Viroid Pathogenicity: One Process, Many Faces
Abstract
:1. Introduction
2. Identification of structural motifs modulating viroid pathogenicity
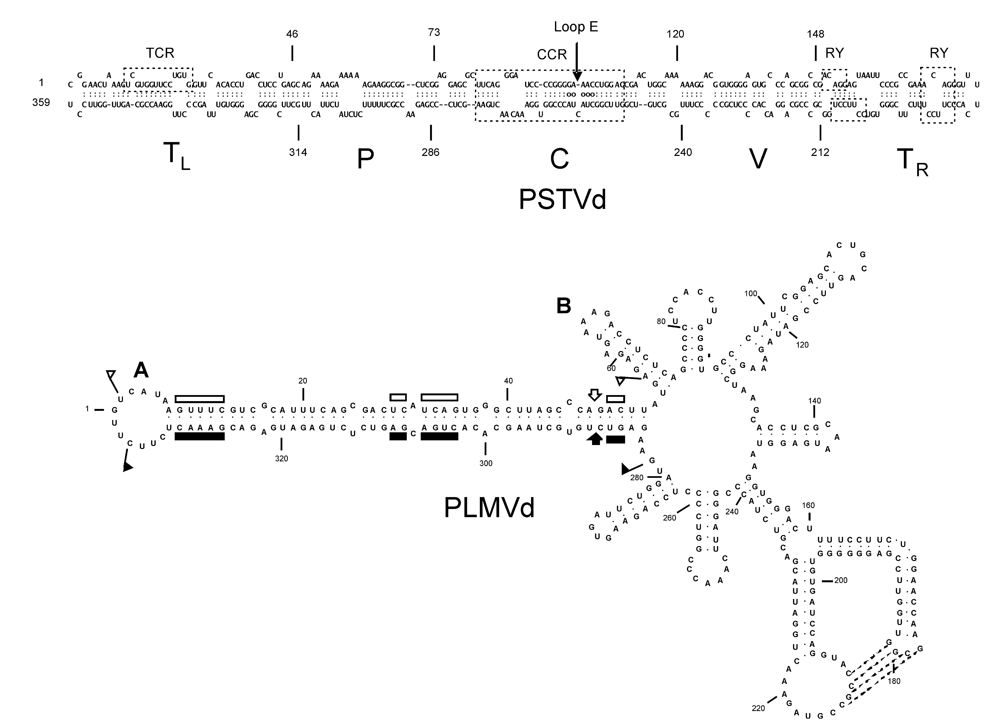

3. Viroid-protein interaction as a potential trigger for symptom induction
4. Host responses to viroid infection
5. Role(s) of RNA silencing in viroid pathogenesis
| miRNA synthesis | siRNA synthesis | |||
|---|---|---|---|---|
| Dicer | DCL1 | DCL2 | DCL3 | DCL4 |
| Precursor | Pri-miRNA | |||
| Primary product (size) | miRNA (21-nt) | siRNA (22-nt) | siRNA (24-nt) | siRNA (21-nt) |
| Downstream events | Transcript cleavage | DNA and histone methylation | ||
| Additional factors involved | RDR6; DCL4 > DCL2 | RDR2; RNA pol IV | ||
| Secondary product | Ta-siRNAs | |||

6. Potential targets of viroid-mediated RNA silencing
7. Beyond genes and pathways…
References
- Diener, T.O. Potato spindle tuber “virus” IV. A replicating, low molecular weight RNA. Virology 1971, 45, 411–428. [Google Scholar] [CrossRef] [PubMed]
- Gross, H.J.; Domdey, H.; Lossow, C.; Jank, P.; Raba, M.; Alberty, H.; Sänger, H-L. Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature 1978, 273, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Dickson, E.; Robertson, H.D.; Niblett, C.L.; Horst, R.K.; Zaitlin, M. Minor differences between nucleotide sequences of mild and severe strains of potato spindle tuber viroid. Nature 1979, 277, 60–62. [Google Scholar] [CrossRef]
- Keese, P.; Symons, R.H. Domains in viroids: Evidence of intermolecular RNA rearrangements and their contribution to viroid evolution. Proc. Natl. Acad. Sci. USA 1985, 82, 4582–4586. [Google Scholar] [CrossRef]
- Schnölzer, M.; Haas, B.; Ramm, K.; Hofmann, H.; Sänger, H.-L. Correlation between structure and pathogenicity of potato spindle tuber viroid (PSTV). EMBO J. 1985, 4, 2181–2190. [Google Scholar] [PubMed]
- Wang, M.B.; Bian, X.Y.; Wu, L.M.; Liu, L.X.; Smith, N.A.; Isenegger, D.; Wu, R.M.; Masuta, C.; Vance, V.B.; Watson, J.M.; Rezaian, A.; Dennis, E.S.; Waterhouse, P.M. On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc. Natl. Acad. Sci. USA 2004, 101, 3275–3280. [Google Scholar] [CrossRef]
- Itaya, A.; Folimonov, A.; Matsuda, Y.; Nelson, R.S.; Ding, B. Potato spindle tuber viroid as inducer of RNA silencing in infected tomato. Mol. Plant–Microbe Interact. 2001, 14, 1332–1334. [Google Scholar] [CrossRef] [PubMed]
- Markarian, N.; Li, H.W.; Ding, S.-W.; Semancik, J.S. RNA silencing as related to viroid induced symptom expression. Arch. Virol. 2004, 149, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Martínez de Alba, A.E.; Flores, R.; Hernández, C. Two chloroplastic viroids induce the accumulation of the small RNAs associated with post-transcriptional gene silencing. J. Virol. 2002, 76, 13094–13096. [Google Scholar] [CrossRef] [PubMed]
