Computer-Aided Diagnosis in Mammography Using Content-Based Image Retrieval Approaches: Current Status and Future Perspectives
Abstract
:1. Introduction
2. Overview of CAD scheme using CBIR approach
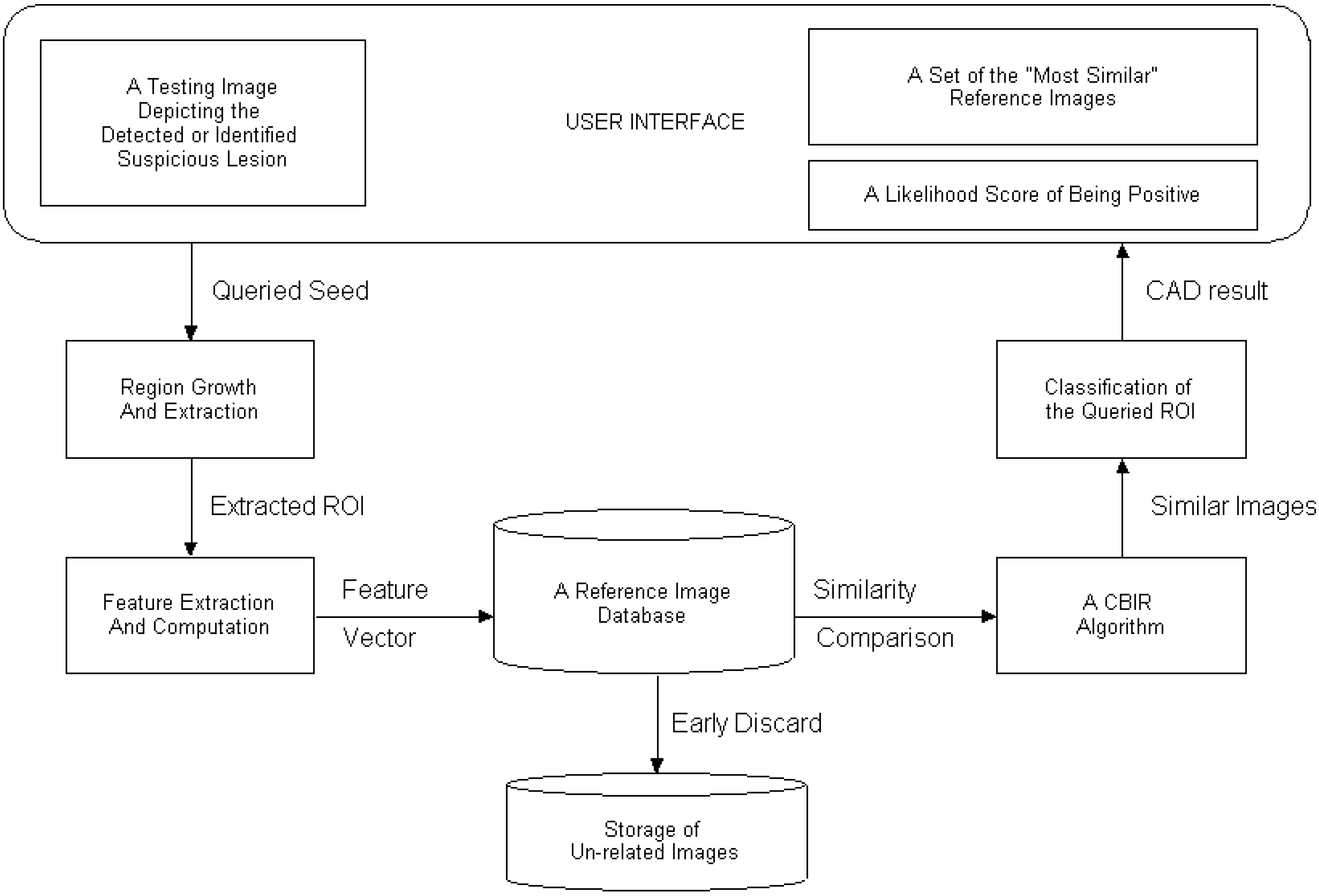
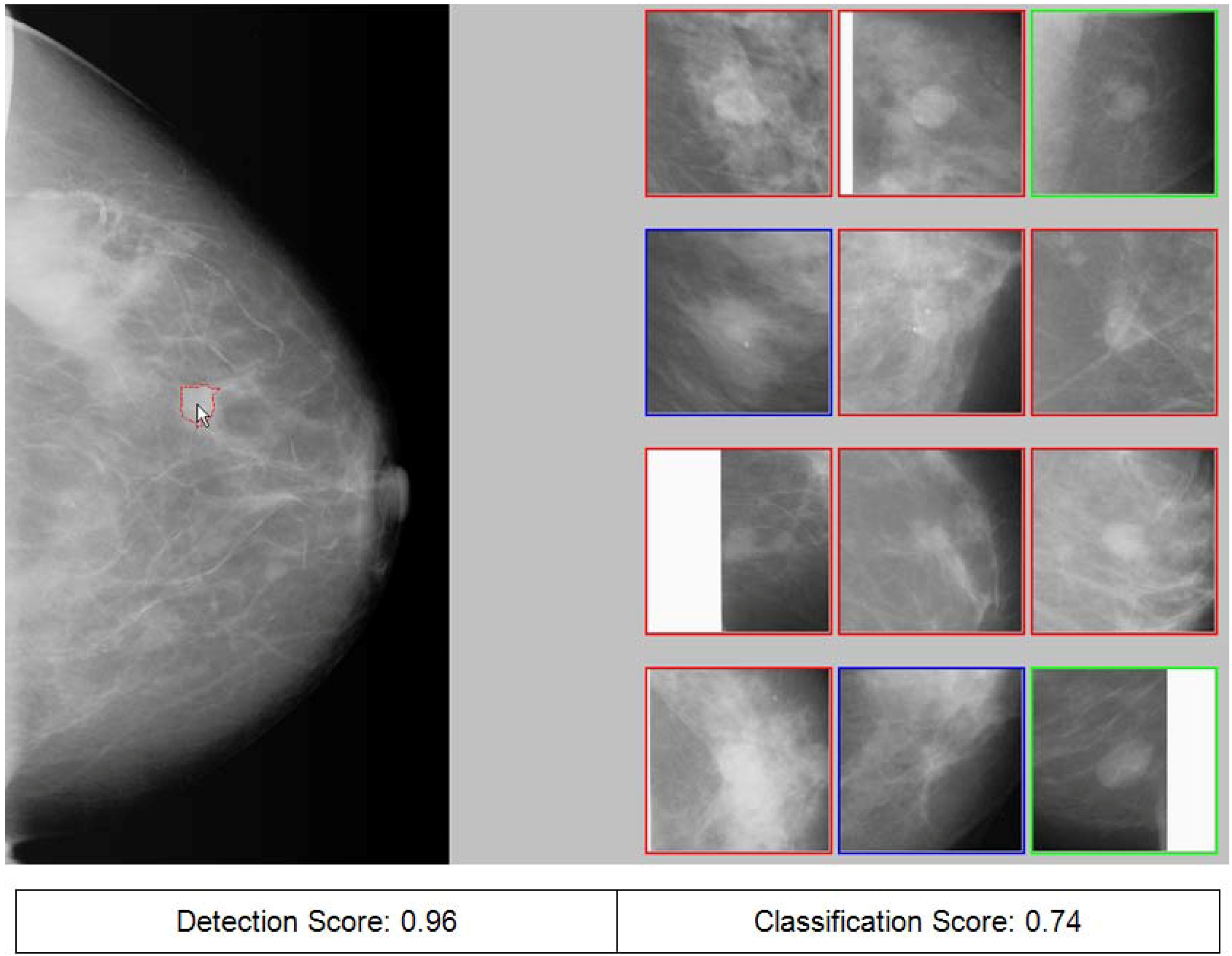
3. Region Segmentation
4. Feature Selection
5. Reference Databases
| Reference Database | Originally (randomly) Selected Database | “Optimized” Database | ||||
| Number of ROIs | 630 | 1262 | 1591 | 2523 | 3153 | 2979 |
| Area under ROC Curve | 0.715 | 0.794 | 0.874 | 0.875 | 0.872 | 0.914 |
| Standard Deviation | 0.026 | 0.023 | 0.017 | 0.017 | 0.017 | 0.012 |
| Threshold on Similarity Level | 0 | 0.1 | 0.2 | 0.3 | 0.4 | 0.5 | 0.6 | 0.7 | 0.8 | 0.9 |
| Area under ROC Curve | 0.854 | 0.859 | 0.859 | 0.864 | 0.877 | 0.888 | 0.908 | 0.911 | 0.919 | 0.932 |
| Standard Deviation | 0.004 | 0.004 | 0.005 | 0.007 | 0.008 | 0.011 | 0.011 | 0.014 | 0.016 | 0.016 |
6. Similarity Searching Methods and Computational Efficiency
7. Assessment of CAD Performance
8. Summary
Acknowledgement
References
- Love, H.J.; Antipow, I.; Hersh, W.; Smith, C.A.; Mailhot, M. Automated semantic indexing of imaging reports to support retrieval of medical images in the multimedia electronic medical record. Meth Inform Med 1999, 38, 303–307. [Google Scholar]
- El-Kwae, E.; Xu, H.; Kabuka, M.R. Content-based retrieval in picture archiving and communication systems. J Digit Imaging 2000, 13, 70–81. [Google Scholar] [CrossRef]
- Ogiela, M.R.; Tadeusiewicz, R. Semantic-oriented syntactic algorithms for content recognition and understanding of images in medical database. In Proceedings of the second International Conference on Multimedia and Exposition, IEEE Computer Society, Tokyo, Japan, 2001; pp. 621–624.
