Identification of a Novel Recombinant Type 2 Porcine Reproductive and Respiratory Syndrome Virus in China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Virus Isolation
2.2. RT-PCR Amplification and Genome Sequencing
2.3. Sequence Comparison and Phylogenetic Analysis
2.4. Putative Recombination Analysis
3. Results
3.1. Genome Characterization and Homology Analysis
3.2. Phylogenetic Analysis
3.3. Amino Acid Analysis of Nsp2 and GP5
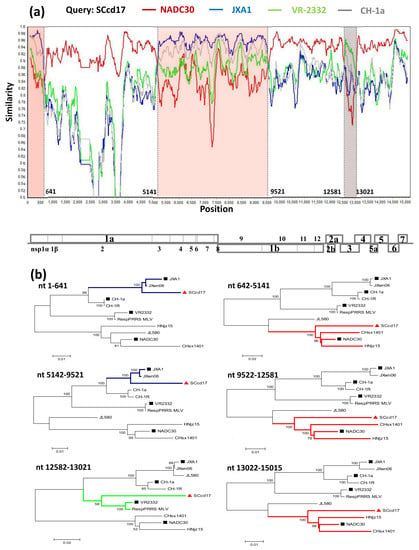
3.4. Recombination Analysis
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Keffaber, K. Reproductive failure of unknown etiol-ogy. Am. Assoc. Swine Pract. Newsl. 1989, 1, 1–9. [Google Scholar]
- Benfield, D.A.; Nelson, E.; Collins, J.E.; Harris, L.; Goyal, S.M.; Robison, D.; Christianson, W.T.; Morrison, R.B.; Gorcyca, D.; Chladek, D. Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR-2332). J. Vet. Diagn. Investig. 1992, 4, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Cavanagh, D. Nidovirales: A new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 1997, 142, 629–633. [Google Scholar] [PubMed]
- Adams, M.J.; Lefkowitz, E.J.; King, A.M.Q.; Harrach, B.; Harrison, R.L.; Knowles, N.J.; Kropinski, A.M.; Krupovic, M.; Kuhn, J.H.; Mushegian, A.R.; et al. Changes to taxonomy and the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses (2017). Arch. Virol. 2017, 162, 2505–2538. [Google Scholar] [CrossRef] [PubMed]
- Meulenberg, J.J.; Petersen-den Besten, A.; de Kluyver, E.P.; Moormann, R.J.; Schaaper, W.M.; Wensvoort, G. Characterization of proteins encoded by ORFs 2 to 7 of Lelystad virus. Virology 1995, 206, 155–163. [Google Scholar] [CrossRef]
- Van Dinten, L.C.; Wassenaar, A.L.; Gorbalenya, A.E.; Spaan, W.J.; Snijder, E.J. Processing of the equine arteritis virus replicase ORF1b protein: Identification of cleavage products containing the putative viral polymerase and helicase domains. J. Virol. 1996, 70, 6625–6633. [Google Scholar] [PubMed]
- Johnson, C.R.; Griggs, T.F.; Gnanandarajah, J.; Murtaugh, M.P. Novel structural protein in porcine reproductive and respiratory syndrome virus encoded by an alternative ORF5 present in all arteriviruses. J. Gen. Virol. 2011, 92, 1107–1116. [Google Scholar] [CrossRef] [PubMed]
- Mardassi, H.; Massie, B.; Dea, S. Intracellular synthesis, processing, and transport of proteins encoded by ORFs 5 to 7 of porcine reproductive and respiratory syndrome virus. Virology 1996, 221, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Kappes, M.A.; Faaberg, K.S. Prrsv structure, replication and recombination: Origin of phenotype and genotype diversity. Virology 2015, 479–480, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Nelson, E.A.; Christopher-Hennings, J.; Drew, T.; Wensvoort, G.; Collins, J.E.; Benfield, D.A. Differentiation of U.S. and European isolates of porcine reproductive and respiratory syndrome virus by monoclonal antibodies. J. Clin. Microbiol. 1993, 31, 3184–3189. [Google Scholar] [PubMed]
