The ND10 Complex Represses Lytic Human Herpesvirus 6A Replication and Promotes Silencing of the Viral Genome
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Immunofluorescence
2.3. Lentivirus Production and Transduction
2.4. Western Blotting
2.5. CellVue Infection Assay
2.6. Quantification of HHV-6A Genome Copies
2.7. Fluorescence In Situ Hybridization (FISH)
2.8. Late Gene Reporter Assay
2.9. Statistical Analyses
3. Results
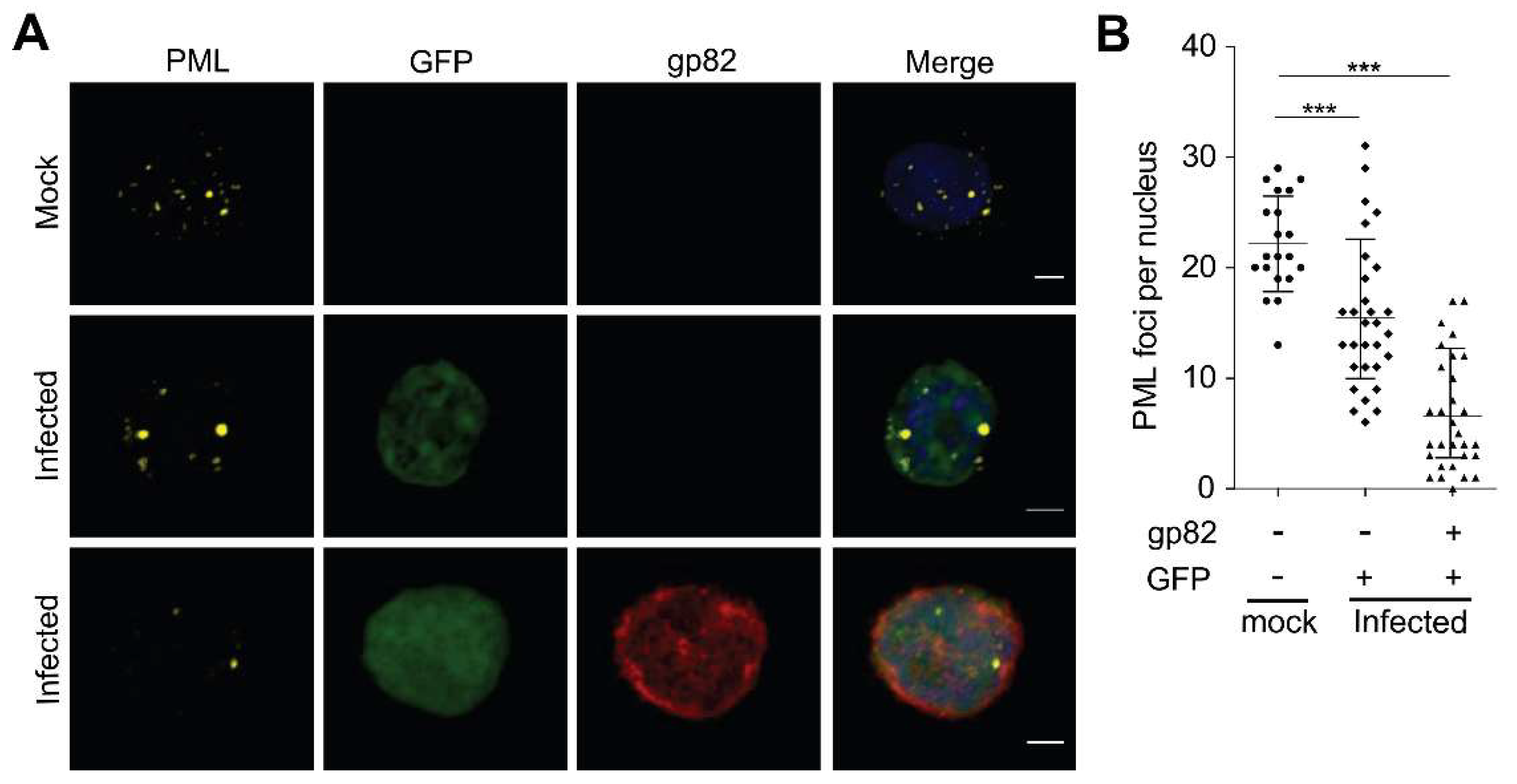
3.1. Analysis of ND10 Complex in HHV-6A Infected Cells
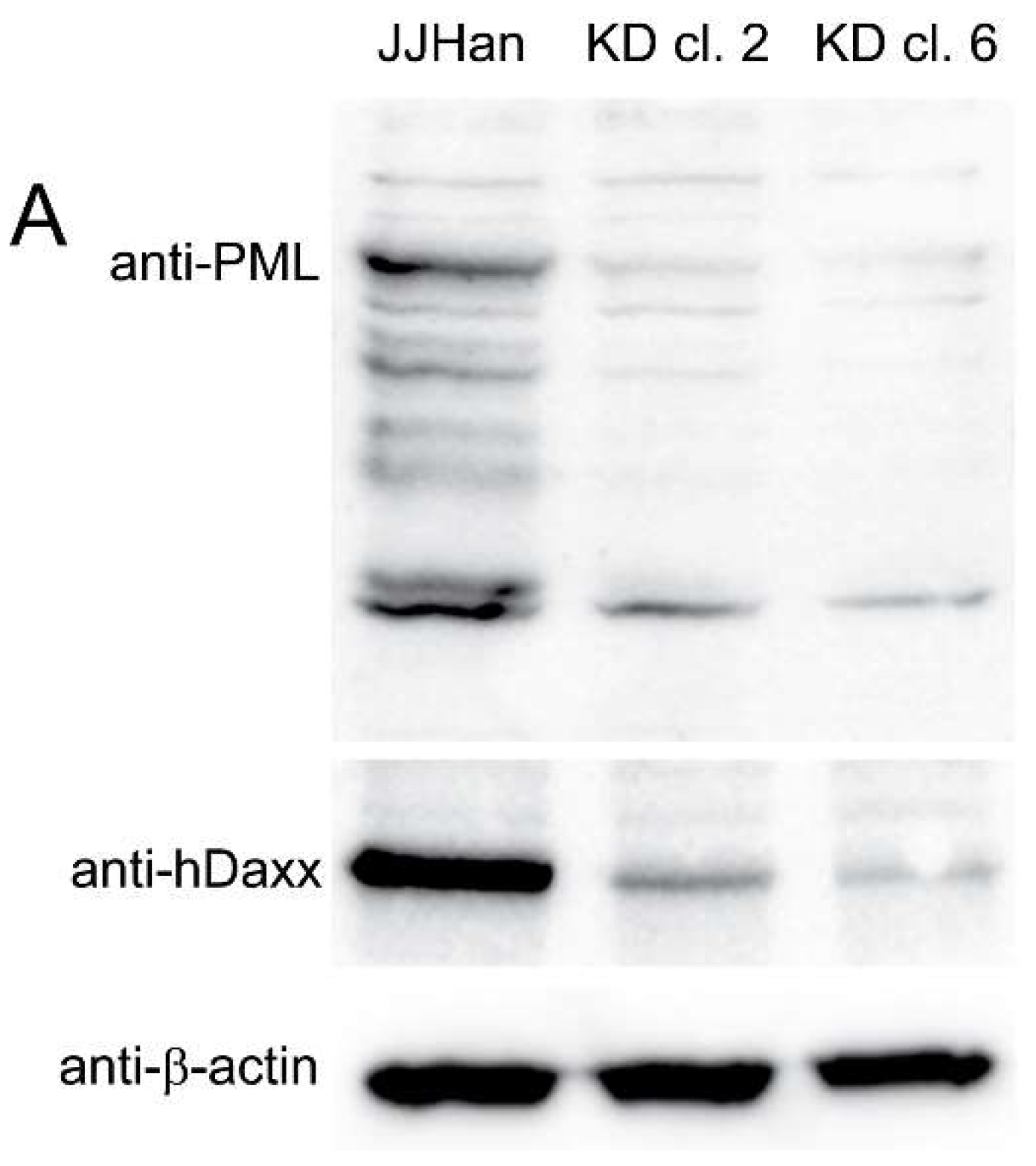
3.2. Generation and Analysis of ND10 Knockdown Cells
3.3. Effect of the ND10 Complex on HHV-6A Replication
3.4. ND10-Mediated Suppression of Viral Protein Expression in Latently Infected Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Levy, J.A.; Ferro, F.; Greenspan, D.; Lennette, E.T. Frequent isolation of HHV-6 from saliva and high seroprevalence of the virus in the population. Lancet 1990, 335, 1047–1050. [Google Scholar] [CrossRef]
- Okuno, T.; Takahashi, K.; Balachandra, K.; Shiraki, K.; Yamanishi, K.; Takahashi, M.; Baba, K. Seroepidemiology of human herpesvirus 6 infection in normal children and adults. J. Clin. Microbiol. 1989, 27, 651–653. [Google Scholar] [PubMed]
- Zerr, D.M.; Meier, A.S.; Selke, S.S.; Frenkel, L.M.; Huang, M.-L.; Wald, A.; Rhoads, M.P.; Nguy, L.; Bornemann, R.; Morrow, R.A.; et al. A Population-Based Study of Primary Human Herpesvirus 6 Infection. N. Engl. J. Med. 2005, 352, 768–776. [Google Scholar] [CrossRef] [PubMed]
- Yamanishi, K.; Shiraki, K.; Kondo, T.; Okuno, T.; Takahashi, M.; Asano, Y.; Kurata, T. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet 1988, 331, 1065–1067. [Google Scholar] [CrossRef]
- Hall, C.B.; Long, C.E.; Schnabel, K.C.; Caserta, M.T.; McIntyre, K.M.; Costanzo, M.A.; Knott, A.; Dewhurst, S.; Insel, R.A.; Epstein, L.G. Human Herpesvirus-6 Infection in Children—A Prospective Study of Complications and Reactivation. N. Engl. J. Med. 1994, 331, 432–438. [Google Scholar] [CrossRef] [PubMed]
- McCullers, J.A.; Lakeman, F.D.; Whitley, R.J. Human herpesvirus 6 is associated with focal encephalitis. Clin. Infect. Dis. 1995, 21, 571–576. [Google Scholar] [CrossRef] [PubMed]