- Hersh, W.; Mailhot, M.; Arnott-Smith, C.; Lowe, H. Selective automated indexing of findings and diagnoses in radiology reports. J Biomed Informatics 2001, 34, 262–273. [Google Scholar] [CrossRef] [PubMed]
- Tagare, H.D.; Jaffe, C.; Duncan, J. Medical image databases: a content-based retrieval approach. J. Am. Med. Informatics Assoc. 1997, 4, 184–198. [Google Scholar] [CrossRef]
- Lehmann, T.M.; Guld, M.O.; Deselaers, T.; Keysers, D.; Schubert, H.; Spitzer, K.; Ney, H.; Wein, B.B. Automatic categorization of medical images for content-based retrieval and data mining. Comput Med Imaging Graph 2005, 29, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Long, L.R.; Antani, S.K.; Thoma, G.R. Image informatics at a national research center. Comput Med Imaging Graph 2005, 29, 171–193. [Google Scholar] [CrossRef] [PubMed]
- Muller, H.; Rosset, A.; Garcia, A.; Vallie, J.; Geissbuhler, A. Benefits of content-based visual data access in radiology. RadioGraphics 2005, 25, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Lam, M.O.; Disney, T.; Raicu, D.S.; Furst, J.; Channin, D.S. BRISC – An open source pulmonary nodule image retrieval framework. J Digit Imaging 2007, 20, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Pourghassem, H.; Ghassemian, H. Content-based medical image classification using a new hierarchical merging scheme. Comput Med Imaging Graph 2008, 32, 651–661. [Google Scholar] [CrossRef] [PubMed]
- Muller, H.; Michoux, N.; Bandon, D.; Geissbuhler, A. A review of content-based image retrieval systems in medical applications – clinical benefits and future directions. Int J Med Inform 2004, 73, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, R.M. Current status and future directions of computer-aided diagnosis in mammography. Comput Med Imaging Graph 2007, 31, 224–235. [Google Scholar] [CrossRef] [PubMed]
- Wiemker, R.; Rogalla, P.; Blaffert, T.; Sifri, D.; Hay, O.; Shah, E.; Truyen, R.; Fleiter, T. Aspects of computer-aided detection (CAD) and volumetry of pulmonary nodules using multi-slice CT. Br J Radiology 2005, 78, S46–S56. [Google Scholar] [CrossRef] [PubMed]
- Sluimer, I.C.; Prokop, M.; Hartmann, I.; Ginneken, B. Automated classification of hyperlucency, fibrosis, ground glass, solid, and focal lesions in high-resolution CT of the lung. Med Phys 2006, 33, 2610–2620. [Google Scholar] [CrossRef]
- Zheng, B.; Leader, J.; McMurray, J.; Park, S.C.; Fuhrman, C.R.; Gur, D.; Sciurba, F.C. Automated detection and quantitative assessment of pulmonary airways depicted on CT images. Med Phys 2007, 34, 2844–2852. [Google Scholar] [CrossRef] [PubMed]
- Buhmann, S.; Liang, J.; Wolf, M.; Salganicoff, M.; Kirchhoff, C.; Reiser, M.; Becker, C.H. Clinical evaluation of a computer-aided diagnosis (CAD) prototype for the detection of pulmonary embolism. Acad Radiol 2007, 14, 651–658. [Google Scholar] [CrossRef] [PubMed]
- Bogoni, L.; Cathier, P.; Dundar, M.; Jerebko, A.; Lakare, S.; Liang, J.; Periaswamy, S.; Baker, M.E.; Macari, M. Computer-aided detection (CAD) for CT colonography: a tool to address a growing need. Br J Radiology 2005, 78, S57–S62. [Google Scholar] [CrossRef] [PubMed]
- Raimondo, F.; Gavrielides, M.A.; Karayannopoulou, G. Automated evaluation of Her-2/neu status in breast tissue from fluorescent in situ hybridization images. IEEE Trans Image Processing 2005, 14, 1288–1299. [Google Scholar] [CrossRef]
- Wang, X.; Zheng, B.; Li, S.; Mulvihill, J.J.; Liu, H. Development and assessment of an integrated computer-aided detection scheme for digital microscopic images of metaphase chromosomes. J Electronic Imaging 2008, 17, 043008-1-9. [Google Scholar]
