FUS Negatively Regulates Kaposi’s Sarcoma-Associated Herpesvirus Gene Expression
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Sirna Knockdowns
2.3. Genome Replication
2.4. Supernatant Transfer
2.5. Cloning, Lentiviral Production and Infection
2.6. Nucleic Acid Isolation and Measurement
2.7. 4sU. Labeling
2.8. Subcellular Fractionation and Western Blotting
2.9. Immunoprecipitation (IP)
2.10. Phosphonoacetic Acid (PAA) Treatment
2.11. Immunofluorescence Microscopy
2.12. Chromatin Immunoprecipitation (ChIP)
3. Results
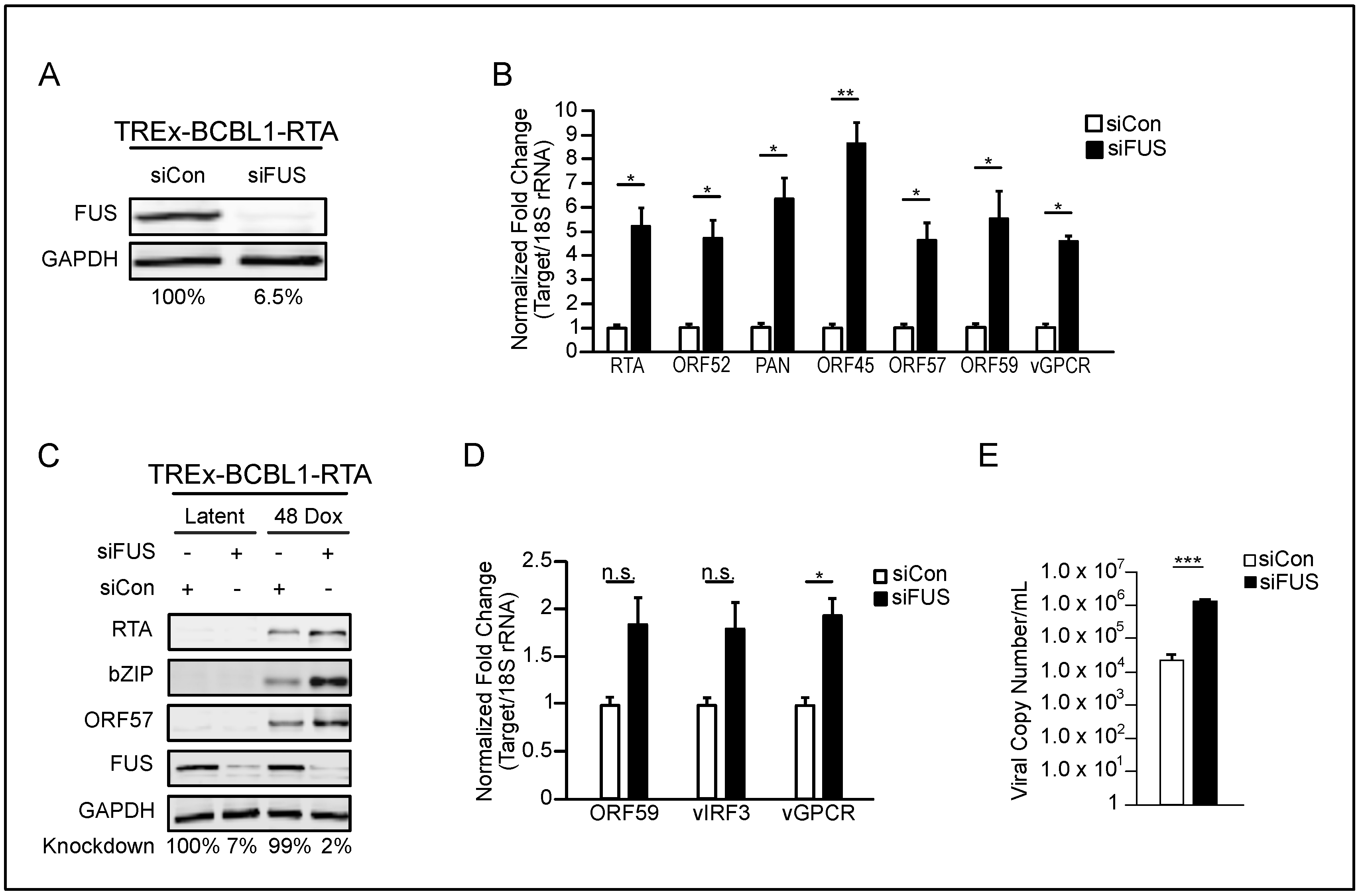
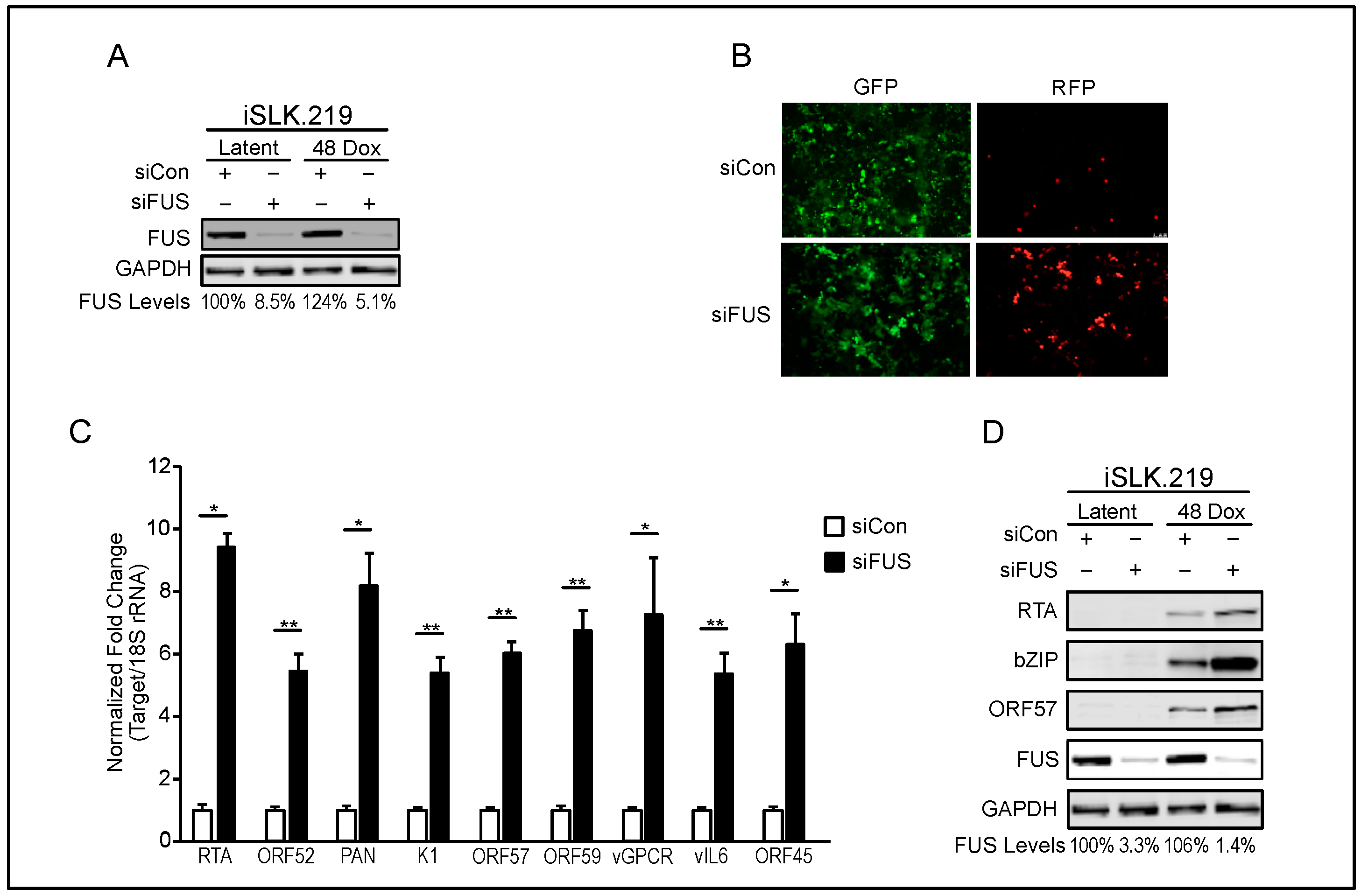
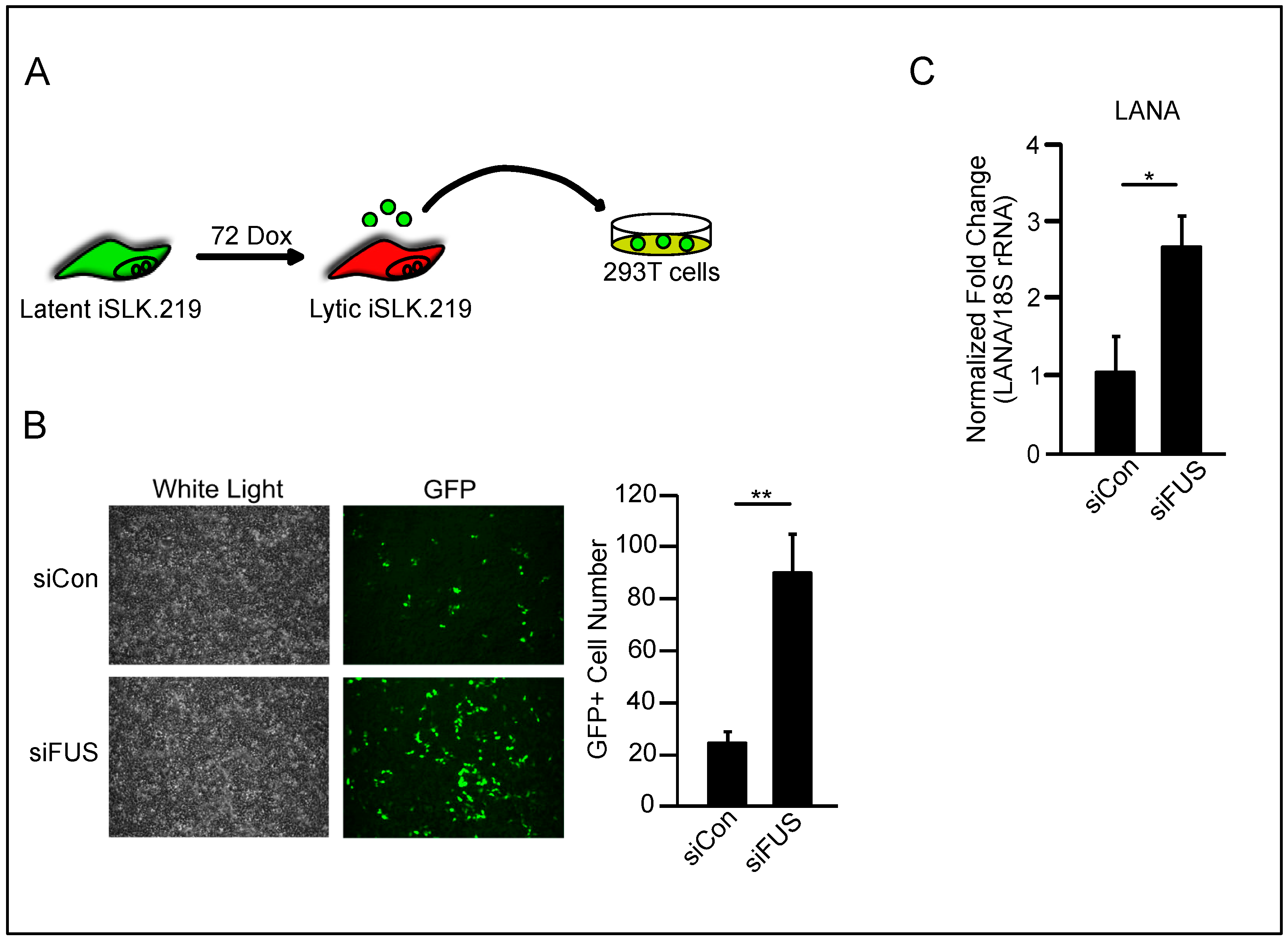
3.1. FUS Knockdown Increases KSHV Lytic Gene Expression and Virion Production
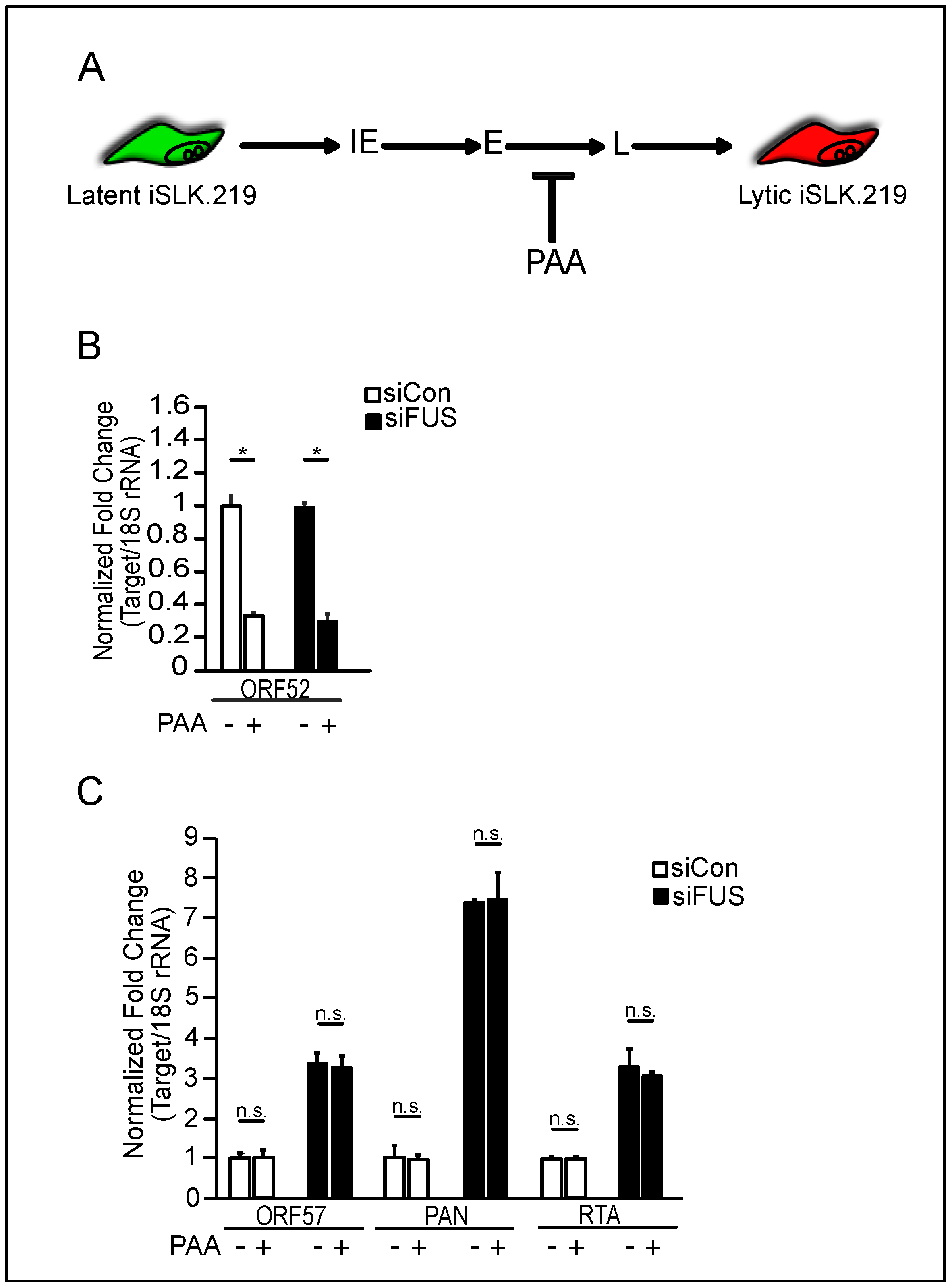
3.2. FUS Depletion Enhances KSHV Gene Expression Prior to Viral DNA Replication
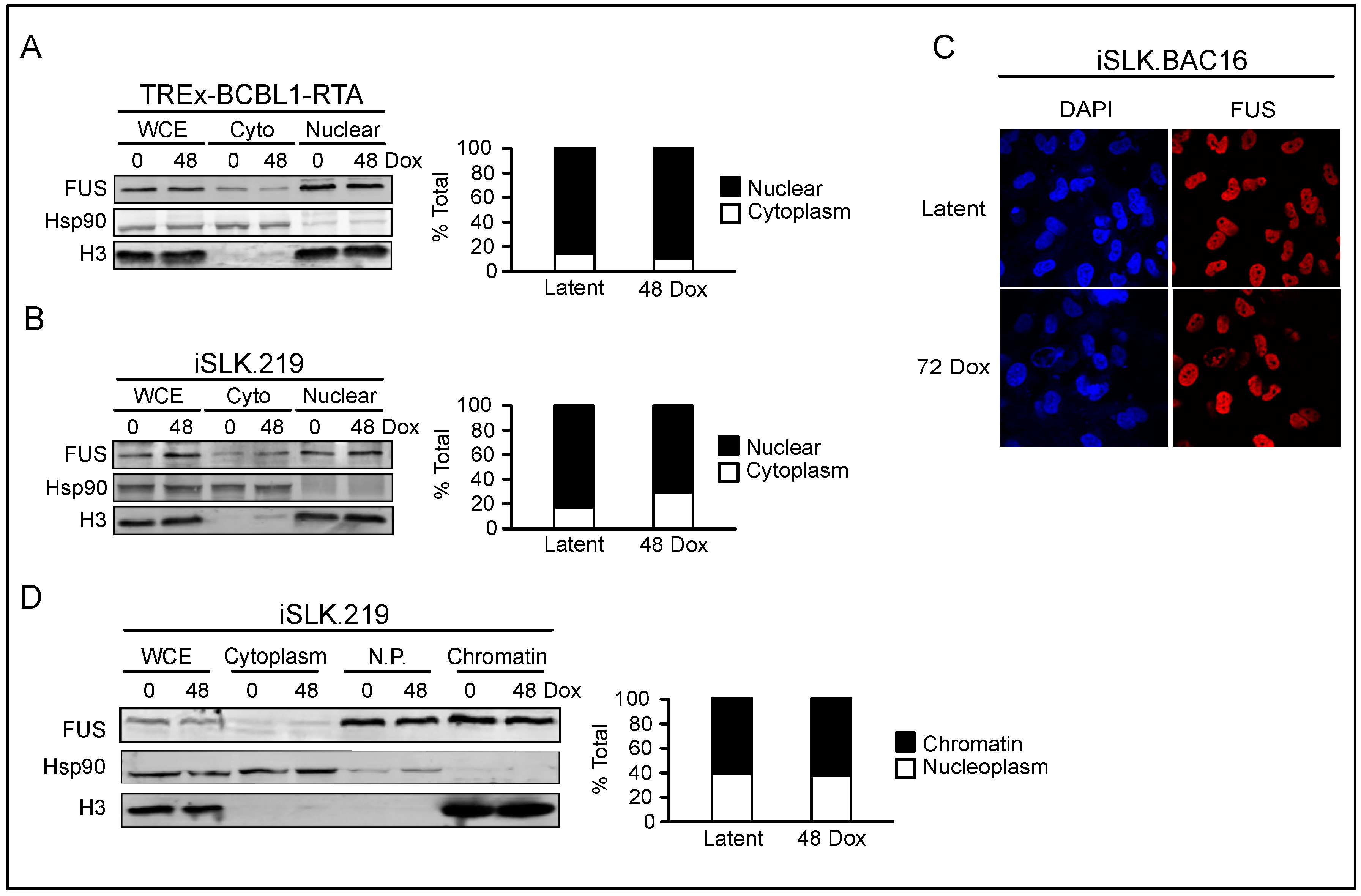
3.3. FUS is Nuclear Localized throughout KSHV Viral Reactivation
3.4. FUS Affects RNA Polymerase II CTD Phosphorylation and Nascent RNA Transcription
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chang, Y.; Cesarman, E.; Pessin, M.S.; Lee, F.; Culpepper, J.; Knowles, D.M.; Moore, P.S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 1994, 266, 1865–1869. [Google Scholar] [CrossRef] [PubMed]
- Kedes, D.H.; Operskalski, E.; Busch, M.; Kohn, R.; Flood, J.; Ganem, D. The seroepidemiology of human herpesvirus 8 (Kaposi’s sarcoma–associated herpesvirus): Distribution of infection in KS risk groups and evidence for sexual transmission. Nat. Med. 1996, 2, 918–924. [Google Scholar] [CrossRef] [PubMed]
- Moore, P.S.; Chang, Y. Detection of herpesvirus-like DNA sequences in Kaposi’s sarcoma in patients with and those without HIV infection. N. Engl. J. Med. 1995, 332, 1181–1185. [Google Scholar] [CrossRef] [PubMed]
- Mesri, E.A.; Feitelson, M.A.; Munger, K. Human Viral Oncogenesis: A Cancer Hallmarks Analysis. Cell Host Microbe 2014, 15, 266–282. [Google Scholar] [CrossRef] [PubMed]