- Luppi, M.; Marasca, R.; Barozzi, P.; Ferrari, S.; Ceccherini-Nelli, L.; Batoni, G.; Merelli, E.; Torelli, G. Three cases of human herpesvirus-6 latent infection: Integration of viral genome in peripheral blood mononuclear cell DNA. J. Med. Virol. 1993, 40, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Arbuckle, J.H.; Medveczky, M.M.; Luka, J.; Hadley, S.H.; Luegmayr, A.; Ablashi, D.; Lund, T.C.; Tolar, J.; De Meirleir, K.; Montoya, J.G.; et al. The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proc. Natl. Acad. Sci. USA 2010, 107, 5563–5568. [Google Scholar] [CrossRef] [PubMed]
- Ward, K.N.; Leong, H.N.; Thiruchelvam, A.D.; Atkinson, C.E.; Clark, D.A. Human Herpesvirus 6 DNA Levels in Cerebrospinal Fluid Due to Primary Infection Differ from Those Due to Chromosomal Viral Integration and Have Implications for Diagnosis of Encephalitis. J. Clin. Microbiol. 2007, 45, 1298–1304. [Google Scholar] [CrossRef] [PubMed]
- Daibata, M.; Taguchi, T.; Nemoto, Y.; Taguchi, H.; Miyoshi, I. Inheritance of chromosomally integrated human herpesvirus 6 DNA. Blood 1999, 94, 1545–1549. [Google Scholar] [PubMed]
- Osterrieder, N.; Wallaschek, N.; Kaufer, B.B. Herpesvirus Genome Integration into Telomeric Repeats of Host Cell Chromosomes. Annu. Rev. Virol. 2014, 1, 215–235. [Google Scholar] [CrossRef] [PubMed]
- Pellett, P.E.; Ablashi, D.V.; Ambros, P.F.; Agut, H.; Caserta, M.T.; Descamps, V.; Flamand, L.; Gautheret-Dejean, A.; Hall, C.B.; Kamble, R.T.; et al. Chromosomally integrated human herpesvirus 6: Questions and answers. Rev. Med. Virol. 2012, 22, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Caselli, E.; Di Luca, D. Molecular biology and clinical associations of Roseoloviruses human herpesvirus 6 and human herpesvirus 7. New Microbiol. 2007, 30, 173–187. [Google Scholar] [PubMed]
- De Bolle, L.; Naesens, L.; De Clercq, E. Update on human herpesvirus 6 biology, clinical features, and therapy. Clin. Microbiol. Rev. 2005, 18, 217–245. [Google Scholar] [CrossRef] [PubMed]
- Tavalai, N.; Stamminger, T. Interplay between Herpesvirus Infection and Host Defense by PML Nuclear Bodies. Viruses 2009, 1, 1240–1264. [Google Scholar] [CrossRef] [PubMed]
