Bombyx mori Nuclear Polyhedrosis Virus (BmNPV) Induces Host Cell Autophagy to Benefit Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells, Viruses, and Plasmids
2.2. Virus Infection and Plasmid Transfection
2.3. SDS-PAGE and Western Blot
2.4. Transmission Electron Microscopy
2.5. Confocal Fluorescence Microscopy
2.6. Real-Time PCR
2.7. Knockout Efficiency Analysis
2.8. Statistical Analysis
3. Results
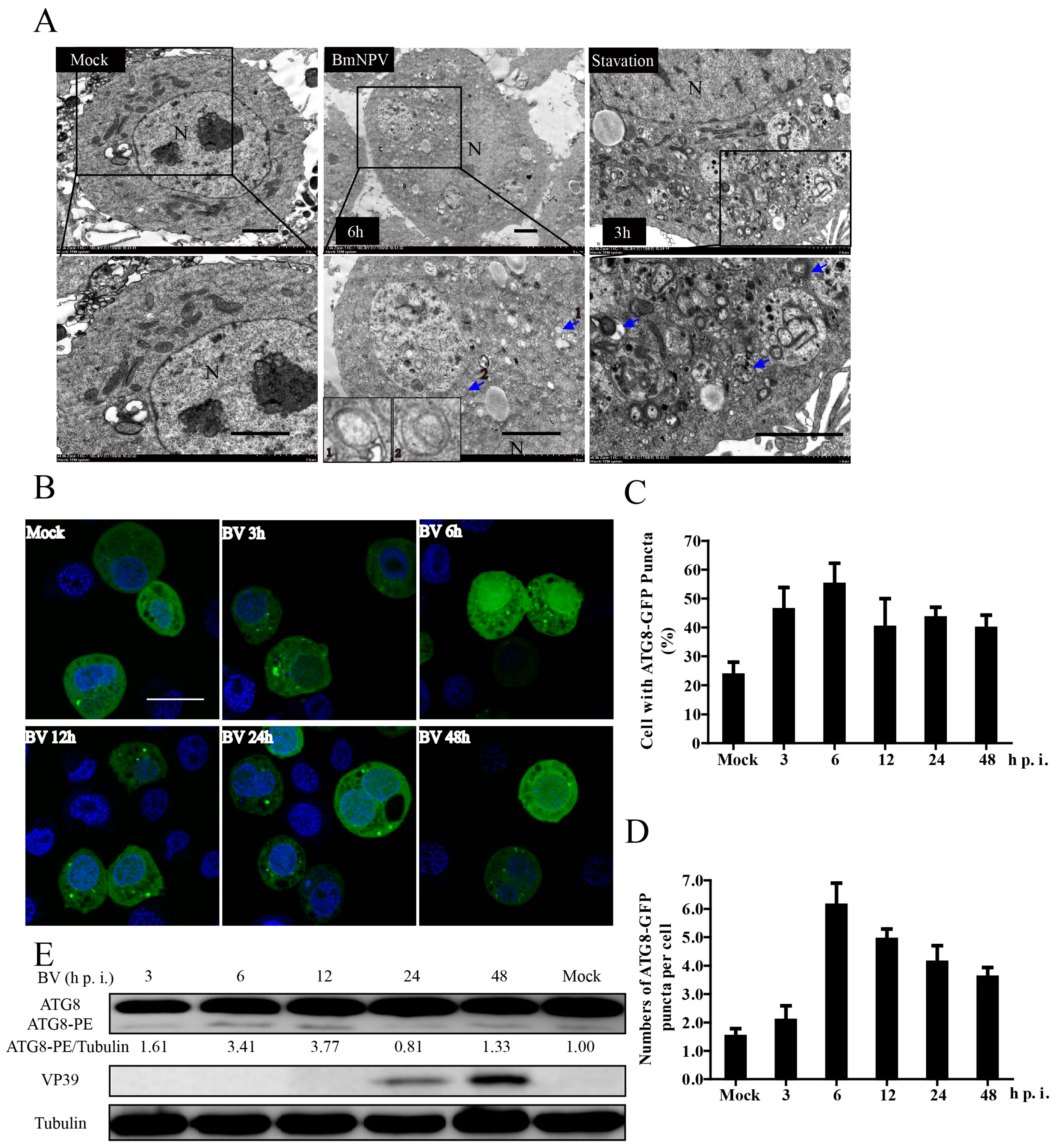
3.1. BmNPV Infection Triggered Autophagy in BmN-SWU1 Cells
3.2. BmNPV Infection Increased the Levels of Autophagic Flux
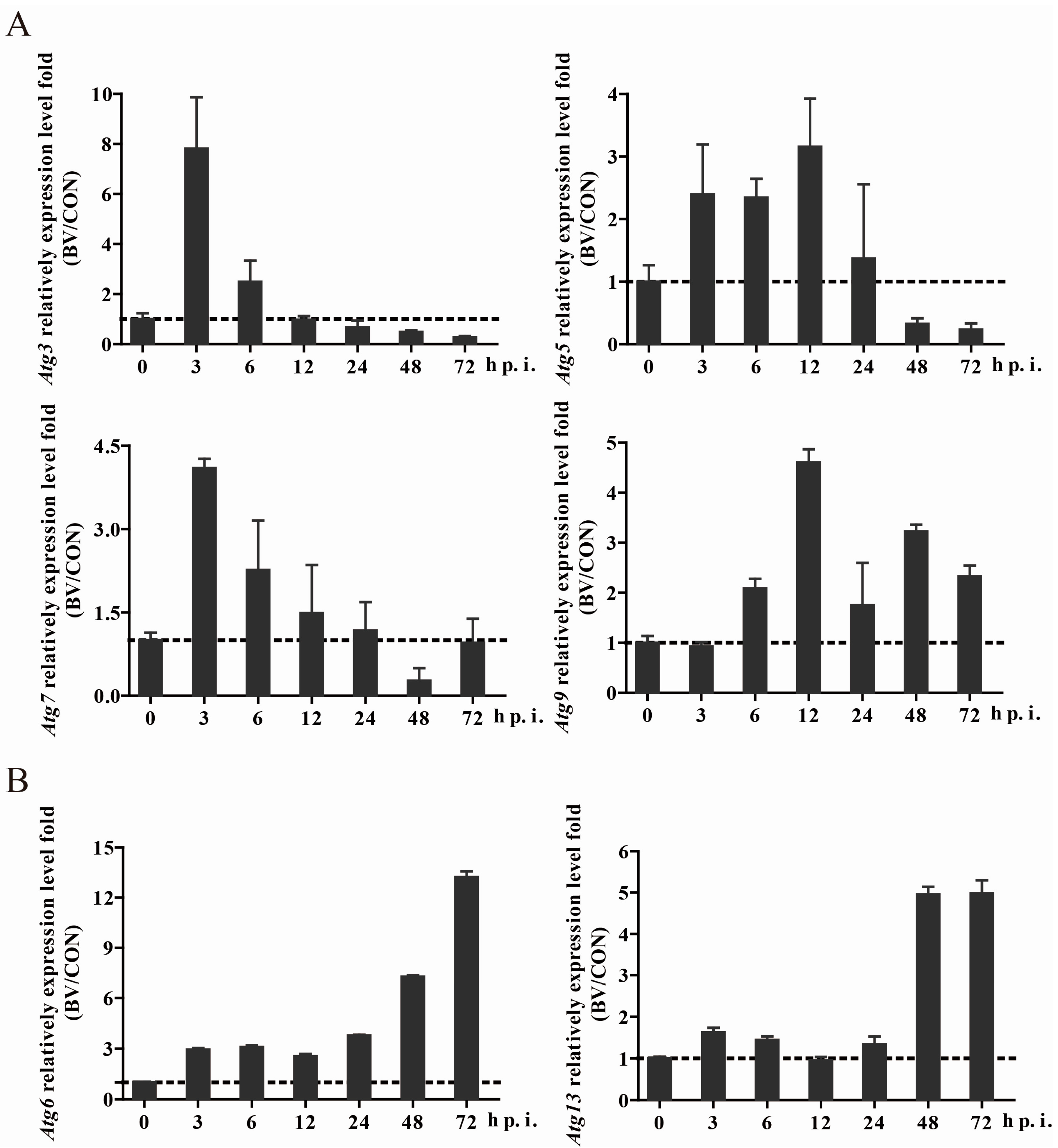
3.3. BmNPV Infection Caused Expression Level Changes of Autophagy-Related Genes
3.4. Atg Genes Influence the Process of BmNPV Infection
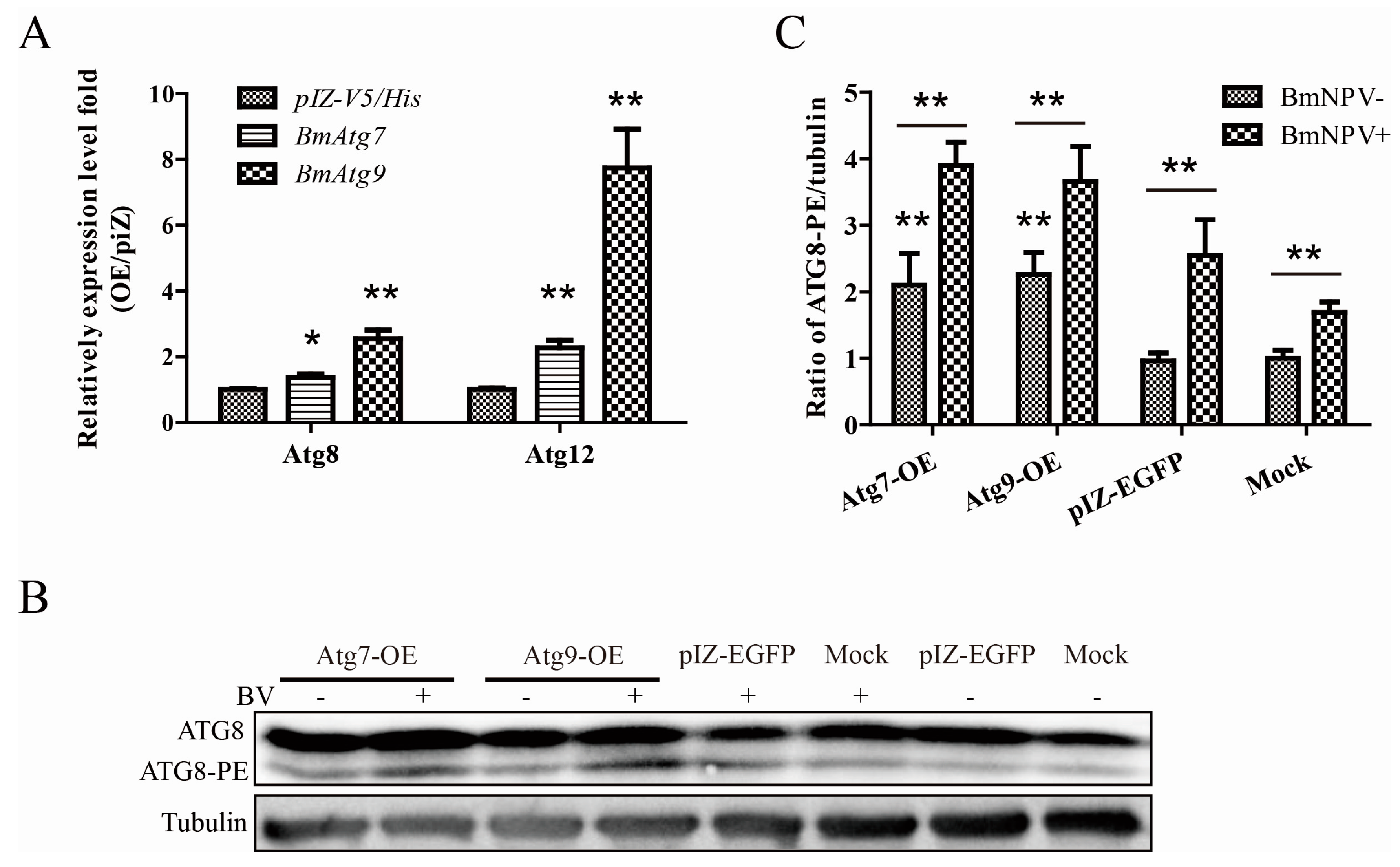
3.5. Atg7 and Atg9 Promote Autophagy Induced by BmNPV In Vitro
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Blissard, G.W.; Rohrmann, G.F. Baculovirus diversity and molecular biology. Annu. Rev. Entomol. 1990, 35, 127–155. [Google Scholar] [CrossRef] [PubMed]
- Kool, M.; Ahrens, C.H.; Vlak, J.M.; Rohrmann, G.F. Replication of baculovirus DNA. J. Gen. Virol. 1995, 76, 2103–2118. [Google Scholar] [CrossRef] [PubMed]
- Keddie, B.A.; Aponte, G.W.; Volkman, L.E. The pathway of infection of autographa californica nuclear polyhedrosis virus in an insect host. Science 1989, 243, 1728–1730. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Sutton, G.; Evans, G.; Axford, D.; Owen, R.; Stuart, D.I. How baculovirus polyhedra fit square pegs into round holes to robustly package viruses. EMBO J. 2010, 29, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Wang, S.; Su, S.; Yao, N.; He, J.; Peng, L.; Sun, J. Construction of a baculovirus-silkworm multigene expression system and its application on producing virus-like particles. PLoS ONE 2012, 7, e32510. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.T.; Dawson, P.W.; Richardson, C.D. Viral interactions with macroautophagy: A double-edged sword. Virology 2010, 402, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of cells and tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Pietrocola, F.; Levine, B.; Kroemer, G. Metabolic control of autophagy. Cell 2014, 159, 1263–1276. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Klionsky, D.J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009, 43, 67–93. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.Y.; Neufeld, T.P. Autophagy takes flight in drosophila. FEBS Lett. 2010, 584, 1342–1349. [Google Scholar] [CrossRef] [PubMed]
- Malagoli, D.; Abdalla, F.C.; Cao, Y.; Feng, Q.; Fujisaki, K.; Gregorc, A.; Matsuo, T.; Nezis, I.P.; Papassideri, I.S.; Sass, M.; et al. Autophagy and its physiological relevance in arthropods: Current knowledge and perspectives. Autophagy 2010, 6, 575–588. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.K.; Lund, J.M.; Ramanathan, B.; Mizushima, N.; Iwasaki, A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science 2007, 315, 1398–1401. [Google Scholar] [CrossRef] [PubMed]
- Shelly, S.; Lukinova, N.; Bambina, S.; Berman, A.; Cherry, S. Autophagy is an essential component of drosophila immunity against vesicular stomatitis virus. Immunity 2009, 30, 588–598. [Google Scholar] [CrossRef] [PubMed]
- Lv, S.; Xu, Q.; Sun, E.; Yang, T.; Li, J.; Feng, Y.; Zhang, Q.; Wang, H.; Zhang, J.; Wu, D. Autophagy activated by bluetongue virus infection plays a positive role in its replication. Viruses 2015, 7, 4657–4675. [Google Scholar] [CrossRef] [PubMed]
- Jordan, T.X.; Randall, G. Manipulation or capitulation: Virus interactions with autophagy. Microbes Infect. 2012, 14, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Feizi, N.; Mehrbod, P.; Romani, B.; Soleimanjahi, H.; Bamdad, T.; Feizi, A.; Jazaeri, E.O.; Targhi, H.S.; Saleh, M.; Jamali, A.; et al. Autophagy induction regulates influenza virus replication in a time-dependent manner. J. Med. Microbiol. 