- Murtaugh, M.P.; Elam, M.R.; Kakach, L.T. Comparison of the structural protein coding sequences of the VR-2332 and Lelystad virus strains of the PRRS virus. Arch. Virol. 1995, 140, 1451–1460. [Google Scholar] [CrossRef] [PubMed]
- Van Nieuwstadt, A.P.; Meulenberg, J.J.; van Essen-Zanbergen, A.; Petersen-den Besten, A.; Bende, R.J.; Moormann, R.J.; Wensvoort, G. Proteins encoded by open reading frames 3 and 4 of the genome of Lelystad virus (Arteriviridae) are structural proteins of the virion. J. Virol. 1996, 70, 4767–4772. [Google Scholar] [PubMed]
- Guo, B.; Chen, Z.; Liu, W. Isolation and identification of porcine reproductory and respiratory syndrome (PRRS) virus from aborted fetuses suspected of PRRS. Chin. J. Prev. Vet. Med. 1996, 2, 1–5. [Google Scholar]
- Tian, K.; Yu, X.; Zhao, T.; Feng, Y.; Cao, Z.; Wang, C.; Hu, Y.; Chen, X.; Hu, D.; Tian, X.; et al. Emergence of fatal PRRSV variants: Unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS ONE 2007, 2, e526. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.J.; Hao, X.F.; Tian, Z.J.; Tong, G.Z.; Yoo, D.; An, T.Q.; Zhou, T.; Li, G.X.; Qiu, H.J.; Wei, T.C.; et al. Highly virulent porcine reproductive and respiratory syndrome virus emerged in China. Transbound. Emerg. Dis. 2008, 55, 152–164. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Fang, L.; Xu, Z.; Liu, S.; Gao, J.; Jiang, Y.; Chen, H.; Xiao, S. Recombination in vaccine and circulating strains of porcine reproductive and respiratory syndrome viruses. Emerg. Infect. Dis. 2009, 15, 2032–2035. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Chen, S.; Zhang, J.; Zeng, J.; Guo, X.; Ge, X.; Zhang, D.; Yang, H. Molecular variation analysis of porcine reproductive and respiratory syndrome virus in China. Virus Res. 2009, 145, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.H.; Tun, H.M.; Sun, B.L.; Mo, J.; Zhou, Q.F.; Deng, Y.X.; Xie, Q.M.; Bi, Y.Z.; Leung, F.C.; Ma, J.Y. Re-emerging of porcine respiratory and reproductive syndrome virus (lineage 3) and increased pathogenicity after genomic recombination with vaccine variant. Vet. Microbiol. 2015, 175, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Ye, C.; Chang, X.B.; Jiang, C.G.; Wang, S.J.; Cai, X.H.; Tong, G.Z.; Tian, Z.J.; Shi, M.; An, T.Q. Importation and recombination are responsible for the latest emergence of highly pathogenic porcine reproductive and respiratory syndrome virus in China. J. Virol. 2015, 89, 10712–10716. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Wang, Z.; Ding, Y.; Ge, X.; Guo, X.; Yang, H. NADC30-like strain of porcine reproductive and respiratory syndrome virus, China. Emerg. Infect. Dis. 2015, 21, 2256–2257. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhuang, J.; Wang, J.; Han, L.; Sun, Z.; Xiao, Y.; Ji, G.; Li, Y.; Tan, F.; Li, X.; et al. Outbreak investigation of NADC30-like PRRSV in South-East China. Transbound. Emerg. Dis. 2016, 63, 474–479. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Fang, L.; Guo, X.; Gao, J.; Song, T.; Bi, J.; He, K.; Chen, H.; Xiao, S. Epidemiology and evolutionary characteristics of the porcine reproductive and respiratory syndrome virus in China between 2006 and 2010. J. Clin. Microbiol. 2011, 49, 3175–3183. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.K.; Wei, C.H.; Yang, X.Y.; Hou, X.L.; Dai, A.L.; Li, X.H.; Wei, M.K.; Pan, X.Z. Genetic diversity and evolutionary characterization of Chinese porcine reproductive and respiratory syndrome viruses based on nsp2 and ORF5. Arch. Virol. 2013, 158, 1811–1816. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Zhu, W.; Chen, Y.; Wei, C.; Zhou, P.; Zhang, M.; Huang, Z.; Sun, L.; Su, S.; Zhang, G. Molecular epidemiology of PRRSV in South China from 2007 to 2011 based on the genetic analysis of ORF5. Microb. Pathog. 2013, 63, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Yang, X.; Tian, Y.; Yin, S.; Geng, G.; Ge, X.; Guo, X.; Yang, H. Genetic diversity analysis of genotype 2 porcine reproductive and respiratory syndrome viruses emerging in recent years in China. BioMed Res. Int. 2014, 2014, 748068. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, J.; Tan, F.; Li, Y.; Ji, G.; Zhuang, J.; Zhai, X.; Tian, K. Genome characterization of two NADC30-like porcine reproductive and respiratory syndrome viruses in China. Springerplus 2016, 5, 1677. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Jiang, P.; Song, Z.; Lv, L.; Li, L.; Bai, J. Pathogenicity and antigenicity of a novel NADC30-like strain of porcine reproductive and respiratory syndrome virus emerged in China. Vet. Microbiol. 2016, 197, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Bian, T.; Sun, Y.; Hao, M.; Zhou, L.; Ge, X.; Guo, X.; Han, J.; Yang, H. A recombinant type 2 porcine reproductive and respiratory syndrome virus between NADC30-like and a MLV-like: Genetic characterization and pathogenicity for piglets. Infect. Genet. Evol. 2017, 54, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Bao, H.; Wang, Y.; Tian, K. Widespread of NADC30-like PRRSV in China: Another Pandora’s box for Chinese pig industry as the outbreak of highly pathogenic PRRSV in 2006? Infect. Genet. Evol. 2017, 49, 12–13. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.J.; Wan, B.; Guo, Z.; Qiao, S.; Li, R.; Xie, S.; Chen, X.X.; Zhang, G. Genomic analysis of a recombinant NADC30-like porcine reproductive and respiratory syndrome virus in China. Virus Genes 2017. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yang, X.; Wang, H.N.; Zhang, A.; Zhang, Z.; Kang, R.; Zeng, F.; Li, H. Molecular characterization of a complete genome and 12 Nsp2 genes of PRRSV of Southwestern China. Food Environ. Virol. 2012, 4, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Kang, R.; Ji, G.; Tian, Y.; Ge, M.; Xie, B.; Yang, X.; Wang, H. Molecular characterization and recombination analysis of porcine reproductive and respiratory syndrome virus emerged in Southwestern China during 2012–2016. Virus Genes 2017. [Google Scholar] [CrossRef] [PubMed]
