1. Introduction
Hepatitis C virus (HCV) infection is a major public health problem, with a recent report estimating that approximately 70 million individuals are chronically infected with HCV worldwide [
1]. There are 7 HCV genotypes, with genotype 1 being the most prevalent globally [
2,
3]. Depending on various risk factors, patients with chronic HCV infection can develop serious liver diseases including cirrhosis and hepatocellular carcinoma. Sustained virologic response resulting from treatment of chronic HCV infection has been shown to be associated with a significant reduction in liver disease progression, development of hepatocellular carcinoma and mortality [
4,
5].
A number of HCV direct acting antivirals (DAAs) have been approved since 2011. These DAAs include inhibitors of HCV NS3/4A protease, NS5A protein, and NS5B polymerase [
6,
7]. Current HCV treatment regimens are generally interferon-sparing, and include 2 or 3 DAAs with different mechanisms of action with and without the addition of ribavirin (RBV). Although newer DAA combinations are highly efficacious in many HCV-infected populations, a number of them are not effective for all HCV genotypes [
8,
9,
10,
11]; some of them have reduced activity against certain baseline resistance-associated polymorphisms [
12,
13,
14,
15], and DAA resistance-conferring substitutions often emerge in patients who experience virologic failure [
11,
16,
17,
18]. There have been major efforts to discover and develop next generation interferon- and RBV-sparing DAA regimens that are convenient to take (once daily oral administration), efficacious in patients infected with all major HCV genotypes, cover broad patient populations, and exhibit a high barrier to the development of DAA resistance. The fixed-dose combination regimen of glecaprevir and pibrentasvir, recently approved in Europe, the United States, Canada, Japan and many other countries for the treatment of chronic HCV infection, is one of the few regimens that has all of these features.
Glecaprevir (ABT-493, an NS3/4A protease inhibitor identified by AbbVie and Enanta) and pibrentasvir (ABT-530, an NS5A inhibitor identified by AbbVie) are next-generation HCV inhibitors with potent antiviral activity against all major HCV genotypes in vitro, with little or no loss in activity against common single-position amino acid substitutions that confer resistance to other NS3/4A protease inhibitors or NS5A inhibitors, respectively [
19,
20]. A Phase 2a clinical study was conducted to assess the antiviral activity, safety, tolerability, and pharmacokinetics of multiple dose levels of glecaprevir or pibrentasvir administered as monotherapy for 3 days in treatment-naïve adults with chronic HCV genotype 1 infection with or without compensated cirrhosis [
21]. Monotherapy with glecaprevir or pibrentasvir was well tolerated, and resulted in mean maximal decreases of approximately 4 log
10 IU/mL from baseline in HCV RNA levels in patients with most of the doses evaluated. The monotherapy was followed by a 12-week treatment with ombitasvir/paritaprevir/ritonavir plus dasabuvir and weight-based RBV. This report describes the prevalence of baseline polymorphisms and treatment-emergent substitutions in NS3 or NS5A in samples from patients who received 3-day monotherapy of either glecaprevir or pibrentasvir, respectively. In addition, the changes in susceptibility conferred by these baseline polymorphisms and treatment-emergent substitutions in NS3 or NS5A to glecaprevir or pibrentasvir, respectively, were determined.