- Everett, R.D. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene 2001, 20, 7266–7273. [Google Scholar] [CrossRef] [PubMed]
- Maul, G.G.; Guldner, H.H.; Spivack, J.G. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0). J. Gen. Virol. 1993, 74 Pt 12, 2679–2690. [Google Scholar] [CrossRef] [PubMed]
- Ishov, A.M.; Sotnikov, A.G.; Negorev, D.; Vladimirova, O.V.; Neff, N.; Kamitani, T.; Yeh, E.T.; Strauss, J.F., 3rd; Maul, G.G. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 1999, 147, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Everett, R.D.; Chelbi-Alix, M.K. PML and PML nuclear bodies: Implications in antiviral defence. Biochimie 2007, 89, 819–830. [Google Scholar] [CrossRef] [PubMed]
- Tavalai, N.; Stamminger, T. New insights into the role of the subnuclear structure ND10 for viral infection. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2008, 1783, 2207–2221. [Google Scholar] [CrossRef] [PubMed]
- Everett, R.D.; Murray, J. ND10 Components Relocate to Sites Associated with Herpes Simplex Virus Type 1 Nucleoprotein Complexes during Virus Infection. J. Virol. 2005, 79, 5078–5089. [Google Scholar] [CrossRef] [PubMed]
- Müller, S.; Dejean, A. Viral Immediate-Early Proteins Abrogate the Modification by SUMO-1 of PML and Sp100 Proteins, Correlating with Nuclear Body Disruption. J. Virol. 1999, 73, 5137–5143. [Google Scholar] [PubMed]
- Bernardi, R.; Pandolfi, P.P. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 2007, 8, 1006–1016. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Salomoni, P.; Pandolfi, P.P. The transcriptional role of PML and the nuclear body. Nat. Cell Biol. 2000, 2, E85. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-R.; Kim, D.-J.; Lee, J.-M.; Choi, C.Y.; Ahn, B.-Y.; Hayward, G.S.; Ahn, J.-H. Ability of the Human Cytomegalovirus IE1 Protein To Modulate Sumoylation of PML Correlates with Its Functional Activities in Transcriptional Regulation and Infectivity in Cultured Fibroblast Cells. J. Virol. 2004, 78, 6527–6542. [Google Scholar] [CrossRef] [PubMed]