- Matoušek, J.; Kozlová, P.; Orctová, L.; Schmitz, A.; Pesina, K.; Bannach, O.; Diermann, N.; Steger, G.; Riesner, D. Accumulation of viroid-specific small RNAs and increase in nucleolytic activities linked to viroid-caused pathogenesis. Biol. Chem. 2007, 388, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Papaefthimiou, I.; Hamilton, A.J.; Denti, M.A.; Baulcombe, D.C.; Tsagris, M.; Tabler, M. Replicating potato spindle tuber viroid RNA is accompanied by short RNA fragments that are characteristic of post-transcriptional gene silencing. Nucleic Acids Res. 2001, 29, 2395–2400. [Google Scholar] [CrossRef] [PubMed]
- Daros, J.A.; Elena, S.F.; Flores, R. Viroids: An Ariadne’s thread into the RNA labyrinth. EMBO Rep. 2006, 7, 593–588. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.; Itaya, A. Viroid: A useful model for studying the basic principles of infection and RNA biology. Mol. Plant-Microbe Inter. 2007, 20, 7–20. [Google Scholar] [CrossRef]
- Flores, R.; Hernández, C.; Martinez de Alba, A.E.; Daròs, J.-A.; Di Serio, F. Viroids and viroid-host interactions. Annu. Rev. Phytopathol. 2005, 43, 117–139. [Google Scholar] [CrossRef] [PubMed]
- Tabler, M.; Tsagris, M. Viroids: petite RNA pathogens with distinguished talents. Trends Plant Sci. 2004, 9, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Maniataki, E.; Tabler, M.; Tsagris, M. Viroid RNA systemic spread may depend on the interaction of a 71-nucleotide bulged hairpin with the host protein VIRP1. RNA 2003, 9, 346–354. [Google Scholar] [CrossRef] [PubMed]
- Visvader, J.E.; Symons, R.H. Eleven new sequence variants of citrus exocortis viroid and the correlation of sequence with pathogencity. Nucleic Acids Res. 1985, 13, 2907–2920. [Google Scholar] [CrossRef] [PubMed]
- Sano, T.; Candresse, T.; Hammond, R.W.; Diener, T.O.; Owens, R.A. Identification of multiple structural domains regulating viroid pathogenicity. Proc. Natl. Acad. Sci. USA 1992, 89, 10104–10108. [Google Scholar] [CrossRef]
- Reanwarakom, K.; Semancik, J.S. Regulation of pathogenicity in hop stunt viroid related group II citrus viroids. J. Gen. Virol. 1998, 79, 3581–3584. [Google Scholar]
- Reanwarakom, K.; Semancik, J.S. Correlation of hop stunt viroid variants to cachexia and xyloporosis diseases of citrus. Phytopathology 1999, 89, 568–574. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, M.J.B.; Randles, J.W. Coconut cadang-cadang viroid (CCCVd) mutants associated with severe disease vary in both the pathogenicity domain and central conserved region. Nuc. Acids Res. 1993, 21, 2771. [Google Scholar] [CrossRef]
- Semancik, J.S.; Szychowski, J.A. Avocado sunblotch disease: a persistent viroid infection in which variants are associated with differential symptoms. J. Gen. Virol. 1994, 75, 1543–1549. [Google Scholar] [CrossRef] [PubMed]
- Schnell, R.J.; Kuhn, D.N.; Olano, C.T.; Quintanilla, W.E. Sequence diversity among avocado sunblotch viroid isolated from single avocado trees. Phytoparasitica 2002, 29, 451–460. [Google Scholar] [CrossRef]
- De la Peña, M.; Navarro, B.; Flores, R. Mapping the molecular determinant of pathogenicity in a hammerhead viroid: a tetraloop within the in vivo branched RNA conformation. Proc. Natl. Acad. Sci. USA 1999, 96, 9960–9965. [Google Scholar] [CrossRef]