- Freer, T.M.; Ulissey, M.J. Screening mammography with computer-aided detection: prospective study of 12,860 patients in a community breast center. Radiology 2001, 220, 781–786. [Google Scholar] [CrossRef] [PubMed]
- Gur, D.; Sumkin, J.H.; Rockette, H.E.; Ganott, M.; Hakim, C.; Hardesty, L.; Poller, W.R.; Shah, R.; Wallace, L. Changes in breast cancer detection and mammography recall rates after the introduction of a computer-aided detection system. J Natl Cancer Inst 2004, 96, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Khoo, L.A.; Taylor, P.; Given-Wilson, R.M. Computer-aided detection in the United Kingdom National Breast Screening Programme: prospective study. Radiology 2005, 237, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.M.; Nicholas, M.J.; Mendel, J.B.; Slanetz, P.J. Prospective assessment of computer-aided detection in interpretation of screening mammograms. Am. J. Roentgenol. 2006, 187, 1483–1491. [Google Scholar]
- Gur, D.; Stalder, J.S.; Hardesty, L.A.; Zheng, B.; Sumkin, J.H.; Chough, D.M.; Shindel, B.E.; Rockette, H.E. Computer-aided detection performance in mammographic examination of masses: assessment. Radiology 2004, 233, 418–423. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Leader, J.K.; Abrams, G.S.; Lu, A.H.; Wallace, L.P.; Maitz, G.S.; Gur, D. Multiview-based computer-aided detection scheme for breast masses. Med Phys 2006, 33, 3135–3143. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, R.M.; Kallergi, M. Computer-aided detection in its present form is not an effective aid for screening mammography. Med Phys 2006, 33, 811–814. [Google Scholar] [CrossRef]
- Fenton, J.J.; Taplin, S.H.; Carney, P.A.; Abraham, L.; Sickles, E.A.; D’Orsi, C.; Berns, E.A.; Cutter, G.; Hendrick, R.E.; Barlow, W.E.; Elmore, J.G. Influence of computer-aided detection on performance of screening mammography. N. Engl. J. Med. 2007, 356, 1399–1409. [Google Scholar] [CrossRef] [PubMed]
- Tourassi, G.D.; Vargas-Voracek, R.; Catarious, D.M.; Floyd, C.E. Computer-assisted detection of mammographic masses: a template matching scheme based on mutual information. Med Phys 2003, 30, 2123–2130. [Google Scholar] [CrossRef] [PubMed]
- El-Napa, I.; Yang, Y.; Galatsanos, N.P.; Nishikawa, R.M.; Wernick, M.N. A similarity learning approach to content-based image retrieval: application to digital mammography. IEEE Trans Med Imaging 2004, 23, 1233–1244. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Li, C.; Wilson, R. A general framework for content-based medical image retrival with its application to mammograms. Proc SPIE 2005, 5748, 134–143. [Google Scholar]
- Alto, H.; Rangayyan, R.M.; Desautels, J.E. Content-based retrieval and analysis of mammographic masses. J. Electron. Imaging 2005, 14, 023016. [Google Scholar] [CrossRef]
- Tao, Y.; Lo, S.B.; Freedman, M.T.; Xuan, J. A preliminary study of content-based mammographic masses retrieval. Proc SPIE 2007, 6514, 65141Z. [Google Scholar]
- Kinoshita, S.K.; de Azevedo-Marques, P.M.; Pereira, R.R.; Rodrigues, J.; Rangayyan, R. Content-based retrieval of mammograms using visual features related to breast density patterns. J Digit Imaging 2007, 20, 172–190. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Mello-Thoms, C.; Wang, X.; Abrams, G.S.; Sumkin, J.H.; Chough, D.M.; Ganott, M.A.; Lu, A.; Gur, D. Interactive computer aided diagnosis of breast masses: computerized selection of visually similar image sets from a reference library. Acad Radiol 2007, 14, 917–927. [Google Scholar]
- Mazurowski, M.A.; Habas, P.A.; Zurada, J.M.; Tourassi, G.D. Decision optimization of case-based computer-aided decision systems using genetic algorithm with application to mammography. Phys Med Biol 2008, 53, 895–908. [Google Scholar] [CrossRef] [PubMed]