- Tavalai, N.; Adler, M.; Scherer, M.; Riedl, Y.; Stamminger, T. Evidence for a Dual Antiviral Role of the Major Nuclear Domain 10 Component Sp100 during the Immediate-Early and Late Phases of the Human Cytomegalovirus Replication Cycle. J. Virol. 2011, 85, 9447–9458. [Google Scholar] [CrossRef] [PubMed]
- Korioth, F.; Maul, G.G.; Plachter, B.; Stamminger, T.; Frey, J. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp. Cell Res. 1996, 229, 155–158. [Google Scholar] [CrossRef] [PubMed]
- Saffert, R.T.; Kalejta, R.F. Human Cytomegalovirus Gene Expression Is Silenced by Daxx-Mediated Intrinsic Immune Defense in Model Latent Infections Established In Vitro. J. Virol. 2007, 81, 9109–9120. [Google Scholar] [CrossRef] [PubMed]
- Cantrell, S.R.; Bresnahan, W.A. Human Cytomegalovirus (HCMV) UL82 Gene Product (pp71) Relieves hDaxx-Mediated Repression of HCMV Replication. J. Virol. 2006, 80, 6188–6191. [Google Scholar] [CrossRef] [PubMed]
- Preston, C.M.; Nicholl, M.J. Role of the cellular protein hDaxx in human cytomegalovirus immediate-early gene expression. J. Gen. Virol. 2006, 87, 1113–1121. [Google Scholar] [CrossRef] [PubMed]
- Wallaschek, N.; Sanyal, A.; Pirzer, F.; Gravel, A.; Mori, Y.; Flamand, L.; Kaufer, B.B. The Telomeric Repeats of Human Herpesvirus 6A (HHV-6A) Are Required for Efficient Virus Integration. PLoS Pathog. 2016, 12, e1005666. [Google Scholar] [CrossRef] [PubMed]
- Tischer, B.K.; von Einem, J.; Kaufer, B.; Osterrieder, N. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 2006, 40, 191–197. [Google Scholar] [PubMed]
- Glass, M.; Everett, R.D. Components of promyelocytic leukemia nuclear bodies (ND10) act cooperatively to repress herpesvirus infection. J. Virol. 2013, 87, 2174–2185. [Google Scholar] [CrossRef] [PubMed]
- Schippers, T.; Jarosinski, K.; Osterrieder, N. The ORF012 Gene of Marek’s Disease Virus Type 1 Produces a Spliced Transcript and Encodes a Novel Nuclear Phosphoprotein Essential for Virus Growth. J. Virol. 2015, 89, 1348–1363. [Google Scholar] [CrossRef] [PubMed]
- Kaufer, B.B. Detection of integrated herpesvirus genomes by fluorescence in situ hybridization (FISH). Methods Mol. Biol. 2013, 1064, 141–152. [Google Scholar] [PubMed]