- De la Peña, M.; Flores, R. Chrysanthemum chlorotic mottle viroid RNA: dissection of the pathogenicity determinant and comparative fitness of symptomatic and non-symptomatic variants. J. Mol. Biol. 2002, 321, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Malfitano, M.; Di Serio, F.; Covelli, L.; Ragozzino, A.; Hernández, C.; Flores, R. Peach latent mosaic viroid variants inducing peach calico (extreme chlorosis) contain a characteristic insertion that is responsible for this symptomatology. Virology 2003, 313, 492–501. [Google Scholar] [CrossRef] [PubMed]
- Correll, C.C.; Swinger, K. Common and distinctive features of GNRA tetraloops based on a GUAA tetraloop structure at 1.4 Å resolution. RNA 2003, 9, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Leontis, N.B.; Westhof, E. A common motif organizes the structure of multi-helix loops in 16 S and 23 S ribosomal RNAs. J. Mol. Biol. 1998, 283, 571–583. [Google Scholar] [CrossRef] [PubMed]
- Branch, A.D.; Benenfeld, B.J.; Robertson, H.D. Ultraviolet light-induced crosslinking reveals a unique region of local tertiary structure in potato spindle tuber viroid and HeLa 5S RNA. Proc. Natl. Acad. Sci. USA 1985, 82, 6590–6594. [Google Scholar] [CrossRef]
- Wassenegger, M.; Spieker, R.L.; Thalmeir, S.; Gast, F.U. A single nucleotide substitution converts potato spindle tuber viroid (PSTVd) from a noninfectious to an infectious RNA for Nicotiana tabacum. Virology 1996, 226, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Ding, B. Inhibition of cell growth and shoot development by a specific nucleotide sequence in a noncoding viroid RNA. Plant Cell 2003, 15, 1360–1374. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Leontis, N.; Qian, S.; Itaya, A.; Qi, Y.; Boris-Lawrie, K.; Ding, B. Tertiary structural and functional analyses of a viroid RNA motif by isostericity matrix and mutagenesis reveal its essential role in replication. J. Virol. 2006, 80, 8566–8581. [Google Scholar] [CrossRef] [PubMed]
- Moazed, D.; Robertson, J.M.; Noller, H.F. Interaction of elongation factors EF-G and EFTu with a conserved loop in 23S RNA. Nature 1988, 372, 68–74. [Google Scholar]
- Sharma, N.; Park, S.W.; Vepachedu, R.; Barbieri, L.; Ciani, M.; Stirpe, F.; Savary, B.J.; Vivanco, J.M. Isolation and characterization of an RIP (ribosome-inactivating protein)-like protein from tobacco with dual enzymatic activity. Plant Physiol. 2004, 134, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Wolff, P.; Gilz, R.; Schumacher, J.; Riesner, D. Complexes of viroids with histones and other proteins. Nucleic Acids Res. 1985, 13, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Klaff, P.; Gruner, R.; Hecker, R.; Sättler, A.; Theissen, G.; Riesner, D. Reconstituted and cellular viroid-protein complexes. J. Gen. Virol. 1989, 70, 2257–2270. [Google Scholar] [CrossRef]
- Goodman, T.C.; Nagel, L.; Rappold, W.; Klotz, G.; Riesner, D. Viroid replication: equilibrium association constant and comparative activity measurements for the viroid-polymerase interaction. Nucleic Acids Res. 1984, 12, 6231–6246. [Google Scholar] [CrossRef] [PubMed]