- An, T.Q.; Tian, Z.J.; Xiao, Y.; Li, R.; Peng, J.M.; Wei, T.C.; Zhang, Y.; Zhou, Y.J.; Tong, G.Z. Origin of highly pathogenic porcine reproductive and respiratory syndrome virus, China. Emerg. Infect. Dis. 2010, 16, 365–367. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zhang, J.; Zeng, J.; Yin, S.; Li, Y.; Zheng, L.; Guo, X.; Ge, X.; Yang, H. The 30-amino-acid deletion in the Nsp2 of highly pathogenic porcine reproductive and respiratory syndrome virus emerging in China is not related to its virulence. J. Virol. 2009, 83, 5156–5167. [Google Scholar] [CrossRef] [PubMed]
- Murtaugh, M.P.; Stadejek, T.; Abrahante, J.E.; Lam, T.T.; Leung, F.C. The ever-expanding diversity of porcine reproductive and respiratory syndrome virus. Virus Res. 2010, 154, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Brar, M.S.; Shi, M.; Murtaugh, M.P.; Leung, F.C. Evolutionary diversification of type 2 porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 2015, 96, 1570–1580. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.K.; Zhou, X.; Zhai, J.Q.; Li, B.; Wei, C.H.; Dai, A.L.; Yang, X.Y.; Luo, M.L. Emergence of a novel highly pathogenic porcine reproductive and respiratory syndrome virus in China. Transbound. Emerg. Dis. 2017. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Han, Q.; Zhang, L.; Zhang, Z.; Wu, Y.; Shen, H.; Jiang, P. Emergence of mosaic recombinant strains potentially associated with vaccine JXA1-R and predominant circulating strains of porcine reproductive and respiratory syndrome virus in different provinces of China. Virol. J. 2017, 14, 67. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Christopher-Hennings, J.; Brown, E.; Liu, H.; Chen, Z.; Lawson, S.R.; Breen, R.; Clement, T.; Gao, X.; Bao, J.; et al. Development of genetic markers in the non-structural protein 2 region of a us type 1 porcine reproductive and respiratory syndrome virus: Implications for future recombinant marker vaccine development. J. Gen. Virol. 2008, 89, 3086–3096. [Google Scholar] [CrossRef] [PubMed]
- Yin, G.; Gao, L.; Shu, X.; Yang, G.; Guo, S.; Li, W. Genetic diversity of the ORF5 gene of porcine reproductive and respiratory syndrome virus isolates in Southwest China from 2007 to 2009. PLoS ONE 2012, 7, e33756. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Cui, T.; Cui, J.; Chen, Y.; Zhang, M.; Zhou, P.; Deng, S.; Su, S.; Zhang, G. Epidemiological and evolutionary characteristics of the PRRSV in Southern China from 2010 to 2013. Microb. Pathog. 