4. Discussion
In this study, resistance analysis was conducted on all baseline and post-baseline samples with sufficient viral titer from genotype 1-infected patients who received 3 days of glecaprevir or pibrentasvir monotherapy. Baseline NS3 or NS5A polymorphisms detected in these patients had no or minimal (≤7-fold reduction in activity) impact on the susceptibility of HCV to glecaprevir or pibrentasvir, respectively, in vitro. The frequency of patients with treatment-emergent resistance-conferring substitutions detected in glecaprevir or pibrentasvir monotherapy study was much lower than those in monotherapy studies with other inhibitors in the respective HCV inhibitor class [
22,
23,
25,
26,
30,
33,
34,
35,
36,
37]. All of the treatment-emergent resistance-conferring NS5A substitutions identified during pibrentasvir monotherapy were linked multiple-position amino acid substitutions. Replicons engineered with the resistance-conferring NS3 or NS5A substitutions that emerged in this monotherapy study had substantially lower replication efficiency relative to the wild-type HCV replicon. Glecaprevir and pibrentasvir demonstrated no loss in activity or less reduction in activity than other members of the respective inhibitor class against common resistance-conferring substitutions in NS3 and NS5A, respectively. Findings in this resistance analysis contributed to the explanation of the favorable HCV viral load declines in patients receiving glecaprevir or pibrentasvir monotherapy.
Baseline samples from 26 out of 47 patients who received glecaprevir monotherapy had NS3 polymorphisms but none of these polymorphisms conferred resistance to glecaprevir in vitro. The most prevalent baseline NS3 polymorphism detected in this study was genotype 1a Q80K, a common NS3 polymorphism that is known to be associated with reduced efficacy with regimens containing simeprevir [
14], voxilaprevir [
38], or other NS3/4A protease inhibitors [
27]; genotype 1a Q80K does not reduce susceptibility to glecaprevir in vitro. Among the 35 patients with post-baseline sequence data available, only 1 treatment-emergent NS3 substitution that conferred resistance to glecaprevir (A156T) was detected in a genotype 1a-infected patient receiving glecaprevir monotherapy. The presence of A156T at 19% prevalence in this patient’s viral population on visit day 3 did not have an apparent impact on the patient’s viral RNA decline [
21]. The NS3 resistance profile of post-baseline samples from genotype 1-infected patients receiving glecaprevir monotherapy was in agreement with the results of in vitro resistance selection study with glecaprevir in HCV genotype 1 replicon cells: The only resistance-conferring NS3 substitutions selected by glecaprevir in vitro were A156T and A156V [
20]. Consistent with the rare emergence of resistance-conferring NS3 substitutions during glecaprevir monotherapy described in this report, only 1 out of 703 (0.14%) treatment-naive genotype 1-infected patients receiving the combination regimen of glecaprevir and pibrentasvir in the Phase 3 ENDURANCE-1 study experienced virologic failure: This patient was infected with genotype 1a HCV and had a treatment-emergent A156V substitution in NS3 [
39].
Treatment-emergent resistance-conferring substitutions in NS3 were less commonly detected in genotype 1-infected patients treated with monotherapy of glecaprevir than other approved HCV NS3/4A protease inhibitors [
22,
34,
35,
36,
37]. Approved HCV protease inhibitors, including telaprevir, boceprevir, simeprevir, asunaprevir, paritaprevir, grazoprevir and voxilaprevir, have reduced activity against genotype 1 HCV with substitutions at NS3 amino acid positions 155, 156 and/or 168 in vitro [
16,
22,
34,
35,
36,
37,
40,
41]. These substitutions also emerged in patients who were treated with each of these protease inhibitors as monotherapy, or patients experiencing virologic failure with combination HCV regimens containing one of these protease inhibitors. For example, in clinical studies with patients infected with HCV genotype 1, substitutions at amino acid positions 155 and 156 emerged in patients treated with voxilaprevir monotherapy [
37] whereas substitutions at amino acid positions 155, 156 and 168 emerged in patients who experienced virologic failure with various regimens containing grazoprevir [
40,
41]. The most common NS3 amino acid substitutions that emerged during paritaprevir monotherapy were R155K and D168V in genotype 1a and D168V in genotype 1b [
22]. A156T was the only resistance-conferring NS3 substitution found to emerge in a single patient in the glecaprevir monotherapy study. The replication efficiency of replicons engineered with substitutions at amino acid A156 was greatly impaired in vitro [
20]. Therefore, the reported prevalence of A156 substitutions as baseline polymorphisms was low [
20] and even if these substitutions emerged in patients treated with regimens containing a protease inhibitor, they became undetectable in as short as 3 months after the patients experienced virologic failure [
41].