- Kolonko, N.; Bannach, O.; Aschermann, K.; Hu, K.H.; Moors, M.; Schmitz, M.; Steger, G.; Riesner, D. Transcription of potato spindle tuber viroid by RNA polymerase II starts in the left terminal loop. Virology 2006, 347, 392–404. [Google Scholar] [CrossRef] [PubMed]
- Martínez de Alba, A.E.; Sägesser, R.; Tabler, M.; Tsagris, M. A bromodomain-containing protein from tomato specifically binds potato spindle tuber viroid RNA in vitro and in vivo. J. Virol. 2003, 77, 9685–9694. [Google Scholar] [CrossRef] [PubMed]
- Gomez, G.; Pallas, V. Identification of an in vitro ribonucleoprotein complex between a viroid RNA and a phloem protein from cucumber plants. Mol. Plant–Microbe Interact. 2001, 14, 910–913. [Google Scholar] [CrossRef] [PubMed]
- Owens, R.A.; Blackburn, M.; Ding, B. Possible involvement of the phloem lectin in long-distance viroid movement. Mol. Plant–Microbe Interact. 2001, 14, 905–909. [Google Scholar] [CrossRef] [PubMed]
- Gomez, G.; Pallas, V. A long-distance translocatable phloem protein from cucumber forms a ribonucleoprotein complex in vivo with hop stunt viroid RNA. J. Virol. 2004, 78, 10104–10110. [Google Scholar] [CrossRef] [PubMed]
- Daròs, J.-A.; Flores, R. A chloroplast protein binds a viroid RNA in vivo and facilitates its hammerhead-mediated self-cleavage. EMBO J. 2002, 21, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Hiddinga, H.J.; Crum, C.J.; Hu, J.; Roth, D.A. Viroid-induced phosphorylation of a host protein related to a dsRNA-dependent protein kinase. Science 1988, 241, 451–453. [Google Scholar] [PubMed]
- Langland, J.O.; Jin, S.; Jacobs, B.L.; Roth, D.A. Identification of a plant-encoded analog of PKR, the mammalian double-stranded RNA-dependent protein kinase. Plant Physiol. 1995, 108, 1259–1267. [Google Scholar] [PubMed]
- Diener, T.O.; Hammond, R.W.; Black, T.; Katze, M.G. Mechanism of viroid pathogenesis: differential activation of the interferon-induced, double-stranded RNA-activated, M(r) 68,000 protein kinase by viroid strains of varying pathogenicity. Biochimie 1993, 75, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Hammond, R.W.; Zhao, Y. Characterization of a tomato protein kinase gene induced by infection by potato spindle tuber viroid. Mol. Plant-Microbe Interact. 2000, 13, 903–910. [Google Scholar] [CrossRef] [PubMed]
- Hammond, R.W.; Zhao, Y. Modification of tobacco plant development by sense and antisense expression of the tomato viroid-induced AGC VIIIa protein kinase PKV suggests involvement in gibberellin signaling. BMC Plant Biology 2009, 9, 108. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.K.; Dagenais, N.; Chory, J.; Weigel, D. Regulation of auxin response by the protein kinase PINOID. Cell 2000, 100, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Devarenne, T.P.; Ekengren, S.K.; Pedley, K.F.; Martin, G.B. Adi3 is a Pdk1-interacting AGC kinase that negatively regulates plant cell death. EMBO J. 2006, 25, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Diener, T.O. Biological properties. In The Viroids; Diener, T.O., Ed.; 1987; Plenum Press: New York, NY, USA. [Google Scholar]