2014, 75, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.X.; Qin, L.T.; Liu, Y.; Liu, X.; Sun, N.; Yang, Y.; Chen, T.; Zhu, H.W.; Ren, J.Q.; Sun, Y.J.; et al. Novel Nsp2 deletion based on molecular epidemiology and evolution of porcine reproductive and respiratory syndrome virus in Shandong Province from 2013 to 2014. Infect. Genet. Evol. 2015, 33, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Brockmeier, S.L.; Loving, C.L.; Vorwald, A.C.; Kehrli, M.E., Jr.; Baker, R.B.; Nicholson, T.L.; Lager, K.M.; Miller, L.C.; Faaberg, K.S. Genomic sequence and virulence comparison of four type 2 porcine reproductive and respiratory syndrome virus strains. Virus Res. 2012, 169, 212–221. [Google Scholar] [CrossRef] [PubMed]
- De Lima, M.; Pattnaik, A.K.; Flores, E.F.; Osorio, F.A. Serologic marker candidates identified among B-cell linear epitopes of Nsp2 and structural proteins of a north American strain of porcine reproductive and respiratory syndrome virus. Virology 2006, 353, 410–421. [Google Scholar] [CrossRef] [PubMed]
- Yoshii, M.; Okinaga, T.; Miyazaki, A.; Kato, K.; Ikeda, H.; Tsunemitsu, H. Genetic polymorphism of the nsp2 gene in north American type—Porcine reproductive and respiratory syndrome virus. Arch. Virol. 2008, 153, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Ostrowski, M.; Galeota, J.A.; Jar, A.M.; Platt, K.B.; Osorio, F.A.; Lopez, O.J. Identification of neutralizing and nonneutralizing epitopes in the porcine reproductive and respiratory syndrome virus GP5 ectodomain. J. Virol. 2002, 76, 4241–4250. [Google Scholar] [CrossRef] [PubMed]
- Ansari, I.H.; Kwon, B.; Osorio, F.A.; Pattnaik, A.K. Influence of n-linked glycosylation of porcine reproductive and respiratory syndrome virus GP5 on virus infectivity, antigenicity, and ability to induce neutralizing antibodies. J. Virol. 2006, 80, 3994–4004. [Google Scholar] [CrossRef] [PubMed]
- Allende, R.; Kutish, G.F.; Laegreid, W.; Lu, Z.; Lewis, T.L.; Rock, D.L.; Friesen, J.; Galeota, J.A.; Doster, A.R.; Osorio, F.A. Mutations in the genome of porcine reproductive and respiratory syndrome virus responsible for the attenuation phenotype. Arch. Virol. 2000, 145, 1149–1161. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhou, R.; Zhang, J.; Zhou, L.; Jiang, Q.; Guo, X.; Ge, X.; Yang, H. Recombination analyses between two strains of porcine reproductive and respiratory syndrome virus in vivo. Virus Res. 2011, 155, 473–486. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhou, L.; Zhang, J.; Ge, X.; Zhou, R.; Zheng, H.; Geng, G.; Guo, X.; Yang, H. Nsp9 and Nsp10 contribute to the fatal virulence of highly pathogenic porcine reproductive and respiratory syndrome virus emerging in China. PLoS Pathog. 2014, 10, e1004216. [Google Scholar] [CrossRef] [PubMed]




| Reference Strains | Country/Year | Accession No. | Reference Strains | Country/Year | Accession No. |
|---|---|---|---|---|---|
| VR-2332 abc | USA/1995 | AY150564 | WUH4 a | CHN/2012 | JQ326271 |
| CH-1a abc | CHN/1996 | AY032626 | HENAN-XINX a | CHN/2013 | KF611905 |