- Cesarman, E.; Chang, Y.; Moore, P.S.; Said, J.W.; Knowles, D.M. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 1995, 332, 1186–1191. [Google Scholar] [CrossRef] [PubMed]
- Soulier, J.; Grollet, L.; Oksenhendler, E.; Cacoub, P.; Cazals-Hatem, D.; Babinet, P.; d’Agay, M.F.; Clauvel, J.P.; Raphael, M.; Degos, L. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood 1995, 86, 1276–1280. [Google Scholar] [PubMed]
- Purushothaman, P.; Dabral, P.; Gupta, N.; Sarkar, R.; Verma, S.C. KSHV Genome Replication and Maintenance. Front. Microbiol. 2016, 7, 54. [Google Scholar] [CrossRef] [PubMed]
- Purushothaman, P.; Uppal, T.; Verma, S.C. Molecular Biology of KSHV Lytic Reactivation. Viruses 2015, 7, 116–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aneja, K.K.; Yuan, Y. Reactivation and Lytic Replication of Kaposi’s Sarcoma-Associated Herpesvirus: An Update. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Friborg, J., Jr.; Kong, W.; Hottiger, M.O.; Nabel, G.J. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 1999, 402, 889–894. [Google Scholar] [CrossRef] [PubMed]
- Radkov, S.A.; Kellam, P.; Boshoff, C. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma–E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 2000, 6, 1121–1127. [Google Scholar] [CrossRef] [PubMed]
- Guasparri, I.; Keller, S.A.; Cesarman, E. KSHV vFLIP Is Essential for the Survival of Infected Lymphoma Cells. J. Exp. Med. 2004, 199, 993–1003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Field, N.; Low, W.; Daniels, M.; Howell, S.; Daviet, L.; Boshoff, C.; Collins, M. KSHV vFLIP binds to IKK-gamma to activate IKK. J. Cell Sci. 2003, 116, 3721–3728. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Eby, M.T.; Rathore, N.; Sinha, S.K.; Kumar, A.; Chaudhary, P.M. The human herpes virus 8-encoded viral FLICE inhibitory protein physically associates with and persistently activates the Ikappa B kinase complex. J. Biol. Chem. 2002, 277, 13745–13751. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, P.M.; Jasmin, A.; Eby, M.T.; Hood, L. Modulation of the NF-kappa B pathway by virally encoded death effector domains-containing proteins. Oncogene 1999, 18, 5738–5746. [Google Scholar] [CrossRef] [PubMed]