2017, 66, 536–541. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Cui, L.; Zhu, E.; Zhou, W.; Wang, Q.; Wu, X.; Wu, B.; Huang, Y.; Liu, H.J. Muscovy duck reovirus sigmans protein triggers autophagy enhancing virus replication. Virol. J. 2017, 14, 53. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.J.; Tao, L.X.; Dong, X.Y.; Yu, M.H.; Tian, T.; Tang, X.D. Characterization of aggregate/aggresome structures formed by polyhedrin of bombyx mori nucleopolyhedrovirus. Sci. Rep. 2015, 5, 14601. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.H.; Cai, X.J.; Liu, M.; Lv, J.; Tang, H.; Tan, J.; Lu, C. Establishment and characterization of an ovarian cell line of the silkworm, Bombyx mori. Tissue Cell 2010, 42, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.E.; Ward, S.L.; Mizushima, N.; Levine, B.; Leib, D.A. Analysis of the role of autophagy in replication of herpes simplex virus in cell culture. J. Virol. 2007, 81, 12128–12134. [Google Scholar] [CrossRef] [PubMed]
- Romanelli, D.; Casartelli, M.; Cappellozza, S.; de Eguileor, M.; Tettamanti, G. Roles and regulation of autophagy and apoptosis in the remodelling of the lepidopteran midgut epithelium during metamorphosis. Sci. Rep. 2016, 6, 32939. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Ma, L.; Guo, E.; Deng, X.; Ma, S.; Xia, Q.; Cao, Y.; Li, S. 20-hydroxyecdysone upregulates ATG genes to induce autophagy in the bombyx fat body. Autophagy 2013, 9, 1172–1187. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Tian, L.; Guo, X.; Li, K.; Li, J.; Deng, X.; Li, Q.; Xia, Q.; Zhong, Y.; Huang, Z.; et al. BmATG5 and BmATG 6 mediate apoptosis following autophagy induced by 20-hydroxyecdysone or starvation. Autophagy 2016, 12, 381–396. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Abeliovich, H.; Agostinis, P.; Agrawal, D.K.; Aliev, G.; Askew, D.S.; Baba, M.; Baehrecke, E.H.; Bahr, B.A.; Ballabio, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 2008, 4, 151–175. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N. Methods for monitoring autophagy. Int. J. Biochem. Cell Biol. 2004, 36, 2491–2502. [Google Scholar] [CrossRef] [PubMed]
- Rusten, T.E.; Lindmo, K.; Juhasz, G.; Sass, M.; Seglen, P.O.; Brech, A.; Stenmark, H. Programmed autophagy in the drosophila fat body is induced by ecdysone through regulation of the pi3k pathway. Dev. Cell 2004, 7, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Scott, R.C.; Schuldiner, O.; Neufeld, T.P. Role and regulation of starvation-induced autophagy in the drosophila fat body. Dev. Cell 2004, 7, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Yano, T.; Mita, S.; Ohmori, H.; Oshima, Y.; Fujimoto, Y.; Ueda, R.; Takada, H.; Goldman, W.E.; Fukase, K.; Silverman, N.; et al. Autophagic control of listeria through intracellular innate immune recognition in drosophila. Nat. Immunol. 2008, 9, 908–916. [Google Scholar] [CrossRef] [PubMed]
- Llopis, J.; McCaffery, J.M.; Miyawaki, A.; Farquhar, M.G.; Tsien, R.Y. Measurement of cytosolic, mitochondrial, and golgi pH in single living cells with green fluorescent proteins. Proc. Natl. Acad. Sci. USA 1998, 95, 6803–6808. [Google Scholar] [CrossRef] [PubMed]
- Evdokimov, A.G.; Pokross, M.E.; Egorov, N.S.; Zaraisky, A.G.; Yampolsky, I.V.; Merzlyak, E.M.; Shkoporov, A.N.; Sander, I.; Lukyanov, K.A.; Chudakov, D.M. Structural basis for the fast maturation of arthropoda green fluorescent protein. EMBO Rep. 2006, 7, 1006–1012. [Google Scholar] [CrossRef] [PubMed]
- Rusten, T.E.; Stenmark, H. P62, an autophagy hero or culprit? Nat. Cell Biol. 2010, 12, 207–209. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Hu, Z.Y.; Li, W.F.; Li, Q.R.; Deng, X.J.; Yang, W.Y.; Cao, Y.; Zhou, C.Z. Systematic cloning and analysis of autophagy-related genes from the silkworm bombyx mori. BMC Mol. Biol. 2009, 10, 50. [Google Scholar] [CrossRef] [PubMed]