| BJ-4 a | CHN/2000 | AF331831 | JL580 ab | CHN/2013 | KR706343 |
| RespPRRS MLV ab | USA/2005 | AF066183 | XW015 a | USA/2013 | KF724409 |
| JXA1 abc | CHN/2006 | EF112445 | FJ1402 a | CHN/2014 | KX169191 |
| TJ ab | CHN/2006 | EU860248 | CHsx1401 ab | CHN/2014 | KP861625 |
| MN184A a | USA/2006 | DQ176019 | HNjz15 ab | CHN/2015 | KT945017 |
| jiangxi-3 a | CHN/2007 | EU200961 | FJZ03 a | CHN/2015 | KP860909 |
| 07BJ a | CHN/2007 | FJ393459 | HNyc15 a | CHN/2015 | KT945018 |
| CH-1R a | CHN/2008 | EU807840 | FJY04 a | CHN/2015 | KP860910 |
| NADC30 abc | USA/2008 | JN654459 | WUH5 a | CHN/2015 | KU523366 |
| JXwn06 ab | CHN/2008 | EF641008 | HENZMD-9 a | CHN/2015 | KU950374 |
| JXA1 P80 a | CHN/2008 | FJ548853 | HNhx a | CHN/2016 | KX766379 |
| HENAN-HEB ab | CHN/2012 | KJ143621 |
| JXA1 | JXwn06 | TJ | VR-2332 | RespPRRSV MLV | CH-1a | NADC30 | JL580 | CHsx1401 | HENAN-HEB | HNjz15 | LV | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| JXA1-Like | VR-2332-Like | CH-1a-Like | NADC30-Like | Type 1 | ||||||||
| Pairwise % Identity to SCcd17 (nt/aa) | ||||||||||||
| Complete genome | 85.2 | 85.3 | 92.7 | 84.0 | 83.9 | 84.9 | 90.2 | 88.3 | 88.1 | 87.6 | 88.3 | 43.4 |
| 5′UTR | 97.8 | 97.8 | 94.9 | 92.7 | 92.7 | 96.7 | 92.2 | 91.5 | 90.3 | 92.2 | 90.9 | 49.2 |
| ORF1a | 82.1 | 82.4 | 82.1 | 79.9 | 79.8 | 81.0 | 88.5 | 86.6 | 86.1 | 84.1 | 86.1 | 32.5 |
| ORF1b | 94.3 | 94.4 | 94.1 | 89.1 | 89.0 | 92.3 | 85.6 | 84.5 | 84.7 | 84.7 | 83.6 | 50.0 |
| nsp1α | 97.3/97.7 | 98.1/97.7 | 98.1/97.7 | 89.7/96.0 | 89.7/96.0 | 94.8/96.0 | 86.2/94.8 | 86.5/96.0 | 83.5/94.8 | 85.3/94.3 | 85.5/94.8 | 49.4/43.3 |
| nsp1β | 88.4/77.9 | 88.4/77.9 | 88.4/77.9 | 89.1/77.9 | 88.9/77.3 | 87.5/74.8 | 94.4/85.7 | 93.3/83.9 | 93.7/84.5 | 92.6/83.3 | 92.2/85.7 | 31.9/25.6 |
| nsp2 | 67.1/66.3 | 67.0/66.4 | 66.5/66.4 | 71.7/67.4 | 71.6/67.3 | 68.1/64.9 | 92.3/89.9 | 82.7/80.2 | 89.0/85.5 | 85.0/82.0 | 89.6/87.4 | 23.6/20.9 |
| nsp3 | 89.1/92.3 | 88.9/92.8 | 88.8/92.3 | 87.8/92.3 | 87.7/92.3 | 88.1/91.8 | 90.4/94.2 | 93.4/94.2 | 90.0/93.7 | 90.8/93.7 | 90.4/94.2 | 32.8/22.7 |
| nsp4 | 97.8/98.0 | 97.9/98.0 | 97.9/98.0 | 92.4/92.9 | 92.4/92.9 | 95.8/94.4 | 88.1/91.8 | 97.0/97.0 | 87.6/92.3 | 86.9/90.2 | 87.8/91.3 | 44.8/54.9 |
| nsp5 | 96.4/95.8 | 97.0/95.8 | 96.8/95.8 | 89.9/88.1 | 89.9/88.1 | 95.0/92.6 | 91.5/92.6 | 93.3/92.0 | 89.1/88.1 | 89.5/90.7 | 88.0/86.1 | 50.9/38.9 |
| nsp6 | 97.9/100 | 97.9/100 | 95.6/100 | 93.3/93.1 | 93.3/93.1 | 95.5/100 | 95.6/93.1 | 97.9/100 | 82.7/85.7 | 90.6/93.1 | 92.1/93.1 | 58.1/69.0 |
| nsp7α | 96.3/87.3 | 96.8/87.3 | 96.8/87.3 | 87.4/84.4 | 87.4/84.4 | 94.6/95.9 | 80.1/90.8 | 92.3/97.3 | 79.7/89.3 | 77.7/88.6 | 79.2/90.8 | 29.6/42.7 |
| nsp7β | 95.3/96.3 | 96.0/97.2 | 95.6/96.3 | 81.7/76.3 | 81.7/76.3 | 90.9/91.4 | 73.2/67.8 | 73.9/72.7 | 71.9/65.2 | 72.1/66.5 | 68.8/63.9 | 22.8/20.9 |