- Kaufer, B.B.; Jarosinski, K.W.; Osterrieder, N. Herpesvirus telomeric repeats facilitate genomic integration into host telomeres and mobilization of viral DNA during reactivation. J. Exp. Med. 2011, 208, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Rens, W.; Fu, B.; O’Brien, P.C.; Ferguson-Smith, M. Cross-species chromosome painting. Nat. Protoc. 2006, 1, 783–790. [Google Scholar] [CrossRef] [PubMed]
- Gravel, A.; Gosselin, J.; Flamand, L. Human Herpesvirus 6 immediate-early 1 protein is a sumoylated nuclear phosphoprotein colocalizing with promyelocytic leukemia protein-associated nuclear bodies. J. Biol. Chem. 2002, 277, 19679–19687. [Google Scholar] [CrossRef] [PubMed]
- Seeler, J.-S.; Marchio, A.; Sitterlin, D.; Transy, C.; Dejean, A. Interaction of SP100 with HP1 proteins: A link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc. Natl. Acad. Sci. USA 1998, 95, 7316–7321. [Google Scholar] [CrossRef] [PubMed]
- Maul, G.G.; Negorev, D.; Bell, P.; Ishov, A.M. Review: Properties and Assembly Mechanisms of ND10, PML Bodies, or PODs. J. Struct. Biol. 2000, 129, 278–287. [Google Scholar] [CrossRef] [PubMed]
- Wagenknecht, N.; Reuter, N.; Scherer, M.; Reichel, A.; Müller, R.; Stamminger, T. Contribution of the Major ND10 Proteins PML, hDaxx and Sp100 to the Regulation of Human Cytomegalovirus Latency and Lytic Replication in the Monocytic Cell Line THP-1. Viruses 2015, 7, 2884–2907. [Google Scholar] [CrossRef] [PubMed]
- Isaac, A.; Wilcox, K.W.; Taylor, J.L. SP100B, a repressor of gene expression preferentially binds to DNA with unmethylated CpGs. J. Cell. Biochem. 2006, 98, 1106–1122. [Google Scholar] [CrossRef] [PubMed]
- Wilcox, K.W.; Sheriff, S.; Isaac, A.; Taylor, J.L. SP100B is a repressor of gene expression. J. Cell. Biochem. 2005, 95, 352–365. [Google Scholar] [CrossRef] [PubMed]
- Lieberman, P.M. Epigenetics and Genetics of Viral Latency. Cell Host Microbe 2016, 19, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Günther, T.; Schreiner, S.; Dobner, T.; Tessmer, U.; Grundhoff, A. Influence of ND10 Components on Epigenetic Determinants of Early KSHV Latency Establishment. PLoS Pathog. 2014, 10, e1004274. [Google Scholar] [CrossRef] [PubMed]