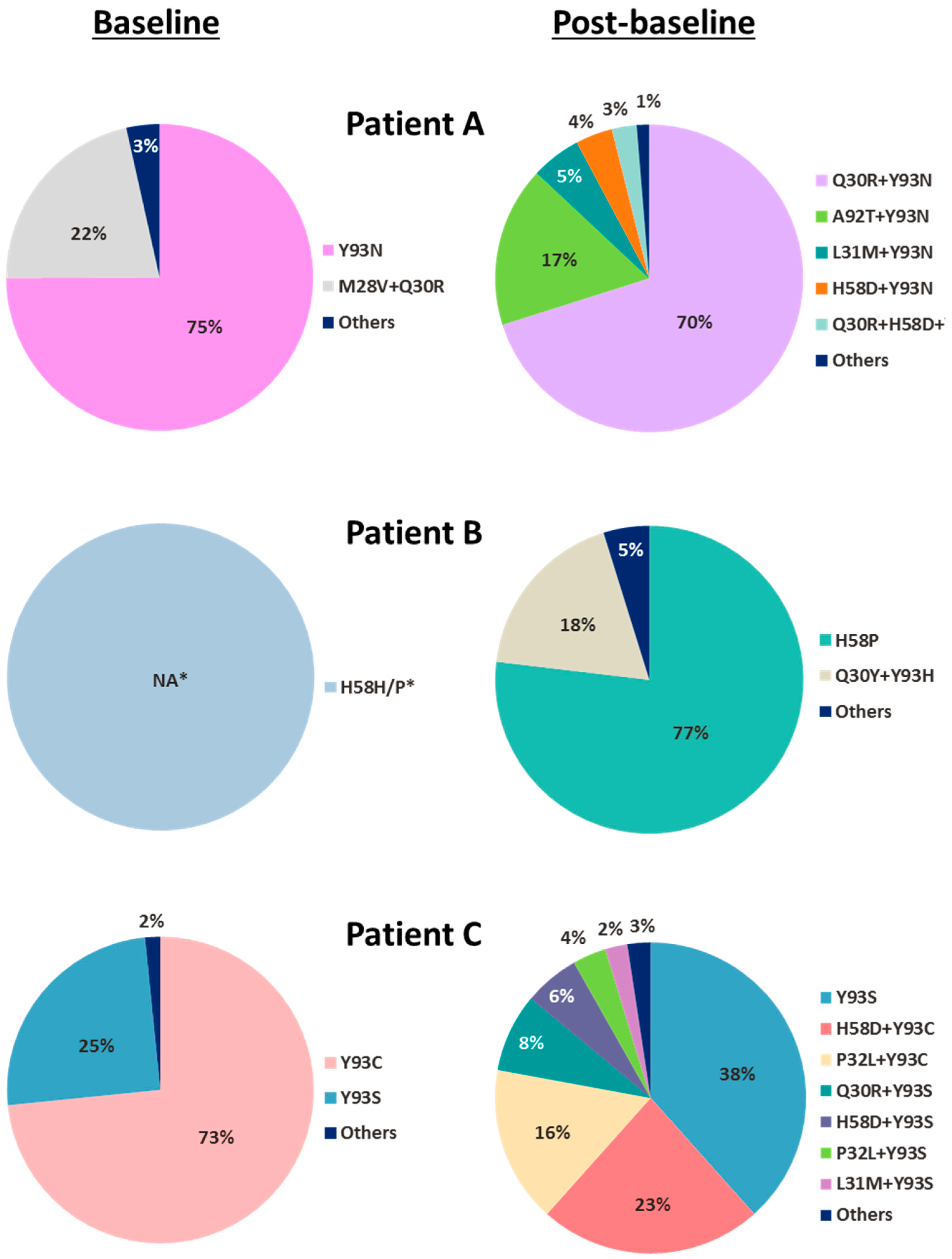
Twelve out of the 40 patients who received pibrentasvir monotherapy had baseline NS5A polymorphisms; 4 of these 12 patients had multiple NS5A polymorphisms. Patients with baseline NS5A polymorphisms had similar viral RNA declines as patients without any NS5A polymorphisms during pibrentasvir monotherapy with the exception of Patient A [
21], whose slightly less robust viral response on visit day 4 was likely due to the presence of treatment-emergent NS5A substitutions rather than that of the baseline NS5A polymorphisms. In studies with HCV genotype 1 replicons, single-position NS5A substitutions did not generally reduce susceptibility to pibrentasvir, and only certain combinations of double-or triple-position NS5A substitutions were shown to confer resistance to pibrentasvir [
19]. Therefore, it is not surprising that almost all of the patients with single or multiple NS5A baseline polymorphisms demonstrated robust viral declines with pibrentasvir monotherapy [
21]. Unlike other NS5A inhibitors, which in monotherapy studies selected both single-and multiple-position NS5A substitutions that conferred resistance to the respective NS5A inhibitors [
23,
25,
30,
33], pibrentasvir did not select any single-position NS5A substitutions during monotherapy. Among the 19 patients with available post-baseline sequence data, 3 genotype 1a-infected patients, all with pre-existing substitutions in NS5A at baseline, had treatment-emergent substitutions in NS5A during pibrentasvir monotherapy. All of the treatment-emergent substitutions found in these 3 patients were linked multiple-position, almost exclusively double-position, NS5A substitutions. The impact of these treatment-emergent NS5A substitutions on viral load declines in these 3 patients receiving pibrentasvir monotherapy appeared to range from low to none at the time points monitored [
21].
The frequency of patients with treatment-emergent NS5A substitutions, all multiple-position substitutions, was low among patients receiving pibrentasvir monotherapy. This could be due to (a) the high genetic barrier to the generation of each of the multiple-position NS5A substitutions, and (b) the low viral fitness of HCV with each of these multiple-position NS5A substitutions. Consistent with the pattern of the emergence of multiple-position NS5A substitutions observed during pibrentasvir monotherapy, the single genotype 1-infected patient who experienced virologic failure in the Phase 3 ENDURANCE-1 study (combination regimen of glecaprevir and pibrentasvir) as mentioned above had the linked triple-position NS5A substitution Q30R + L31M + H58D detectable at the time of virologic failure [
39].