| nsp8 | 96.2/97.7 | 97.0/97.7 | 97.0/97.7 | 92.8/97.7 | 92.8/97.7 | 97.0/97.7 | 86.9/97.7 | 85.9/97.7 | 86.8/95.3 | 82.0/92.9 | 83.0/92.9 | 48.8/65.0 |
| nsp9 | 96.0/98.7 | 96.1/99.0 | 95.8/99.0 | 89.4/97.4 | 89.4/97.4 | 93.6/98.1 | 84.3/97.2 | 83.2/96.3 | 83.9/96.3 | 83.5/96.3 | 82.5/96.6 | 54.0/69.8 |
| nsp10 | 82.9/94.4 | 83.0/95.1 | 82.8/94.6 | 85.8/95.6 | 85.5/95.6 | 84.1/94.4 | 94.6/95.1 | 93.3/97.9 | 92.2/98.2 | 93.5/97.5 | 92.6/97.7 | 46.4/57.8 |
| nsp11 | 88.2/94.4 | 88.6/94.9 | 88.2/94.4 | 86.9/92.5 | 87.6/94.0 | 91.1/94.4 | 94.5/94.9 | 92.2/94.4 | 88.4/93.5 | 94.6/95.4 | 92.3/94.4 | 53.5/74.5 |
| nsp12 | 86.3/95.3 | 86.3/95.3 | 86.3/95.3 | 85.0/93.2 | 85.3/93.2 | 87.2/93.9 | 95.7/97.3 | 94.3/98.7 | 95.0/96.7 | 95.2/98.0 | 95.2/97.3 | 24.3/15.5 |
| ORF2a | 84.3/85.2 | 84.6/85.2 | 84.6/85.2 | 88.3/89.7 | 88.0/89.2 | 86.1/87.9 | 92.8/92.3 | 91.8/91.4 | 90.9/91.0 | 91.6/90.1 | 92.3/93.1 | 52.4/62.0 |
| ORF2b | 86.9/85.3 | 86.9/85.3 | 86.985.3 | 90.7/86.8 | 90.2/86.8 | 86.5/83.7 | 93.9/91.4 | 94.9/94.4 | 92.9/91.4 | 93.4/92.9 | 96.3/94.4 | 65.2/67.1 |
| ORF3 | 83.2/80.0 | 83.0/80.5 | 83.2/80.5 | 88.2/83.8 | 88.6/84.7 | 85.2/81.9 | 84.5/84.3 | 82.7/80.0 | 84.5/85.2 | 93.7/83.8 | 83.2/84.3 | 52.7/47.1 |
| ORF4 | 80.5/82.9 | 81.9/84.9 | 81.7/85.5 | 82.5/82.2 | 82.5/82.2 | 82.7/84.9 | 94.3/92.4 | 84.8/84.9 | 92.9/91.8 | 92.7/90.0 | 92.2/90.6 | 58.5/61.4 |
| ORF5 | 88.8/83.0 | 89.0/83.6 | 89.0/83.6 | 89.1/81.2 | 89.2/81.8 | 90.2/84.2 | 96.4/92.7 | 94.9/88.8 | 95.4/91.6 | 95.3/91.6 | 95.7/93.2 | 46.2/42.2 |
| ORF6 | 87.0/93.5 | 86.8/93.5 | 86.8/93.5 | 88.1/94.1 | 88.4/94.1 | 85.9/92.2 | 96.9/98.3 | 95.6/97.1 | 95.4/98.3 | 96.1/98.3 | 95.6/98.3 | 60.4/78.8 |
| ORF7 | 87.7/87.9 | 85.6/83.2 | 87.7/87.9 | 90.4/91.5 | 90.4/91.5 | 89.0/89.7 | 97.2/96.7 | 95.5/94.1 | 94.3/92.4 | 94.9/94.1 | 95.8/95.9 | 55.0/51.7 |
| 3′UTR | 89.2 | 82.3 | 89.2 | 93.0 | 93.0 | 88.5 | 97.3 | 97.3 | 96.6 | 96.6 | 96.6 | 56.0 |
| Strains | Amino Acid Sites | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 29 | 35 | 76 | 120 | 152 | 193 | 267 | 300 | 314 | 470 | 485 | 534 | 553 | 608 | 610 | 636 | 785 | 846 | 886 | 924 | 958 | 1093 | |
| VR-2332 | A | A | P | E | L | S | E | Q | V | L | S | P | Q | E | S | K | Q | S | F | G | D | R |
| RespPRRSMLV | A | A | P | E | L | S | E | Q | V | M | S | P | Q | E | S | K | Q | S | F | G | D | R |
| CH-1a | A | A | P | E | L | S | E | Q | V | M | G | P | Q | G | S | K | Q | S | F | G | D | R |
| JXA1 | A | A | P | E | L | S | E | Q | V | M | G | DL | DL | E | S | K | Q | S | F | G | D | R |
| JXwn06 | A | A | P | E | L | S | E | Q | V | M | G | DL | DL | E | S | K | Q | S | F | G | D | R |
| TJ | A | A | P | E | L | S | E | Q | V | L | G | DL | DL | E | S | K | Q | S | F | G | D | R |
| NADC30 | A | A | P | E | L | F | E | Q | V | L | G | L | Q | E | S | K | Q | S | F | G | D | R |
| CHsx1401 | A | A | P | E | L | S | E | Q | V | L | G | L | Q | E | S | K | Q | S | F | G | D | R |