- Itakura, E.; Mizushima, N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian ATG proteins. Autophagy 2010, 6, 764–776. [Google Scholar] [CrossRef] [PubMed]
- Kageyama, S.; Omori, H.; Saitoh, T.; Sone, T.; Guan, J.L.; Akira, S.; Imamoto, F.; Noda, T.; Yoshimori, T. The LC3 recruitment mechanism is separate from ATG9l1-dependent membrane formation in the autophagic response against salmonella. Mol. Biol. Cell 2011, 22, 2290–2300. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Yamamoto, A.; Hatano, M.; Kobayashi, Y.; Kabeya, Y.; Suzuki, K.; Tokuhisa, T.; Ohsumi, Y.; Yoshimori, T. Dissection of autophagosome formation using APG5-deficient mouse embryonic stem cells. J. Cell Biol. 2001, 152, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Nakatogawa, H.; Suzuki, K.; Kamada, Y.; Ohsumi, Y. Dynamics and diversity in autophagy mechanisms: Lessons from yeast. Nat. Rev. Mol. Cell Biol. 2009, 10, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.Z.; Xu, L.M.; Liu, M.; Zhang, Z.Y.; Yin, J.S.; Liu, H.B.; Lu, T.Y. Autophagy induced by infectious hematopoietic necrosis virus inhibits intracellular viral replication and extracellular viral yields in epithelioma papulosum cyprini cell line. Dev. Comp. Immunol. 2017, 77, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.D.; Johnsen, I.B.; Stiberg, K.A.; Sherstova, T.; Wakita, T.; Richard, G.M.; Kandasamy, R.K.; Meurs, E.F.; Anthonsen, M.W. Hepatitis c virus triggers golgi fragmentation and autophagy through the immunity-related gtpase m. Proc. Natl. Acad. Sci. USA 2017, 114, E3462–E3471. [Google Scholar] [CrossRef] [PubMed]
- Ke, P.Y.; Chen, S.S. Autophagy: A novel guardian of HCV against innate immune response. Autophagy 2011, 7, 533–535. [Google Scholar] [CrossRef] [PubMed]
