Differential Susceptibility of Fetal Retinal Pigment Epithelial Cells, hiPSC- Retinal Stem Cells, and Retinal Organoids to Zika Virus Infection. Viruses 2023, 15, 142. https://doi.org/10.3390/v15010142
Contreras D, Garcia G Jr., Jones MK, Martinez LE, Jayakarunakaran A, Gangalapudi V, Tang J, Wu Y, Zhao JJ, Chen Z, et al. Differential Susceptibility of Fetal Retinal Pigment Epithelial Cells, hiPSC- Retinal Stem Cells, and Retinal Organoids to Zika Virus Infection. Viruses. 2023; 15(1):142. https://doi.org/10.3390/v15010142
Chicago/Turabian StyleContreras, Deisy, Gustavo Garcia, Jr., Melissa Kaye Jones, Laura E. Martinez, Akshaya Jayakarunakaran, Vineela Gangalapudi, Jie Tang, Ying Wu, Jiagang J. Zhao, Zhaohui Chen, and et al. 2023. "Differential Susceptibility of Fetal Retinal Pigment Epithelial Cells, hiPSC- Retinal Stem Cells, and Retinal Organoids to Zika Virus Infection" Viruses 15, no. 1: 142. https://doi.org/10.3390/v15010142




