Tumor Necrosis Factor Receptor-Associated Factor 5 Interacts with the NS3 Protein and Promotes Classical Swine Fever Virus Replication
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Virus
2.2. Plasmid Construction
2.3. Real-Time Quantitative PCR (RT-qRCR)
2.4. Western Blot
2.5. Yeast Two-Hybrid Screening
2.6. Co-Immunoprecipitation Assays
2.7. GST Pull-Down Experiment
2.8. Cell Viability Assay
2.9. Statistical Analysis
3. Results
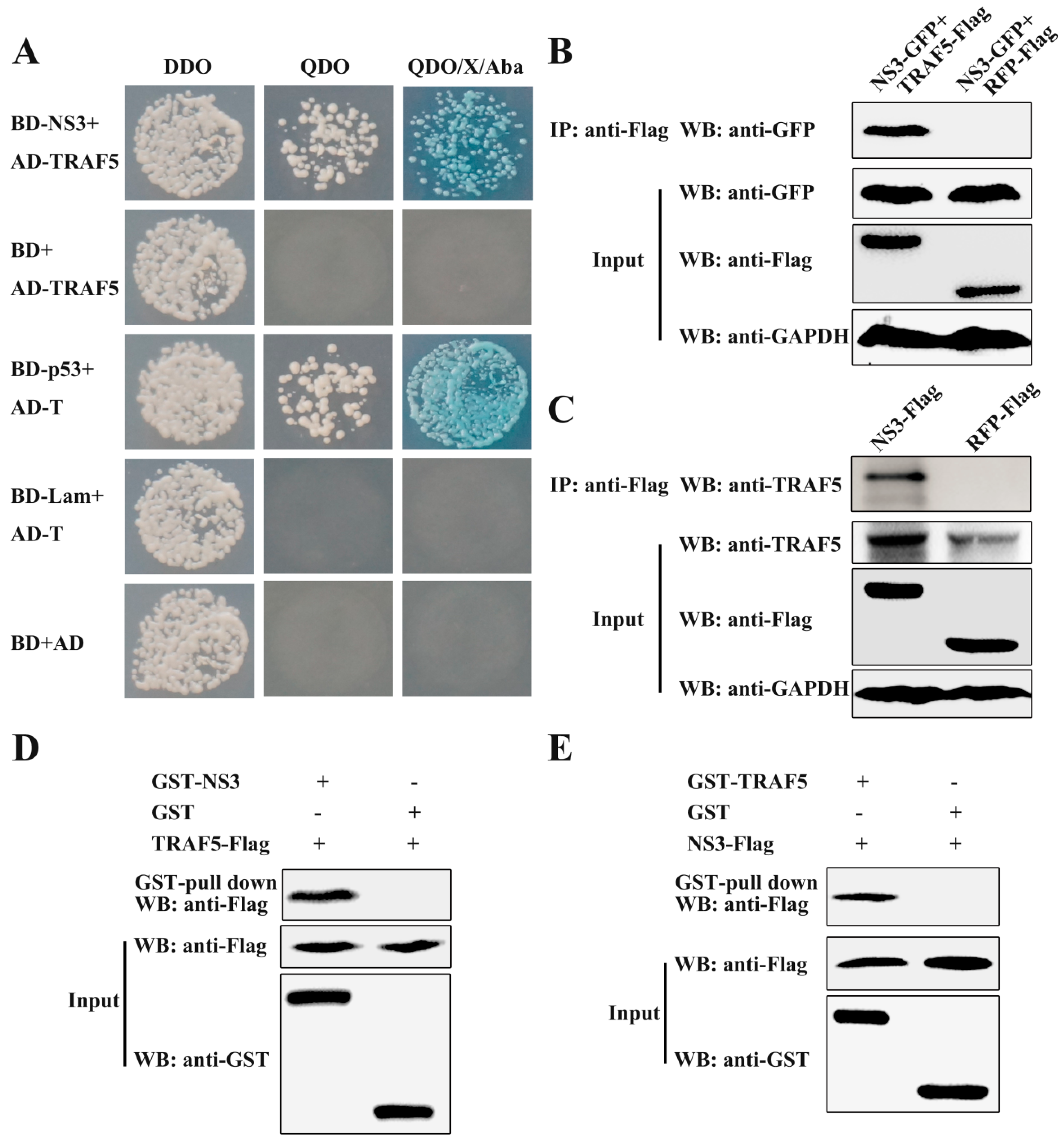
3.1. CSFV NS3 Interacts with TRAF5
3.2. TRAF5 Knockdown Inhibits CSFV Replication
3.3. TRAF5 Overexpression Promotes CSFV Replication
3.4. CSFV or NS3 Promotes TRAF5 Expression
3.5. CSFV Infection Promotes p38 MAPK Activation in PAMs
3.6. TRAF5 Promotes p38 MAPK Activation in PAMs
3.7. Inhibition of p38 MAPK Activation Suppresses CSFV Replication in PAMs
3.8. TRAF5 Does not Promote CSFV Replication Following Inhibition of p38 MAPK Activation
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Simmonds, P.; Becher, P.; Bukh, J.; Gould, E.A.; Meyers, G.; Monath, T.; Muerhoff, S.; Pletnev, A.; Rico-Hesse, R.; Smith, D.B.; et al. ICTV virus taxonomy profile: Flaviviridae. J. Gen. Virol. 2017, 98, 2–3. [Google Scholar] [CrossRef] [PubMed]
- Tautz, N.; Tews, B.A.; Meyers, G. The molecular biology of pestiviruses. Adv. Virus Res. 2015, 93, 47–160. [Google Scholar] [PubMed]
- Tamura, J.K.; Warrener, P.; Collett, M.S. RNA-stimulated ntpase activity associated with the p80 protein of the pestivirus bovine viral Diarrhea virus. Virology 1993, 193, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Warrener, P.; Collett, M.S. Pestivirus NS3 (p80) protein possesses RNA helicase activity. J. Virol. 1995, 69, 1720–1726. [Google Scholar] [PubMed]
- Tautz, N.; Elbers, K.; Stoll, D.; Meyers, G.; Thiel, H.J. Serine protease of pestiviruses: Determination of cleavage sites. J. Virol. 1997, 71, 5415–5422. [Google Scholar] [PubMed]
- Wang, Y.; Zhu, Z.; Wang, P.; Yu, J.; Wan, L.; Chen, J.; Xiao, M. Characterisation of interaction between NS3 and NS5B protein of classical swine fever virus by deletion of terminal sequences of NS5B. Virus Res. 2011, 156, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wang, Y.; Zhao, Y.; Zhu, Z.; Yu, J.; Wan, L.; Chen, J.; Xiao, M. Classical swine fever virus NS3 enhances RNA-dependent RNA polymerase activity by binding to NS5B. Virus Res. 2010, 148, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Wang, Y.; Yu, J.; Wan, L.; Chen, J.; Xiao, M. Classical swine fever virus NS3 is an IRES-binding protein and increases IRES-dependent translation. Virus Res. 2010, 153, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Hong, H.X.; Zhang, Y.M.; Guo, K.K.; Deng, X.M.; Ye, G.S.; Yang, X.Y. Cytopathic effect of classical swine fever virus NS3 protein on pk-15 cells. Intervirology 2007, 50, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.; Dong, W.; Cao, Z.; Li, X.; Wang, J.; Qian, G.; Lv, Q.; Wang, C.; Guo, K.; Zhang, Y. TRAF6 is a novel NS3-interacting protein that inhibits classical swine fever virus replication. Sci. Rep. 2017, 7, 6737. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Reichardt, A.; Liang, H.; Aliyari, R.; Cheng, D.; Wang, Y.; Xu, F.; Cheng, G.; Liu, Y. Single amino acid substitutions confer the antiviral activity of the TRAF3 adaptor protein onto TRAF5. Sci. Signal. 2012, 5, ra81. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Sun, A.; Wang, W.; He, J.; Hou, J.; Zhou, P.; Chen, Z. TRAF1 is involved in the classical NF-κB activation and CD30-induced alternative activity in hodgkin’s lymphoma cells. Mol. Immunol. 2009, 46, 2441–2448. [Google Scholar] [CrossRef] [PubMed]
- Buchta, C.M.; Bishop, G.A. TRAF5 negatively regulates TLR signaling in B lymphocytes. J. Immunol. 2014, 192, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Au, P.Y.; Yeh, W.C. Physiological roles and mechanisms of signaling by TRAF2 and TRAF5. Adv. Exp. Med. Biol. 2007, 597, 32–47. [Google Scholar] [PubMed]
- Liu, S.; Chen, J.; Cai, X.; Wu, J.; Chen, X.; Wu, Y.T.; Sun, L.; Chen, Z.J. Mavs recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. eLife 2013, 2, e00785. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Deng, X.; Deng, J.; Zhou, J.; Ren, Y.; Liu, S.; Prusak, D.J.; Wood, T.G.; Bao, X. Functional motifs responsible for human metapneumovirus M2-2-mediated innate immune evasion. Virology 2016, 499, 361–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, E.D.; Wang, C.Y. TRAF5 is a downstream target of MAVS in antiviral innate immune signaling. PLoS ONE 2010, 5, e9172. [Google Scholar] [CrossRef] [PubMed]
- Lawan, A.; Bennett, A.M. Mitogen-activated protein kinase regulation in hepatic metabolism. Trends Endocrinol. Metab. 2017, 28, 868–878. [Google Scholar] [CrossRef] [PubMed]
- Selim, K.A.; Abdelrasoul, H.; Aboelmagd, M.; Tawila, A.M. The role of the MAPK signaling, topoisomerase and dietary bioactives in controlling cancer incidence. Diseases 2017, 5, 13. [Google Scholar] [CrossRef] [PubMed]