- Rosa, N.A.; Felipe, J.C.; Traina, A.J.; Rangayyan, R.M.; Azevedo-Marques, P.M. Using relevance feedback to reduce the semantic gap in content-based image retrieval of mammographic masses. Conf Proc IEEE Med Biol Soc 2008, 406–409. [Google Scholar]
- Park, S.C.; Pu, J.; Zheng, B. Improving performance of computer-aided detection scheme by combining results from two machine learning classifiers. Acad Radiol 2009, 16, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Giger, M.L.; Huo, Z.; Vyborny, C.J.; Lan, L.; Bonta, I.R.; Horsch, K.; Nishikawa, R.M.; Rosenbourgh, I. Intelligent CAD workstation for breast imaging using similarity to known lesions and multiple visual prompt aides. Proc SPIE 2002, 4684, 768–773. [Google Scholar]
- Zheng, B.; Abrams, G.; Britton, C.A.; Hakim, C.M.; Lu, A.; Clearfield, R.J.; Drescher, J.; Maitz, G.S.; Gur, D. Evaluation of an interactive computer-aided diagnosis scheme for mammography: a pilot study. Proc SPIE 2007, 6515, 65151M. [Google Scholar]
- Zheng, B.; Sumkin, J.H.; Good, W.F.; Maitz, G.S.; Chang, Y.H.; Gur, D. Applying computer-assisted detection schemes to digitized mammograms after JPEG data compression: an assessment. Acad Radiol 2000, 7, 595–602. [Google Scholar] [CrossRef]
- Zheng, B.; Lu, A.; Hardesty, L.A.; Sumkin, J.H.; Hakim, C.M.; Ganott, M.A.; Gur, D. A method to improve visual similarity of breast masses for an interactive computer-aided diagnosis environment. Med Phys 2006, 33, 111–117. [Google Scholar] [CrossRef]
- Jiang, Y.; Nishikawa, R.M.; Papaioannou, J. Dependence of computer classification of clustered microcalcifications on the correct detection of microcalcifications. Med Phys 2001, 28, 1949–1957. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Pu, J.; Park, S.C.; Zuley, M.; Gur, D. Assessment of the relationship between lesion segmentation accuracy and computer-aided diagnosis scheme performance. Proc SPIE 2008, 6915, 6915–30. [Google Scholar]
- Hill, P.R.; Canagarajah, N.; Bull, D.R. Image segmentation using a texture gradient based watershed transform. IEEE Trans Image Processing 2003, 12, 1618–1633. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Chang, Y.H.; Gur, D. Computerized detection of masses in digitized mammograms using single image segmentation and multi-layer topographic feature extraction. Acad Radiol 1995, 2, 959–966. [Google Scholar]
- Eltonsy, N.H.; Tourassi, G.D.; Elmaghraby, A.S. A concentric morphology model for the detection of masses in mammography. IEEE Trans Med Imaging 2007, 26, 880–889. [Google Scholar] [CrossRef] [PubMed]
- Lobregt, S.; Viergever, M.A. A discrete dynamic contour model. IEEE Trans. Med Imaging 1995, 14, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Sahiner, B.; Petrick, N.; Chan, H.P.; Hadjiiski, L.M.; Paramagul, C.; Helvie, M.A.; Gurcan, M.N. Computer-aided characterization of mammographic masses: accuracy of mass segmentation and its effects on characterization. IEEE Trans. Med Imaging 2001, 20, 1275–1284. [Google Scholar] [CrossRef] [PubMed]
- Brake, G.M.; Karssemeijer, N. Segmentation of suspicious densities in digital mammograms. Med Phys 2001, 28, 259–266. [Google Scholar] [CrossRef]
- Yuan, Y.; Giger, M.L.; Li, H.; Suzuki, K.; Sennett, C. A dual-stage method for lesion segmentation on digital mammograms. Med Phys 2007, 34, 4180–4193. [Google Scholar] [CrossRef]
- Dominguez, A.R.; Nandi, A.K. Improved dynamic-programming-based algorithms for segmentation of masses in mammograms. Med Phys 2007, 34, 4265–4268. [Google Scholar] [CrossRef]