When tested against HCV replicons engineered with NS5A substitutions known to confer resistance to other members of the NS5A inhibitor class, pibrentasvir demonstrated no loss in activity or less reduction in activity than other approved NS5A inhibitors which included daclatasvir, ombitasvir, elbasvir, ledipasvir and velpatasvir. In monotherapy studies in genotype 1-infected patients, significantly fewer patients had treatment-emergent NS5A substitutions with pibrentasvir monotherapy than with other NS5A inhibitors [
23,
25,
26,
30,
33]. For example, treatment-emergent NS5A substitutions at amino acids M28, Q30, L31 and/or Y93 were detected by population sequencing in the majority of available post-baseline samples from genotype 1-infected patients (
n = 20) receiving 5-day elbasvir monotherapy [
26]. In particular, treatment-emergent substitutions at amino acid Y93 were the most prevalent substitutions (present in approximately 80% of available post-baseline samples from these patients) with elbasvir monotherapy, including Y93H and Y93N in genotype 1a (201- to 605-fold resistant to elbasvir,
Table 6) as well as Y93H in genotype 1b (5-fold resistant to elbasvir,
Table 6) [
26]. In a 3-day monotherapy study with ledipasvir, treatment-emergent NS5A substitutions were detected by population sequencing on Day 4 in 100% of genotype 1-infected patients receiving ≥3 mg of ledipasvir (
n = 41), with the most prevalent substitutions being Q30R and L31M in genotype 1a, as well as Y93H in genotype 1b [
25], each of which confers >300-fold resistance to ledipasvir (
Table 6). For the 3-day monotherapy study with ombitasvir, treatment-emergent NS5A amino acid substitutions were detected by clonal sequencing in 100% of available post-baseline samples from day 3 and day 6 visits of genotype 1a-infected patients (
n = 8) receiving different doses of ombitasvir [
23]. The predominant treatment-emergent NS5A substitutions were M28T, M28V and Q30R (58- to 8965-fold resistant to ombitasvir), while Y93C and Y93H (both >1600-fold resistant to ombitasvir) were the minor substitutions [
23]. In addition, in a 3-day monotherapy with velpatasvir in genotype 1-infected patients, treatment-emergent substitutions in NS5A were detected by next-generation sequencing (NGS) at 1% sensitivity cutoff in 100% (10/10) or 100% (40/40) available “Days 2–10” post-baseline samples from patients with or without baseline NS5A polymorphisms, respectively [
30]. The predominant treatment-emergent genotype 1 NS5A substitutions detected in velpatasvir monotherapy were genotype 1a Y93H (≥400-fold resistant to velpatasvir,
Table 6) and Y93N (≥3000-fold resistant to velpatasvir,