- Granato, M.; Santarelli, R.; Farina, A.; Gonnella, R.; Lotti, L.V.; Faggioni, A.; Cirone, M. Epstein-barr virus blocks the autophagic flux and appropriates the autophagic machinery to enhance viral replication. J. Virol. 2014, 88, 12715–12726. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.T.; Xiao, L.F.; Hu, J.J.; Zhao, X.X.; Liu, J.X.; Zhang, Y. Itraq proteomic analysis of the interactions between bombyx mori nuclear polyhedrosis virus and silkworm. J. Proteom. 2017, 166, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.Y.; Xia, Y.Q.; Zhou, J.; Chen, Z.W.; Lu, D.; Zhang, N.Z.; Liu, X.S.; Ai, H.; Zhou, L.L. Molecular characterization of autophagy-related gene 5 from spodoptera exigua and expression analysis under various stress conditions. Arch. Insect Biochem. Physiol. 2016, 92, 225–241. [Google Scholar] [CrossRef] [PubMed]
- Jackson, W.T. Viruses and the autophagy pathway. Virology 2015, 479–480, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Crawford, S.E.; Hyser, J.M.; Utama, B.; Estes, M.K. Autophagy hijacked through viroporin-activated calcium/calmodulin-dependent kinase kinase-beta signaling is required for rotavirus replication. Proc. Natl. Acad. Sci. USA 2012, 109, E3405–E3413. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Gai, Z.; Ai, H.; Wu, W.; Yang, Y.; Peng, J.; Hong, H.; Li, Y.; Liu, K. Baculovirus infection triggers a shift from amino acid starvation-induced autophagy to apoptosis. PLoS ONE 2012, 7, e37457. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Parnell, L.A.; Diamond, M.S.; Mysorekar, I.U. Inhibition of autophagy limits vertical transmission of zika virus in pregnant mice. J. Exp. Med. 2017, 214, 2303–2313. [Google Scholar] [CrossRef] [PubMed]


| Plasmid Name | Primer Sequence |
|---|---|
| pIZ/V5-Flag-BmAtg8-Enhanced Green Fluorescent Protein (EGFP); pIZ/V5-Flag-BmAtg8-EGFP-RFP (RFP: Red Fluorescent Protein) | 5′-CGCGGATCCGATTACAAGGATGACGACGATAAGAAATTCCAATACAAAG-3′; 5′-CCGCTCGAGTTAATTTCCATAGACAT-3′ |