- Domingo, C.; Conejero, V.; Vera, P. Genes encoding acidic and basic class III β-1,3-glucanases are expressed in tomato plants upon viroid infection. Plant Mol. Biol. 1994, 24, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Tornero, P.; Conejero, V.; Vera, P. A gene encoding a novel isoform of the PR-1 protein family from tomato is induced upon viroid infection. Mol. Gen. Genet. 1994, 243, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Vera, P.; Hernandez-Yago, J.; Conejero, V. “Pathogenesis-related” P1 (p14) protein. Vacuolar and apoplastic localization in leaf tissue from tomato plants infected with citrus exocortis viroid: In vitro synthesis and processing. J. Gen. Virol. 1989, 70, 1933–1942. [Google Scholar] [CrossRef]
- Vidal, A.M.; Ben-Cheikh, W.; Talón, M.; García-Martínez, J.L. Regulation of gibberellin 20-oxidase gene expression and gibberellin content in citrus by temperature and citrus exocortis viroid. Planta. 2003, 216, 442–448. [Google Scholar] [CrossRef]
- Fleet, C.M.; Sun, T-P. A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr. Opin. Plant Biol. 2005, 8, 77–85. [Google Scholar] [CrossRef]
- Sun, T.P.; Gubler, F. Molecular mechanisms of gibberrellin signaling in plants. Ann. Rev. Plant Biol. 2004, 55, 197–223. [Google Scholar] [CrossRef]
- Droge-Laser, W.; Kaiser, A.; Lindsay, W.P.; Halkier, B.A.; Loake, G.J.; Doerner, P.; Dixon, R.A.; Lamb, C. Rapid stimulation of a soybean protein-serine kinase that phosphorylates a novel bZIP DNA-binding protein, G/HBF-1, during the induction of early transcription-dependent defenses. EMBO J. 1997, 16, 726–738. [Google Scholar] [CrossRef] [PubMed]
- Pastori, G.M.; Foyer, C.H. Common components, networks, and pathways of cross-tolerance to stress. The central role of âredoxâ and abscisic acid-mediated controls . Plant Phys. 2002, 129, 460–468. [Google Scholar] [CrossRef]
- Itaya, A.; Matsuda, Y.; Gonzales, R.A.; Nelson, R.S.; Ding, B. Potato spindle tuber viroid strains of different pathogenicity induces and suppresses expression of common and unique genes in infected tomato. Mol. Plant–Microbe Interact. 2002, 15, 990–999. [Google Scholar] [CrossRef] [PubMed]
- Tessitori, M.; Maria, G.; Capasso, C.; Catara, G.; Rizza, S.; De Luca, V.; Catara, A.; Capasso, A.; Carginale, V. Differential display analysis of gene expression in Etrog citron leaves infected by Citrus viroid III. Biochim. Biophys. Acta 2007, 1769, 228–235. [Google Scholar] [PubMed]
- Ascencio-Ibáñez, J.T.; Sozzani, R.; Lee, T.-J.; Chu, T.-M.; Wolfinger, R.D.; Cella, R.; Hanley-Bowdoin. L. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 2008, 148, 436–454. [Google Scholar] [CrossRef] [PubMed]
- Brodersen, P.; Voinnet, O. The diversity of RNA silencing pathways in plants. Trends Genet. 2006, 22, 268–280. [Google Scholar] [CrossRef] [PubMed]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.W.; Voinnet, O. Antiviral immunity directed by small RNAs. Cell 2007, 130, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Carthew, R.W.; Sontheimer, E.J. Origin and mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ferrer, V.; Voinnet, O. Roles of plant small RNAs in biotic stress responses. Annu. Rev. Plant Biol. 2009, 60, 485–510. [Google Scholar] [CrossRef] [PubMed]