- Casper, C.; Nichols, W.G.; Huang, M.-L.; Corey, L.; Wald, A. Remission of HHV-8 and HIV-associated multicentric Castleman disease with ganciclovir treatment. Blood 2004, 103, 1632–1634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crum-Cianflone, N.F.; Wallace, M.R.; Looney, D. Successful secondary prophylaxis for primary effusion lymphoma with human herpesvirus 8 therapy. AIDS Lond. Engl. 2006, 20, 1567–1569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hocqueloux, L.; Agbalika, F.; Oksenhendler, E.; Molina, J.M. Long-term remission of an AIDS-related primary effusion lymphoma with antiviral therapy. AIDS Lond. Engl. 2001, 15, 280–282. [Google Scholar] [CrossRef]
- Luppi, M.; Trovato, R.; Barozzi, P.; Vallisa, D.; Rossi, G.; Re, A.; Ravazzini, L.; Potenza, L.; Riva, G.; Morselli, M.; et al. Treatment of herpesvirus associated primary effusion lymphoma with intracavity cidofovir. Leukemia 2005, 19, 473–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, D.F.; Kuppermann, B.D.; Wolitz, R.A.; Palestine, A.G.; Li, H.; Robinson, C.A. Oral ganciclovir for patients with cytomegalovirus retinitis treated with a ganciclovir implant. Roche Ganciclovir Study Group. N. Engl. J. Med. 1999, 340, 1063–1070. [Google Scholar] [CrossRef] [PubMed]
- Bais, C.; Santomasso, B.; Coso, O.; Arvanitakis, L.; Raaka, E.G.; Gutkind, J.S.; Asch, A.S.; Cesarman, E.; Gershengorn, M.C.; Mesri, E.A.; et al. G-protein-coupled receptor of Kaposi’s sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature 1998, 391, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Staskus, K.A.; Zhong, W.; Gebhard, K.; Herndier, B.; Wang, H.; Renne, R.; Beneke, J.; Pudney, J.; Anderson, D.J.; Ganem, D.; et al. Kaposi’s sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J. Virol. 1997, 71, 715–719. [Google Scholar] [PubMed]