Table 6), seen in 67% (6/9) and 56% (5/9), respectively, of Day 5 samples collected from patients without baseline NS5A polymorphisms. In the current study, after the 3-day monotherapy with pibrentasvir in genotype 1-infected patients, treatment-emergent NS5A substitutions were detected by population sequencing in only 16% (3/19) of patients with post-baseline samples that had sufficient viral titer to be sequenced. The presence of treatment-emergent NS5A substitutions, if any, in post-baseline samples from the other 21 patients receiving pibrentasvir monotherapy could not be determined due to the rapid declines of HCV viral RNA in these patients. Importantly, while NS5A Y93H in genotype 1, especially in genotype 1a, is a common treatment-emergent single-position substitution observed in patients receiving monotherapy of other NS5A inhibitors or patients who experienced virologic failure with regimens containing other NS5A inhibitors [
11,
16,
17,
23,
25,
26,
27,
28,
29,
30,
33], the Y93H substitution did not emerge in genotype 1-infected patients receiving pibrentasvir monotherapy.
Both population sequencing and NGS with 15% sensitivity cutoff are regarded as specific and sensitive sequencing methods to identify substitutions that can impact clinical responses to HCV DAAs [
29,
42,
43]. Monotherapy resistance analyses for NS5A inhibitors were generally done by population sequencing [
25,
26,
33] and the analysis for velpatasvir monotherapy was done by NGS [
30]. Despite the differences in study designs and/or sequence analysis methods between the monotherapy studies with pibrentasvir and the other approved NS5A inhibitors, the low number of genotype 1-infected patients with treatment-emergent NS5A substitutions for pibrentasvir detected by population sequencing in monotherapy samples is consistent with the high cure rate observed in HCV-infected patients treated with the combination regimen of glecaprevir and pibrentasvir as discussed below.
The favorable monotherapy resistance profiles of glecaprevir and pibrentasvir in genotype 1-infected patients have translated into robust clinical efficacy with the combination of glecaprevir and pibrentasvir in patients chronically infected with HCV genotypes 1–6. In Phase 2b/3 registration studies, the overall rate of sustained virologic response 12 weeks after the end of treatment (SVR12) was 98% with the combination of 300 mg glecaprevir and 120 mg pibrentasvir for 8, 12 or 16 weeks in 2256 patients infected with HCV genotypes 1–6, including patients who were treatment-naïve or treatment-experienced (pegIFN, RBV and/or sofosbuvir) with compensated cirrhosis or without cirrhosis [
44]. Additional studies also showed that the combination regimen of glecaprevir and pibrentasvir was efficacious in patients with HIV-1 co-infection [
39] or chronic kidney disease [
45].