- Panahi, G.; Pasalar, P.; Zare, M.; Rizzuto, R.; Meshkani, R. High glucose induces inflammatory responses in HepG2 cells via the oxidative stress-mediated activation of NF-κB, and MAPK pathways in HepG2 cells. Arch. Physiol. Biochem. 2018, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Huang, G.; Wang, Z.; Qin, H.; Mo, B.; Wang, C. Elongation factor-2 kinase acts downstream of p38 MAPK to regulate proliferation, apoptosis and autophagy in human lung fibroblasts. Exp. Cell Res. 2018, 363, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Shen, G.N.; Luo, Y.H.; Piao, X.J.; Jiang, X.Y.; Meng, L.Q.; Wang, Y.; Zhang, Y.; Wang, J.R.; Wang, H.; et al. Novel 1,4-naphthoquinone derivatives induce apoptosis via ROS-mediated p38/MAPK, AKT and STAT3 signaling in human hepatoma HEP3B cells. Int. J. Biochem. Cell Biol. 2018, 96, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Nick, J.A.; Young, S.K.; Brown, K.K.; Avdi, N.J.; Arndt, P.G.; Suratt, B.T.; Janes, M.S.; Henson, P.M.; Worthen, G.S. Role of p38 mitogen-activated protein kinase in a murine model of pulmonary inflammation. J. Immunol. 2000, 164, 2151–2159. [Google Scholar] [CrossRef] [PubMed]
- Nishiura, H.; Zhao, R.; Yamamoto, T. The role of the ribosomal protein S19 C-terminus in altering the chemotaxis of leucocytes by causing functional differences in the C5A receptor response. J. Biochem. 2011, 150, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Senger, K.; Pham, V.C.; Varfolomeev, E.; Hackney, J.A.; Corzo, C.A.; Collier, J.; Lau, V.W.C.; Huang, Z.; Hamidzhadeh, K.; Caplazi, P.; et al. The kinase TPL2 activates ERK and p38 signaling to promote neutrophilic inflammation. Sci. Signal. 2017, 10. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Long, C.; Yang, G.; Luo, Y.; Du, H. MIR-26B inhibits melanoma cell proliferation and enhances apoptosis by suppressing TRAF5-mediated MAPK activation. Biochem. Biophys. Res. Commun. 2016, 471, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Esparza, E.M.; Lindsten, T.; Stockhausen, J.M.; Arch, R.H. Tumor necrosis factor receptor (TNFR)-associated factor 5 is a critical intermediate of costimulatory signaling pathways triggered by glucocorticoid-induced TNFR in t cells. J. Biol. Chem. 2006, 281, 8559–8564. [Google Scholar] [CrossRef] [PubMed]
- Shirakata, M.; Imadome, K.I.; Okazaki, K.; Hirai, K. Activation of TRAF5 and TRAF6 signal cascades negatively regulates the latent replication origin of epstein-barr virus through p38 mitogen-activated protein kinase. J. Virol. 2001, 75, 5059–5068. [Google Scholar] [CrossRef] [PubMed]
