Visualizing the Nucleotide Addition Cycle of Viral RNA-Dependent RNA Polymerase
Abstract
:1. Introduction
2. Visualizing the Viral RNA-Dependent RNA Polymerases Nucleotide Addition Cycle States and Related Key Conformational Changes
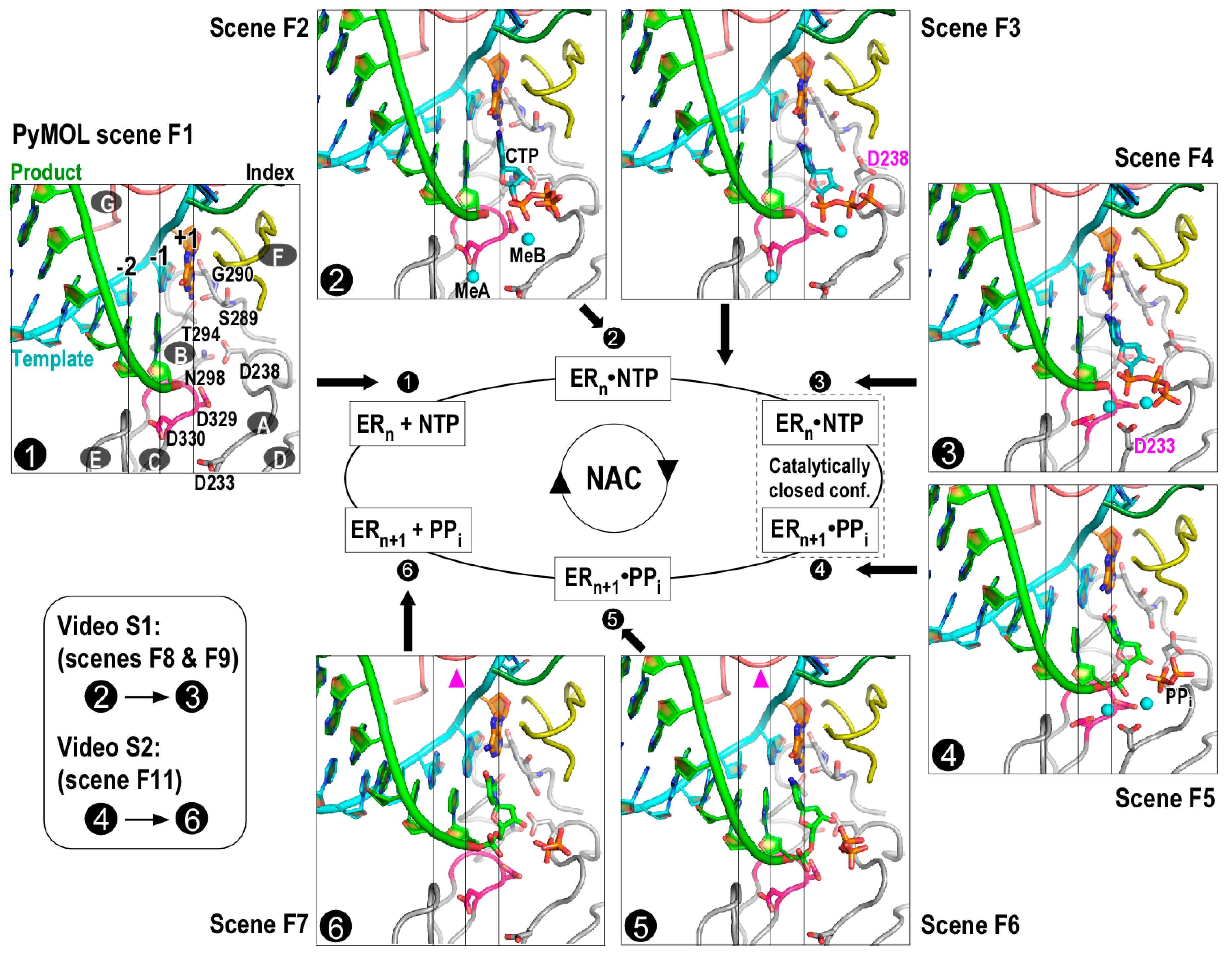
2.1. Model Generation of Reference Nucleotide Addition Cycle States
2.2. Generation of Videos Illustrating Key Conformation Changes
3. Correlation between Structural and Biochemical Data
4. Perspectives
Supplementary Materials
Acknowledgments
Conflicts of Interest
References
- Bruenn, J.A. A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucleic Acids Res. 2003, 31, 1821–1829. [Google Scholar] [CrossRef] [PubMed]
- Poch, O.; Sauvaget, I.; Delarue, M.; Tordo, N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989, 8, 3867–3874. [Google Scholar] [PubMed]
- Gorbalenya, A.E.; Pringle, F.M.; Zeddam, J.L.; Luke, B.T.; Cameron, C.E.; Kalmakoff, J.; Hanzlik, T.N.; Gordon, K.H.; Ward, V.K. The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J. Mol. Biol. 2002, 324, 47–62. [Google Scholar] [CrossRef]
- Te Velthuis, A.J. Common and unique features of viral RNA-dependent polymerases. Cell Mol. Life Sci. 2014, 71, 4403–4420. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, W.; Gong, P. A Structural Overview of RNA-Dependent RNA Polymerases from the Flaviviridae Family. Int. J. Mol. Sci. 2015, 16, 12943–12957. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Orta, C.; Arias, A.; Escarmis, C.; Verdaguer, N. A comparison of viral RNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 2006, 16, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Peersen, O.B. Picornaviral polymerase structure, function, and fidelity modulation. Virus Res. 2017, 234, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Kohlstaedt, L.A.; Wang, J.; Friedman, J.M.; Rice, P.A.; Steitz, T.A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 1992, 256, 1783–1790. [Google Scholar] [CrossRef] [PubMed]
- Beese, L.S.; Derbyshire, V.; Steitz, T.A. Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science 1993, 260, 352–355. [Google Scholar] [CrossRef] [PubMed]
- Sousa, R.; Chung, Y.J.; Rose, J.P.; Wang, B.C. Crystal structure of bacteriophage T7 RNA polymerase at 3.3 A resolution. Nature 1993, 364, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Shu, B.; Gong, P. The uncoupling of catalysis and translocation in the viral RNA-dependent RNA polymerase. RNA Biol. 2017, 14, 1314–1319. [Google Scholar] [CrossRef] [PubMed]
- Gong, P.; Peersen, O.B. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. USA 2010, 107, 22505–22510. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.W.; Steitz, T.A. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell 2004, 116, 393–404. [Google Scholar] [CrossRef]
- Gong, P.; Kortus, M.G.; Nix, J.C.; Davis, R.E.; Peersen, O.B. Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts. PLoS ONE 2013, 8, e60272. [Google Scholar] [CrossRef] [PubMed]
- Shu, B.; Gong, P. Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation. Proc. Natl. Acad. Sci. USA 2016, 113, E4005–E4014. [Google Scholar] [CrossRef] [PubMed]