| pIZ/V5–Flag-BmAtg3 | 5′-CGGGGTACCATGGATTACAAGGATGACGACGATAAGCAAAGTGTAATAAACACAGTAAAG-3′; 5′-CTAGTCTAGACGGTTAGTTAATCGAGAAATTCTG-3′ |
| pIZ/V5–Flag-BmAtg4 | 5′-CGGGGTACCATGGATTACAAGGATGACGACGATAAGAATGGTTCAAGCGGTT-3′; 5′-CTAGTCTAGAGCGGCTAAACTAAAACAAATTCTTC-3′ |
| pIZ/V5–HA-BmAtg5 | 5′-CGCGGATCCATGTACCCATACGATGTTCCAGATTACGAATGGTTCAAGCGGTT-3′; 5′-CCGCTCGAGCGGTCAGCATAAACACAAATG-3′ |
| pIZ/V5–Flag-BmAtg7 | 5′-CCGGAATTCATGGATTACAAGGATGACGACGATAAGTTGGAAGCACAAAAC-3′; 5′-CTAGTCTAGACGCTAATCTTCTTCATCGGTTAATG-3′ |
| pIZ/V5–Flag-BmAtg9 | 5′-CCGGAATTCATGGATTACAAGGATGACGACGATAAGGCTTACTTCGGAAG-3′; 5′-CTAGTCTAGACTTAGGGACGGTGTAGGAGAG-3′ |
| pIZ/V5–Flag-BmAtg12 | 5′-GATCCATGGATTACAAGGATGACGACGATAAGAGTGATGAGAAACTTATAGATG-3′; 5′-CCGCTCGAGTCAGCCCCAAGCTTGGCTT-3′ |
| pSL1180-Cas9-Flag-sgAtg3; Atg3-T | 5′-AAGTGCAACACCGGTGTCAAGTAAC-3′; 5′-AAACGTTACTTGACACCGGTGTTGC-3′; 5′-ATGCAAAGTGTAATAAACACAGTAAAG-3′; 5′-TACATCGTCTGTAGCATGGC-3′ |
| pSL1180-Cas9-Flag-sgAtg4; Atg4-T | 5′-AAGTGGAACTAAACGATTTAAATAA-3′; 5′-AAACTTATTTAAATCGTTTAGTTCC-3′; 5′-ATGAATGGTTCAAGCGGTT-3′; 5′-CAACCAAACCGGAGATTCTTTAG-3′ |
| pSL1180-Cas9-Flag-sgAtg5; Atg5-T | 5′-AAGTGGTACTTCGAGAAATATGGGA-3′; 5′-AAACTCCCATATTTCTCGAAGTACC-3′; 5′-ATGGCCAACGACAGGGAG-3′; 5′-CGATGACAAGAGGAAAGTAGC-3′ |
| pSL1180-Cas9-Flag-sgAtg7; Atg7-T | 5′-AAGTGGTGAGGGTATGCCAGAATGA-3′; 5′-AAACTCATTCTGGCATACCCTCACC-3′; 5′-ATGTTGGAAGCACAAAAC-3′; 5′-GTTTTTGTTCATTATTGTACCCATC-3′ |
| pSL1180-Cas9-Flag-sgAtg9; Atg9-T | 5′-AAGTGTCAGCCAATCCACCTAAAGG-3′; 5′-AAACCCTTTAGGTGGATTGGCTGAC-3′; 5′-ATGGCTTACTTCGGAAG-3′; 5′-ACAGTGCACGAGAAATGTTGAG-3′ |
| pSL1180-Cas9-Flag-sgAtg12; Atg12-T | 5′-AAGTGGTTGATGCTGAAAAGCCTAT-3′; 5′-AAACATAGGCTTTTCAGCATCAACC-3′; 5′-ATGAGTGATGAGAAACTTATAGATG-3′; 5′-TCAGCCCCAAGCTTGGCTT-3′ |
| Primers Name | Primer Sequence(RT-PCR) |
|---|---|
| BmAtg1 | 5′-CATCGTCCACCGTGACTTGA-3′; 5′-GTCTGCTTTGGCGTCGTATTT-3′ |