- Horie, R.; Ishida, T.; Maruyama-Nagai, M.; Ito, K.; Watanabe, M.; Yoneyama, A.; Higashihara, M.; Kimura, S.; Watanabe, T. Traf activation of C/EBPβ (NF-IL6) via p38 MAPK induces HIV-1 gene expression in monocytes/macrophages. Microbes Infect 2007, 9, 721–728. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.; Dong, W.; Qian, G.; Wang, J.; Li, X.; Cao, Z.; Lv, Q.; Wang, C.; Guo, K.; Zhang, Y. US10, a novel NPRO-interacting protein, inhibits classical swine fever virus replication. J. Gen. Virol. 2017, 98, 1679–1692. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Lamp, B.; Riedel, C.; Wentz, E.; Tortorici, M.A.; Rumenapf, T. Autocatalytic cleavage within classical swine fever virus NS3 leads to a functional separation of protease and helicase. J. Virol. 2013, 87, 11872–11883. [Google Scholar] [CrossRef] [PubMed]
- Tautz, N.; Kaiser, A.; Thiel, H.J. NS3 serine protease of bovine viral diarrhea virus: Characterization of active site residues, NS4A cofactor domain, and protease-cofactor interactions. Virology 2000, 273, 351–363. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Wang, C.; Liang, W.; Zhang, J.; Zhang, L.; Lv, H.; Dong, W.; Zhang, Y. RAB1A is required for assembly of classical swine fever virus particle. Virology 2018, 514, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Wang, C.; Zhang, L.; Wang, T.; Zhang, J.; Liang, W.; Li, C.; Qian, G.; Ouyang, Y.; Guo, K.; et al. Rab5 enhances classical swine fever virus proliferation and interacts with viral NS4B protein to facilitate formation of NS4B related complex. Front. Microbiol. 2017, 8, 1468. [Google Scholar] [CrossRef] [PubMed]
- Khan, K.A.; Abbas, W.; Varin, A.; Kumar, A.; Di Martino, V.; Dichamp, I.; Herbein, G. HIV-1 NEF interacts with HCV core, recruits TRAF2, TRAF5 and TRAF6, and stimulates HIV-1 replication in macrophages. J. Innate Immun. 2013, 5, 639–656. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Guo, K.; Zheng, M.; Ning, P.; Li, H.; Kang, K.; Lin, Z.; Zhang, C.; Liang, W.; Zhang, Y. A comparison of the impact of Shimen and C strains of classical swine fever virus on toll-like receptor expression. J. Gen. Virol. 2015, 96, 1732–1745. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, S.; Liao, Y.; Zhang, E.; Feng, S.; Yu, S.; Li, L.F.; He, W.R.; Li, Y.; Luo, Y.; et al. Mitogen-activated protein kinase kinase 2 (MEK2), a novel E2-interacting protein, promotes the growth of classical swine fever virus via attenuation of the JAK-STAT signaling pathway. J. Virol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Qadri, I.; Iwahashi, M.; Capasso, J.M.; Hopken, M.W.; Flores, S.; Schaack, J.; Simon, F.R. Induced oxidative stress and activated expression of manganese superoxide dismutase during hepatitis C virus replication: Role of JNK, p38 MAPK and AP-1. Biochem. J. 2004, 378, 919–928. [Google Scholar] [CrossRef] [PubMed]