- Zamyatkin, D.F.; Parra, F.; Alonso, J.M.; Harki, D.A.; Peterson, B.R.; Grochulski, P.; Ng, K.K. Structural insights into mechanisms of catalysis and inhibition in Norwalk virus polymerase. J. Biol. Chem. 2008, 283, 7705–7712. [Google Scholar] [CrossRef] [PubMed]
- Appleby, T.C.; Perry, J.K.; Murakami, E.; Barauskas, O.; Feng, J.; Cho, A.; Fox, D., 3rd; Wetmore, D.R.; McGrath, M.E.; Ray, A.S.; et al. Viral replication. Structural basis for RNA replication by the hepatitis C virus polymerase. Science 2015, 347, 771–775. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Orta, C.; Arias, A.; Perez-Luque, R.; Escarmis, C.; Domingo, E.; Verdaguer, N. Structure of foot-and-mouth disease virus RNA-dependent RNA polymerase and its complex with a template-primer RNA. J. Biol. Chem. 2004, 279, 47212–47221. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Orta, C.; Arias, A.; Perez-Luque, R.; Escarmis, C.; Domingo, E.; Verdaguer, N. Sequential structures provide insights into the fidelity of RNA replication. Proc. Natl. Acad. Sci. USA 2007, 104, 9463–9468. [Google Scholar] [CrossRef] [PubMed]
- Beese, L.S.; Steitz, T.A. Structural basis for the 3′–5′ exonuclease activity of Escherichia coli DNA polymerase I: A two metal ion mechanism. EMBO J. 1991, 10, 25–33. [Google Scholar] [PubMed]
- Arnold, J.J.; Cameron, C.E. Poliovirus RNA-dependent RNA polymerase (3Dpol): Pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+. Biochemistry 2004, 43, 5126–5137. [Google Scholar] [CrossRef] [PubMed]
- Arnold, J.J.; Gohara, D.W.; Cameron, C.E. Poliovirus RNA-dependent RNA polymerase (3Dpol): Pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mn2+. Biochemistry 2004, 43, 5138–5148. [Google Scholar] [CrossRef] [PubMed]
- Arnold, J.J.; Cameron, C.E. Poliovirus RNA-dependent RNA polymerase (3Dpol). Assembly of stable elongation-competent complexes by using a symmetrical primer-template substrate (sym/sub). J. Biol. Chem. 2000, 275, 5329–5336. [Google Scholar] [CrossRef] [PubMed]
- Sholders, A.J.; Peersen, O.B. Distinct conformations of a putative translocation element in poliovirus polymerase. J. Mol. Biol. 2014, 426, 1407–1419. [Google Scholar] [CrossRef] [PubMed]
| NAC State | PDB(s) for Model Construction | Reference PDB(s) | Active Site Conformation |
|---|---|---|---|
| 1 | 5F8G (EV71) | 3OL6, 4K4S, 4K4T, 4K4U, 4K4V, 4K4W, 4K4X, 4K4Z, 4K50, 5F8G, 5F8L [12,14,15] | Open |
| 2 | 5F8H (EV71), 3OLB (PV) a | 3OLA (chains I/M), 3OLB, 4K4Y, 5F8H [12,15] | Open |
| 2/3 | 5F8I (EV71) | 5F8I [15] | Partially closed |
| 3 | 5F8J (EV71), 3BSO (NV) b | 3BSO [16] | Closed |
| 4 | 5F8M (EV71) | 3OL7, 3OL8, 5F8J, 5F8M [12,15] | Closed |
| 5 | 5F8G (EV71), 3OL9 (PV) c | 3OL9, 3OLA (chains A/E) [12] | Open |
| 6 | 5F8N (EV71) | 5F8N [15] | Open |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, J.; Gong, P. Visualizing the Nucleotide Addition Cycle of Viral RNA-Dependent RNA Polymerase. Viruses 2018, 10, 24. https://doi.org/10.3390/v10010024
Wu J, Gong P. Visualizing the Nucleotide Addition Cycle of Viral RNA-Dependent RNA Polymerase. Viruses. 2018; 10(1):24. https://doi.org/10.3390/v10010024
Chicago/Turabian StyleWu, Jiqin, and Peng Gong. 2018. "Visualizing the Nucleotide Addition Cycle of Viral RNA-Dependent RNA Polymerase" Viruses 10, no. 1: 24. https://doi.org/10.3390/v10010024