| BmAtg2 | 5′-GACGACTCACCGATTTACTTCAGA-3′; 5′-CTCAGTGCCCAACAATCCAAG-3′ |
| BmAtg3 | 5′-AACTCAAAGCCGATAAGAAACA-3′; 5′-TTTTAGCGTGATCTTGGGAC-3′ |
| BmAtg4 | 5′-TACCTCAGGGTGTATCATCA-3′; 5′-TAAGTCTGTATCGCTGTCTTG-3′ |
| BmAtg5 | 5′-AAGTTCCCTGAAGACATTCT-3′; 5′-ATTTTGTAATCCAAGCCATA-3′ |
| BmAtg6 | 5′-GGGCTTTTGTCTTCCGTA-3′; 5′-TGTGGCTCAGATTTGTCCT-3′ |
| BmAtg7 | 5′-GAGGCGAGATGGCTGC-3′; 5′-CGAGGTGCTAATTCCGTG-3′ |
| BmAtg8 | 5′-AAGGCTAGGCTTGGAGAC-3′; 5′-CAGATGTGGGTGGAATGA-3′ |
| BmAtg9 | 5′-TTTAGGTGGATTGGCTGA-3′; 5′-TTGGCACTTCGTGGATG-3′ |
| BmAtg11 | 5′-TAAGTCCCATAGTAGAGC-3′; 5′-ACAAACTTCCACTTCAT-3′ |
| BmAtg12 | 5′-AGACGCAGAGCCAATCA-3′; 5′-AACTCCATAATCCATCCAATA-3′ |
| BmAtg13 | 5′-AGAGTTTACAGTGGCGAG-3′; 5′-TGTCTGAACGGTTAGGAG-3′ |
| BmAtg16 | 5′-CACATTCGGTAGAGTTAGTTTCGG-3′; 5′-CGGCATACACTTCTCCATCGT-3′ |
| BmAtg18 | 5′-TTGCTGTTGGCGGTCT-3′; 5′-CAAGCGATATGGCGGA-3′ |
| BmNPVie-1 | 5′-AAGAAGGAGGACGGCAGCAT-3′; 5′-ATCTCGCCAGAAATCCAATAAAAC-3′ |
| BmNPVvp39 | 5′-CTAATGCCCGTGGGTATGG-3′; 5′-TTGATGAGGTGGCTGTTGC-3′ |
| BmNPVp10 | 5′-TAGACGCCATTGCGGAAA-3′; 5′-CGGGCAAACCGTCCAAA-3′ |
| BmRPL3 | 5′-CGGTGTTGTTGGATACATTGAG-3′; 5′-GCTCATCCTGCCATTTCTTACT-3′ |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Xiao, Q.; Zhou, X.-L.; Zhu, Y.; Dong, Z.-Q.; Chen, P.; Pan, M.-H.; Lu, C. Bombyx mori Nuclear Polyhedrosis Virus (BmNPV) Induces Host Cell Autophagy to Benefit Infection. Viruses 2018, 10, 14. https://doi.org/10.3390/v10010014
Wang L, Xiao Q, Zhou X-L, Zhu Y, Dong Z-Q, Chen P, Pan M-H, Lu C. Bombyx mori Nuclear Polyhedrosis Virus (BmNPV) Induces Host Cell Autophagy to Benefit Infection. Viruses. 2018; 10(1):14. https://doi.org/10.3390/v10010014
Chicago/Turabian StyleWang, La, Qin Xiao, Xiao-Lin Zhou, Yan Zhu, Zhan-Qi Dong, Peng Chen, Min-Hui Pan, and Cheng Lu. 2018. "Bombyx mori Nuclear Polyhedrosis Virus (BmNPV) Induces Host Cell Autophagy to Benefit Infection" Viruses 10, no. 1: 14. https://doi.org/10.3390/v10010014