| JL580 | A | A | P | E | L | F | E | Q | V | L | G | L | Q | E | S | K | Q | S | F | G | D | R |
| HENAN-HEB | A | A | P | E | L | S | E | Q | V | L | G | L | Q | E | S | K | P | S | F | G | D | R |
| HNjz15 | A | A | P | E | L | S | E | Q | V | L | G | L | Q | E | S | K | Q | N | F | G | D | R |
| SCcd17 | T | V | S | R | S | I | D | R | A | S | A | S | R | K | A | R | R | I | L | S | E | K |
| Strains | Potential N-glycosylation Sites | |||||||
|---|---|---|---|---|---|---|---|---|
| 30 | 32 | 33 | 34 | 35 | 44 | 51 | 58 | |
| VR-2332 | + | - | + | - | - | + | + | - |
| RespPRRS MLV | + | - | + | - | - | + | + | - |
| CH-1a | - | - | - | + | - | + | + | - |
| JXA1 | + | - | - | + | + | + | + | - |
| JXwn06 | + | - | - | + | + | + | + | - |
| TJ | + | - | - | + | + | + | + | - |
| NADC30 | - | - | - | + | - | + | + | - |
| CHsx1401 | - | - | - | + | - | + | + | - |
| JL580 | - | - | - | + | - | + | + | - |
| HENAN-HEB | - | + | - | - | - | + | + | - |
| HNzj15 | - | - | - | + | - | + | + | - |
| SCcd17 | - | + | - | + | - | + | + | + |
| Strains | Isolation Date | Recombination with | Recombination Regions | Accession No. | References |
|---|---|---|---|---|---|
| HENAN-HEB | 2013 | JXA1 | nsp2 | KJ143621 | [29] |
| HENAN-XINX | 2013 | VR-2332 | nsp2–5 | KF611905 | [29] |
| JL580 | 2014 | 09NEN1 (JXA1-like) | nsp2, nsp3, nsp7, ORF2a, ORF4 | KR706343 | [19] |
| Chsx1401 | 2014 | VR-2332 | nsp11 | KP861625 | [29] |
| FJ1402 | 2014 | GD (JXA1-like) | nsp2–3, nsp12, ORF3 | KX169191 | [27] |
| HENZMD-9 | 2015 | JXA1 | 5′-UTR-nsp2, nsp4–9 | KU950374 | [30] |
| HNyc15 | 2015 | VR-2332/CH-1a | ORF2–4 | KT945018 | [26] |
| 15HEN1 | 2015 | JXA1-R a | nsp3–9 | KX815413 | [38] |
| 15LN3 | 2015 | JXA1-R a | nsp3–9 | KX815425 | [38] |
| 15SC3 | 2015 | JXA1-R a | 5′-UTR-nsp2, nsp7–nsp9 | KX815428 | [38] |
| TJnh1501 | 2015 | TJbd14-1(JXA1-like) | nsp2 | KX510269 | [28] |
| SCnj16 | 2016 | JXA1 | 5′-UTR, nsp1–2, nsp3–9 | MF196906 | [32] |
| HNhx | 2016 | JXA1 | nsp4–9 | KX766379 | [30] |
| SCcd17 | 2017 | JXA1+VR-2332 | JXA1(5′-UTR-nsp1α, nsp3–9) VR-2332 (ORF3–4) | MG914067 | This study |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, L.; Kang, R.; Xie, B.; Tian, Y.; Wu, X.; Lv, X.; Yang, X.; Wang, H. Identification of a Novel Recombinant Type 2 Porcine Reproductive and Respiratory Syndrome Virus in China. Viruses 2018, 10, 151. https://doi.org/10.3390/v10040151
Zhou L, Kang R, Xie B, Tian Y, Wu X, Lv X, Yang X, Wang H. Identification of a Novel Recombinant Type 2 Porcine Reproductive and Respiratory Syndrome Virus in China. Viruses. 2018; 10(4):151. https://doi.org/10.3390/v10040151
Chicago/Turabian StyleZhou, Long, Runmin Kang, Bo Xie, Yiming Tian, Xuan Wu, Xuebin Lv, Xin Yang, and Hongning Wang. 2018. "Identification of a Novel Recombinant Type 2 Porcine Reproductive and Respiratory Syndrome Virus in China" Viruses 10, no. 4: 151. https://doi.org/10.3390/v10040151