- Wassenegger, M.; Heimes, S.; Riedel, L.; Sänger, H.L. RNA-directed de novo methylation of genomic sequences in plants. Cell 1994, 76, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Gómez ,G.; Martínez, G.; Pallás, V. Interplay between viroid-induced pathogenesis and RNA silencing pathways. Trends Plant Sci. 2009, 14, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Machida, S.; Yamahata, N.; Watanuki, H.; Owens, R.A.; Sano, T. Successive accumulation of two size classes of viroid-specific small RNA in potato spindle tuber viroid-infected tomato plants. J. Gen. Virol. 2007, 88, 3452–3457. [Google Scholar] [CrossRef] [PubMed]
- Itaya, A.; Zhong, X.-H.; Bundschuh, R.; Qi, Y.-J; Wang, Y.; Takeda, R.; Harris, A.R.; Molina, C.; Nelson, R.S.; Ding, B. A structured viroid RNA serves as a substrate for Dicer-like cleavage to produce biologically active small RNAs but is resistant to RNA-induced silencing complex-mediated degradation. J. Virol. 2007, 81, 2980–2994. [Google Scholar] [CrossRef] [PubMed]
- Landry, P.; Perreault, J.P. Identification of a peach latent mosaic viroid hairpin able to act as a Dicer-like substrate. J. Virol. 2005, 79, 6540–6543. [Google Scholar] [CrossRef] [PubMed]
- Martín, R.; Arenas, C.; Daròs, J.A.; Covarrubias, A.; Reyes, J.L.; Chua, N.H. Characterization of small RNAs derived from Citrus exocortis viroid in infected tomato plants. Virology 2007, 367, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Schwind, N.; Zweibel, M.; Itaya, A.; Ding, B.; Wang, M-B.; Krczal, G.; Wassenegger, M. RNAi-mediated resistance to Potato spindle tuber viroid in transgenic tomato expressing a viroid hairpin RNA construct . Mol. Plant Path. 2009, 10, 459–469. [Google Scholar] [CrossRef]
- Qi, Y.; Ding, B. Differential subnuclear localization of RNA strands of opposite polarity derived from an autonomously replicating viroid. Plant Cell 2003, 15, 2566–2577. [Google Scholar] [CrossRef] [PubMed]
- Vogt, U.; Pelissier, T.; Putz, A.; Razvi, F.; Fischer, R.; Wassenegger, M. Viroid-induced RNA silencing of GFP-viroid fusion transgenes does not induce extensive spreading of methylation or transitive silencing. Plant J. 2004, 38, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, A.; Martínez de Alba, A.-E.; Flores, R.; Gago, S. Double-stranded RNA interferes in a sequence-specific manner with the infection of representative members of the two viroid families. Virology 2008, 371, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Gómez, G.; Martinez, G.; Pallás, V. Viroid-induced symptoms in Nicotiana benthamiana plants are dependent of RDR6 activity. Plant Physiol. 2008, 148, 414–423. [Google Scholar] [CrossRef] [PubMed]
- Qu, F.; Ye, X.-H.; Hou, G.-C.; Sato, S.; Clemente, T.E.; Morris, T.J. RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in N. benthamiana . J. Virology 2005, 79, 15209–15217. [Google Scholar] [CrossRef]
- Gas, M.E.; Hernańdez, C.; Flores, R.; Daròs, J.A. Processing of nuclear viroids in vivo: An interplay between RNA conformations. PLoS Pathog. 2007, 3, 1813–1826. [Google Scholar] [CrossRef]