- Staudt, M.R.; Kanan, Y.; Jeong, J.H.; Papin, J.F.; Hines-Boykin, R.; Dittmer, D.P. The tumor microenvironment controls primary effusion lymphoma growth in vivo. Cancer Res. 2004, 64, 4790–4799. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Lin, S.-F.; Gradoville, L.; Yuan, Y.; Zhu, F.; Miller, G. A viral gene that activates lytic cycle expression of Kaposi’s sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 1998, 95, 10866–10871. [Google Scholar] [CrossRef] [PubMed]
- Lukac, D.M.; Renne, R.; Kirshner, J.R.; Ganem, D. Reactivation of Kaposi’s sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 1998, 252, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; AuCoin, D.P.; Huete, A.R.; Cei, S.A.; Hanson, L.J.; Pari, G.S. A Kaposi’s Sarcoma-Associated Herpesvirus/Human Herpesvirus 8 ORF50 Deletion Mutant Is Defective for Reactivation of Latent Virus and DNA Replication. J. Virol. 2005, 79, 3479–3487. [Google Scholar] [CrossRef] [PubMed]
- Lukac, D.M.; Garibyan, L.; Kirshner, J.R.; Palmeri, D.; Ganem, D. DNA Binding by Kaposi’s Sarcoma-Associated Herpesvirus Lytic Switch Protein Is Necessary for Transcriptional Activation of Two Viral Delayed Early Promoters. J. Virol. 2001, 75, 6786–6799. [Google Scholar] [CrossRef] [PubMed]
- Carroll, K.D.; Khadim, F.; Spadavecchia, S.; Palmeri, D.; Lukac, D.M. Direct Interactions of Kaposi’s Sarcoma-Associated Herpesvirus/Human Herpesvirus 8 ORF50/Rta Protein with the Cellular Protein Octamer-1 and DNA Are Critical for Specifying Transactivation of a Delayed-Early Promoter and Stimulating Viral Reactivation. J. Virol. 2007, 81, 8451–8467. [Google Scholar] [CrossRef] [PubMed]