- Waris, G.; Turkson, J.; Hassanein, T.; Siddiqui, A. Hepatitis C virus (HCV) constitutively activates STAT-3 via oxidative stress: Role of STAT-3 in HCV replication. J. Virol. 2005, 79, 1569–1580. [Google Scholar] [CrossRef] [PubMed]
- Kaminska, B. Mapk signalling pathways as molecular targets for anti-inflammatory therapy—From molecular mechanisms to therapeutic benefits. Biochim. Biophys. Acta 2005, 1754, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Bi, J.; Wang, D.; Fang, L.; Zhang, L.; Li, F.; Chen, H.; Xiao, S. Porcine reproductive and respiratory syndrome virus infection activates IL-10 production through NF-kappab and p38 MAPK pathways in porcine alveolar macrophages. Dev. Comp. Immunol. 2013, 39, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; Huang, Y.; Wang, T.; Zhang, X.; Chen, Y.; Cui, B.; Li, D.; Zhao, X.; Zhang, W.; Chang, L.; et al. Porcine circovirus type 2 activates PI3K/AKT and p38 MAPK pathways to promote interleukin-10 production in macrophages via cap interaction of GC1QR. Oncotarget 2016, 7, 17492–17507. [Google Scholar] [CrossRef] [PubMed]
- Mittal, S.K.; Roche, P.A. Suppression of antigen presentation by IL-10. Curr. Opin. Immunol. 2015, 34, 22–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conti, P.; Kempuraj, D.; Kandere, K.; Di Gioacchino, M.; Barbacane, R.C.; Castellani, M.L.; Felaco, M.; Boucher, W.; Letourneau, R.; Theoharides, T.C. IL-10, an inflammatory/inhibitory cytokine, but not always. Immunol. Lett. 2003, 86, 123–129. [Google Scholar] [CrossRef]
- Redpath, S.; Ghazal, P.; Gascoigne, N.R. Hijacking and exploitation of IL-10 by intracellular pathogens. Trends Microbiol. 2001, 9, 86–92. [Google Scholar] [CrossRef]
- Breen, E.C. Pro- and anti-inflammatory cytokines in human immunodeficiency virus infection and acquired immunodeficiency syndrome. Pharmacol. Ther. 2002, 95, 295–304. [Google Scholar] [CrossRef]
- Diaz-San Segundo, F.; Rodriguez-Calvo, T.; de Avila, A.; Sevilla, N. Immunosuppression during acute infection with foot-and-mouth disease virus in swine is mediated by IL-10. PLoS ONE 2009, 4, e5659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Mu, G.; Dang, R.; Yang, Z. Up-regulation of IL-10 upon PRRSV vaccination impacts on the immune response against CSFV. Vet. Microbiol. 2016, 197, 68–71. [Google Scholar] [CrossRef] [PubMed]




| Primers | Sequence (5′ → 3′) | Purpose |
|---|---|---|
| BD-NS3-F | GGAATTCCATATGCCTAAGAAAAAGCGCAAAGTTGGGCCTGCCGTTTGCAAGAAG | Amplification of NS3 |
| BD-NS3-R | AACTGCAGTAGCTCCTTCAATTCTGTCTCCTTCCCCTC | |
| NS3-GFP-F | CATGCTAGCGATGGGGCCTGCCGTTTGCAAGAAG | Amplification of NS3 |