- Knipe, D.M. Nuclear Sensing of Viral DNA, Epigenetic Regulation of Herpes Simplex Virus Infection, and Innate Immunity. Virology 2015, 479–480, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Merkl, P.E.; Orzalli, M.H.; Knipe, D.M. Mechanisms of Host IFI16, PML, and Daxx Protein Restriction of Herpes Simplex Virus 1 Replication. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [PubMed]



| Name | Sequence (5′→3′) |
|---|---|
| B2M forward | CCAGCAGAGAATGGAAAGTCAA |
| B2M reverse | TCTCCATTCTTCAGTAAGTCAACTTCA |
| B2M probe | ATGTGTCTGGGTTTCATCCATCCGACA |
| HHV-6 U86 forward | TGTACATGGGCTGTAGGAGTTGA |
| HHV-6 U86 reverse | ACATCCTCTGCTTCCAATCTACAATC |
| HHV-6 U86 probe | TTCCGAAGCAAAGCGCACCTGG |
| HHV-6 U57-P2a-GFP forward | GTTTGTGATCGAAAGTGCAGTAGACGGTTTCCATTTTACTTGTACAGCTCGTCCATGCCG |
| HHV-6 U57-P2a-GFP reverse | GAGAAACCATACCTTTCCAACTCATTATCGAATCATCCATAGGATCTGGAGCGACCAATT |
| HHV-6 U57 sequencing forward | CTTTGTTGGAGGAGACGATGG |
| HHV-6 U57 sequencing reverse | GCCTCTTCACTGTTCATCCAA |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanyal, A.; Wallaschek, N.; Glass, M.; Flamand, L.; Wight, D.J.; Kaufer, B.B. The ND10 Complex Represses Lytic Human Herpesvirus 6A Replication and Promotes Silencing of the Viral Genome. Viruses 2018, 10, 401. https://doi.org/10.3390/v10080401
Sanyal A, Wallaschek N, Glass M, Flamand L, Wight DJ, Kaufer BB. The ND10 Complex Represses Lytic Human Herpesvirus 6A Replication and Promotes Silencing of the Viral Genome. Viruses. 2018; 10(8):401. https://doi.org/10.3390/v10080401
Chicago/Turabian StyleSanyal, Anirban, Nina Wallaschek, Mandy Glass, Louis Flamand, Darren J. Wight, and Benedikt B. Kaufer. 2018. "The ND10 Complex Represses Lytic Human Herpesvirus 6A Replication and Promotes Silencing of the Viral Genome" Viruses 10, no. 8: 401. https://doi.org/10.3390/v10080401
APA StyleSanyal, A., Wallaschek, N., Glass, M., Flamand, L., Wight, D. J., & Kaufer, B. B. (2018). The ND10 Complex Represses Lytic Human Herpesvirus 6A Replication and Promotes Silencing of the Viral Genome. Viruses, 10(8), 401. https://doi.org/10.3390/v10080401