- Whitham, S.A.; Yang, C.; Goodin, M.M. Global impact: elucidating plant responses to viral infection. Mol. Plant Microbe Interact. 2006, 19, 207–215. [Google Scholar] [CrossRef]
- Wise, R.P.; Moscou, M.J.; Bogdanove, A.J.; Whitham, S.A. Transcript profiling in host-pathogen interactions. Annu. Rev. Phytopathol. 2007, 45, 329–369. [Google Scholar] [CrossRef] [PubMed]
- Boller, T.; He, S.Y. Innate immunity in plants: An arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 2009, 324, 742–744. [Google Scholar] [CrossRef] [PubMed]
- Daròs, J.A.; Flores, R. Arabidopsis thaliana has the enzymatic machinery for replicating representative viroid species of the family Pospiviroidae. Proc. Natl. Acad. Sci. USA 2004, 101, 6792–6797. [Google Scholar] [CrossRef]
- Harders, J.; Lukacs, N.; Robert-Nicoud, M.; Jovin, T. M.; Riesner, D. Imaging of viroids in nuclei from tomato leaf tissue by in situ hybridization and confocal laser scanning microscopy. EMBO J. 1989, 8, 3941–3949. [Google Scholar] [PubMed]
- Hiscox, J.A. RNA viruses: hijacking the dynamic nucleolus. Nature Rev. Microbiol. 2007, 5, 119–127. [Google Scholar] [CrossRef]
- Senthil, G.; Liu, H.; Puram, V.G.; Clark, A.; Stromberg, A.; Goodin, M.M. Specific and common changes in Nicotiana benthamiana gene expression in response to infection by enveloped viruses. J. Gen. Virol. 2005, 86, 2615–2625. [Google Scholar] [CrossRef] [PubMed]
- Culver, J.N.; Padmanabhan, M.S. Virus-induced disease: altering host physiology one interaction at a time. Annu. Rev. Phytopathol. 2007, 45, 221–243. [Google Scholar] [CrossRef] [PubMed]
- Farag, M.A.; Huhman, D.V.; Dixon, R.A.; Sumner, L.W. Metabolomics reveals novel pathways and differential mechanistic and elicitor-specific responses in phenylpropanoid and isoflavonoid biosynthesis in Medicago truncatula cell cultures. Plant Physiol. 2009, 146, 387–402. [Google Scholar] [CrossRef]
- Marks, F.; Klingmüller, U.; Müller-Decker, K. Cellular signal processing: an introduction to the molecular mechanisms of signal transduction. 2009; Garland Science: New York, NY, USA. [Google Scholar]
- Dangl, J.L.; Dietrich, R.A.; Thomas, H. Senescence and programmed cell death. In: Biochemistry & Mo. , Ed.; 2000; Am. Soc. Plant Biol.: Rockville, MD, USA. [Google Scholar]
- Grant, M.R.; Jones, J.D.G. Hormone (dis)harmony moulds plant health and disease. Science 2009, 324, 750–752. [Google Scholar] [CrossRef] [PubMed]
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Share and Cite
Owens, R.A.; Hammond, R.W. Viroid Pathogenicity: One Process, Many Faces. Viruses 2009, 1, 298-316. https://doi.org/10.3390/v1020298
Owens RA, Hammond RW. Viroid Pathogenicity: One Process, Many Faces. Viruses. 2009; 1(2):298-316. https://doi.org/10.3390/v1020298
Chicago/Turabian StyleOwens, Robert A., and Rosemarie W. Hammond. 2009. "Viroid Pathogenicity: One Process, Many Faces" Viruses 1, no. 2: 298-316. https://doi.org/10.3390/v1020298
APA StyleOwens, R. A., & Hammond, R. W. (2009). Viroid Pathogenicity: One Process, Many Faces. Viruses, 1(2), 298-316. https://doi.org/10.3390/v1020298