| NS3-GFP-R | AACTGCAGTAGACCAACTACTTGTTTTAGTGCTCTGCC | |
| CMV-NS3-F | CTAGCTAGCATGGGGCCTGCCGTTTGC | Amplification of NS3 |
| CMV-NS3-R | ATAAGAATGCGGCCGCTTACTTATCGTCGTCATCCTTGTAATCTAGACCAACTACTTG | |
| GST-NS3-F | ACGCGTCGACTCGGGCCTGCCGTTTGCA | Amplification of NS3 |
| GST-NS3-R | ATAAGAATGCGGCCGCTCATAGACCAACTACTTGTTTTAGTGC | |
| RFP-F | GGAATTCATGGTGCGCTCCTCCAAGAAC | Amplification of RFP |
| RFP-R | CGGGATCCCTACTTATCGTCGTCATCCTTGTAATCCAGGAACAGGTGGTGGCGG | |
| AD-TRAF5-F | GGAATTCCATATGATGGCCTCTTCTGAGGAGCAAG | Amplification of TRAF5 |
| AD-TRAF5-R | CGGGATCCTTAGAGATCCTCCAGGTCAGTTAAATCC | |
| CMV-TRAF5-F | GCTCTAGAATGGCCTCTTCTGAGGAGCAAG | Amplification of TRAF5 |
| CMV-TRAF5-R | CGGGATCCGAGATCCTCCAGGTCAGTTAAATCC | |
| GST-TRAF5-F | CGGGATCCATGGCCTCTTCTGAGGAGCAAG | Amplification of TRAF5 |
| GST-TRAF5-R | CCGCTCGAGTTAGAGATCCTCCAGGTCAGTTAAATCC | |
| β-actin-F | CAAGGACCTCTACGCCAACAC | RT-qPCR for detection of β-actin |
| β-actin-R | TGGAGGCGCGATGATCTT | |
| CSFV-F | GATCCTCATACTGCCCACTTAC | RT-qPCR for detection of CSFV RT-qPCR for detection of TRAF5 |
| CSFV-R | GTATACCCCTTCACCAGCTTG | |
| TRAF5-F | GGGGAGACTAACAAACATGATG | |
| TRAF5-R | GTAGAAGGGCTGGCTGAAGA | |
| TRAF5-sh1-F | GATCCGGTGTACGGCCAAGATCATTCTCAAGAGGAATGATCTTGGCCGTACACCTTTTTG | Knockdown of TRAF5 |
| TRAF5-sh1-R | AATTCAAAAAGGTGTACGGCCAAGATCATTCCTCTTGAGAATGATCTTGGCCGTACACCG | |
| TRAF5-sh2-F | GATCCGGAAGGTGACAGACTACAAGCTCAAGAGGCTTGTAGTCTGTCACCTTCCTTTTTG | Knockdown of TRAF5 |
| TRAF5-sh2-R | AATTCAAAAAGGAAGGTGACAGACTACAAGCCTCTTGAGCTTGTAGTCTGTCACCTTCCG | |
| TRAF5-sh3-F | GATCCGCACCTGTCGCTGTACTTTGTTCAAGAGACAAAGTACAGCGACAGGTGCTTTTTG | Knockdown of TRAF5 |
| TRAF5-sh3-R | AATTCAAAAAGCACCTGTCGCTGTACTTTGTCTCTTGAACAAAGTACAGCGACAGGTGCG |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lv, H.; Dong, W.; Guo, K.; Jin, M.; Li, X.; Li, C.; Zhang, Y. Tumor Necrosis Factor Receptor-Associated Factor 5 Interacts with the NS3 Protein and Promotes Classical Swine Fever Virus Replication. Viruses 2018, 10, 305. https://doi.org/10.3390/v10060305
Lv H, Dong W, Guo K, Jin M, Li X, Li C, Zhang Y. Tumor Necrosis Factor Receptor-Associated Factor 5 Interacts with the NS3 Protein and Promotes Classical Swine Fever Virus Replication. Viruses. 2018; 10(6):305. https://doi.org/10.3390/v10060305
Chicago/Turabian StyleLv, Huifang, Wang Dong, Kangkang Guo, Mingxing Jin, Xiaomeng Li, Cunfa Li, and Yanming Zhang. 2018. "Tumor Necrosis Factor Receptor-Associated Factor 5 Interacts with the NS3 Protein and Promotes Classical Swine Fever Virus Replication" Viruses 10, no. 6: 305. https://doi.org/10.3390/v10060305