- Sakakibara, S.; Ueda, K.; Chen, J.; Okuno, T.; Yamanishi, K. Octamer-Binding Sequence Is a Key Element for the Autoregulation of Kaposi’s Sarcoma-Associated Herpesvirus ORF50/Lyta Gene Expression. J. Virol. 2001, 75, 6894–6900. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Chang, J.; Lynch, S.J.; Lukac, D.M.; Ganem, D. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jκ (CSL), the target of the Notch signaling pathway. Genes Dev. 2002, 16, 1977–1989. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wood, C. The Transcriptional Repressor K-RBP Modulates RTA-Mediated Transactivation and Lytic Replication of Kaposi’s Sarcoma-Associated Herpesvirus. J. Virol. 2007, 81, 6294–6306. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, S.; Wu, M.-H.; Geng, Y.; Wood, C. Identification of a Cellular Protein That Interacts and Synergizes with the RTA (ORF50) Protein of Kaposi’s Sarcoma-Associated Herpesvirus in Transcriptional Activation. J. Virol. 2001, 75, 11961–11973. [Google Scholar] [CrossRef] [PubMed]
- Gwack, Y.; Baek, H.J.; Nakamura, H.; Lee, S.H.; Meisterernst, M.; Roeder, R.G.; Jung, J.U. Principal role of TRAP/mediator and SWI/SNF complexes in Kaposi’s sarcoma-associated herpesvirus RTA-mediated lytic reactivation. Mol. Cell. Biol. 2003, 23, 2055–2067. [Google Scholar] [CrossRef] [PubMed]
- Gwack, Y.; Nakamura, H.; Lee, S.H.; Souvlis, J.; Yustein, J.T.; Gygi, S.; Kung, H.-J.; Jung, J.U. Poly(ADP-Ribose) Polymerase 1 and Ste20-Like Kinase hKFC Act as Transcriptional Repressors for Gamma-2 Herpesvirus Lytic Replication. Mol. Cell. Biol. 2003, 23, 8282–8294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, P.-C.; Fitzgerald, L.D.; Van Geelen, A.; Izumiya, Y.; Ellison, T.J.; Wang, D.-H.; Ann, D.K.; Luciw, P.A.; Kung, H.-J. KRAB domain-associated protein-1 as a latency regulator for Kaposi’s sarcoma-associated herpesvirus and its modulation by the viral protein kinase. Cancer Res. 2009, 69, 5681–5689. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, J.C.; Ebmeier, C.C.; Podell, E.R.; Heimiller, J.; Taatjes, D.J.; Cech, T.R. FUS binds the CTD of RNA polymerase II and regulates its phosphorylation at Ser2. Genes Dev. 2012, 26, 2690–2695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, R.; Yu, J.; Zhang, Z.; Gygi, M.P.; Krainer, A.R.; Gygi, S.P.; Reed, R. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol. Cell 2007, 26, 867–881. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Gal, J.; Chen, J.; Zhu, H. Self-assembled FUS binds active chromatin and regulates gene transcription. Proc. Natl. Acad. Sci. USA 2014, 111, 17809–17814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sama, R.R.K.; Ward, C.L.; Kaushansky, L.J.; Lemay, N.; Ishigaki, S.; Urano, F.; Bosco, D.A. FUS/TLS assembles into stress granules and is a prosurvival factor during hyperosmolar stress. J. Cell. Physiol. 2013, 228, 2222–2231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Myoung, J.; Ganem, D. Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: Maintenance of tight latency with efficient reactivation upon induction. J. Virol. Methods 2011, 174, 12–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brulois, K.F.; Chang, H.; Lee, A.S.-Y.; Ensser, A.; Wong, L.-Y.; Toth, Z.; Lee, S.H.; Lee, H.-R.; Myoung, J.; Ganem, D.; et al. Construction and Manipulation of a New Kaposi’s Sarcoma-Associated Herpesvirus Bacterial Artificial Chromosome Clone. J. Virol. 2012, 86, 9708–9720. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Lu, M.; Gwack, Y.; Souvlis, J.; Zeichner, S.L.; Jung, J.U. Global Changes in Kaposi’s Sarcoma-Associated Virus Gene Expression Patterns following Expression of a Tetracycline-Inducible Rta Transactivator. J. Virol. 2003, 77, 4205–4220. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Bose, P.; Leong-Quong, R.Y.; Fujita, D.J.; Riabowol, K. REAP: A two minute cell fractionation method. BMC Res. Notes 2010, 3, 294. [Google Scholar] [CrossRef] [PubMed]
- Karijolich, J.; Zhao, Y.; Peterson, B.; Zhou, Q.; Glaunsinger, B. Kaposi’s Sarcoma-Associated Herpesvirus ORF45 Mediates Transcriptional Activation of the HIV-1 Long Terminal Repeat via RSK2. J. Virol. 2014, 88, 7024–7035. [Google Scholar] [CrossRef] [PubMed]
- Hicks, G.G.; Singh, N.; Nashabi, A.; Mai, S.; Bozek, G.; Klewes, L.; Arapovic, D.; White, E.K.; Koury, M.J.; Oltz, E.M.; et al. Fus deficiency in mice results in defective B-lymphocyte development and activation, high levels of chromosomal instability and perinatal death. Nat. Genet. 2000, 24, ng0200_175. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.Y.; Riley, T.R.; Coady, T.; Bussemaker, H.J.; Manley, J.L. TLS/FUS (translocated in liposarcoma/fused in sarcoma) regulates target gene transcription via single-stranded DNA response elements. Proc. Natl. Acad. Sci. USA 2012, 109, 6030–6035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vieira, J.; O’Hearn, P.M. Use of the red fluorescent protein as a marker of Kaposi’s sarcoma-associated herpesvirus lytic gene expression. Virology 2004, 325, 225–240. [Google Scholar] [CrossRef] [PubMed]
- Zinszner, H.; Sok, J.; Immanuel, D.; Yin, Y.; Ron, D. TLS (FUS) binds RNA in vivo and engages in nucleo-cytoplasmic shuttling. J. Cell Sci. 1997, 110 Pt 15, 1741–1750. [Google Scholar] [PubMed]
- Phatnani, H.P.; Greenleaf, A.L. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006, 20, 2922–2936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, Y.; Blechingberg, J.; Fernandes, A.M.; Li, S.; Fryland, T.; Børglum, A.D.; Bolund, L.; Nielsen, A.L. EWS and FUS bind a subset of transcribed genes encoding proteins enriched in RNA regulatory functions. BMC Genom. 2015, 16, 929. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Schwartz, J.C.; Cech, T.R. Nucleic acid-binding specificity of human FUS protein. Nucleic Acids Res. 2015, 43, 7535–7543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peterlin, B.M.; Price, D.H. Controlling the Elongation Phase of Transcription with P-TEFb. Mol. Cell 2006, 23, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Bartkowiak, B.; Liu, P.; Phatnani, H.P.; Fuda, N.J.; Cooper, J.J.; Price, D.H.; Adelman, K.; Lis, J.T.; Greenleaf, A.L. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 2010, 24, 2303–2316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Vona, C.; Bezdan, D.; Islam, A.B.M.M.K.; Salichs, E.; López-Bigas, N.; Ossowski, S.; de la Luna, S. Chromatin-wide Profiling of DYRK1A Reveals a Role as a Gene-Specific RNA Polymerase II CTD Kinase. Mol. Cell 2015, 57, 506–520. [Google Scholar] [CrossRef] [PubMed]
- Toth, Z.; Brulois, K.F.; Wong, L.-Y.; Lee, H.-R.; Chung, B.; Jung, J.U. Negative Elongation Factor-Mediated Suppression of RNA Polymerase II Elongation of Kaposi’s Sarcoma-Associated Herpesvirus Lytic Gene Expression. J. Virol. 2012, 86, 9696–9707. [Google Scholar] [CrossRef] [PubMed]
- Tsai, W.-H.; Wang, P.-W.; Lin, S.-Y.; Wu, I.-L.; Ko, Y.-C.; Chen, Y.-L.; Li, M.; Lin, S.-F. Ser-634 and Ser-636 of Kaposi’s Sarcoma-Associated Herpesvirus RTA are Involved in Transactivation and are Potential Cdk9 Phosphorylation Sites. Front. Microbiol. 2012, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dormann, D.; Rodde, R.; Edbauer, D.; Bentmann, E.; Fischer, I.; Hruscha, A.; Than, M.E.; Mackenzie, I.R.A.; Capell, A.; Schmid, B.; et al. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 2010, 29, 2841–2857. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dunker, W.; Song, Y.; Zhao, Y.; Karijolich, J. FUS Negatively Regulates Kaposi’s Sarcoma-Associated Herpesvirus Gene Expression. Viruses 2018, 10, 359. https://doi.org/10.3390/v10070359
Dunker W, Song Y, Zhao Y, Karijolich J. FUS Negatively Regulates Kaposi’s Sarcoma-Associated Herpesvirus Gene Expression. Viruses. 2018; 10(7):359. https://doi.org/10.3390/v10070359
Chicago/Turabian StyleDunker, William, Yu Song, Yang Zhao, and John Karijolich. 2018. "FUS Negatively Regulates Kaposi’s Sarcoma-Associated Herpesvirus Gene Expression" Viruses 10, no. 7: 359. https://doi.org/10.3390/v10070359