- Yuan, Y.; Giger, M.L.; Li, H.; Suzuki, K.; Sennett, C. A dual-stage method for lesion segmentation on digital mammograms. Med Phys 2007, 34, 4180–4193. [Google Scholar] [CrossRef]
- Elter, M.; Horsch, A. CADx of mammographic masses and clustered microcalcifications: a review. Med Phys 2009, 36, 2052–2068. [Google Scholar] [CrossRef] [PubMed]
- Vyborny, C.J.; Doi, T.; O’Shaughnessy, K.F. Breast cancer: importance of spiculation in computer-aided detection. Radiology 2000, 215, 703–707. [Google Scholar] [CrossRef] [PubMed]
- Kegelmeyer, W.P.; Pruneda, J.M.; Bourland, P.D.; Hillis, A.; Riggs, M.W.; Nipper, M.L. Computer-aided mammographic screening for spiculated lesions. Radiology 1994, 191, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Rangayyan, R.M.; Mudigonda, N.R.; Desautels, J.E. Boundary modeling and shape analysis methods for classification of mammographic masses. Med Biol Eng Comput 2000, 38, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Sahiner, B.; Chan, H.P.; Petrick, N.; Helvie, M.A.; Hadjiiski, L.M. Improvement of mammographic mass characterization using spiculation measures and morphological features. Med Phys 2001, 28, 1455–1465. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Song, E.; Xu, X.; Ma, G.; Zheng, B. Automated detection of breast mass spiculation levels and evaluation of scheme performance. Acad Radiol 2008, 15, 1534–1544. [Google Scholar] [CrossRef] [PubMed]
- Tourassi, G.D.; Harrawood, B.; Singh, S.; Lo, J.Y.; Floyd, C.E. Evaluation of information-theoretic similarity measures for content-based retrieval and detection of masses in mammograms. Med Phys 2007, 34, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Park, S.C.; Zheng, B. Improving performance of content-based image retrieval schemes in searching for similar breast mass regions: an assessment. Phys Med Biol 2009, 54, 949–961. [Google Scholar] [CrossRef] [PubMed]
- Deserno, T.M.; Antani, S.; Long, R. Ontology of gaps in content-based image retrieval. J Digit Imaging 2009, 22, 202–215. [Google Scholar] [CrossRef] [PubMed]
- Filev, P.; Hadjiiski, L.; Sahiner, B.; Chan, H.P.; Helvie, M.A. Comparison of similarity measures for the task of template matching of masses on serial mammograms. Med Phys 2005, 32, 515–529. [Google Scholar] [CrossRef]
- Felipe, J.C.; Traina, C.; Traina, A.J. A new family of distance functions for perceptual similarity retrieval of medical images. J Digit Imaging 2009, 22, 183–201. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Manjunath, B. Texture features and learning similarity. In Proc IEEE Conference on Computer Vision and Pattern Recognition, San Francisco, CA, 1996; pp. 425–430.
- Ortega, M.; Rui, Y.; Chakrabarti, K. Supporting ranked Boolean similarity queries in MARS. IEEE Trans Knowledge Data Eng 1998, 10, 905–925. [Google Scholar] [CrossRef]
- Daugman, J.G. High confidence visual recognition of persons by a test of statistical independence. IEEE Trans Pattern Anal. Machine Intel. 1993, 15, 1148–1161. [Google Scholar] [CrossRef]
- Kuo, W.; Chang, R.; Lee, C.C.; Moon, W.K.; Chen, D.R. Retrieval technique for the diagnosis of solid breast tumors on sonogram. Ultrasound Med. Biol. 2002, 28, 903–909. [Google Scholar] [CrossRef]
- Milanese, R.; Cherbuliez, M. A rotation, translation and scale-invariant approach to content-based image retrieval. J. Visual Commun. Image Represent 1999, 10, 186–196. [Google Scholar] [CrossRef]
- Park, S.C.; Wang, X.; Zheng, B. Assessment of performance improvement in content-based medical image retrieval schemes using fractal dimension. Acad Radiol 2009, 16. in press. [Google Scholar] [CrossRef] [PubMed]
- Velanovich, V. Fractal analysis of mammographic lesions: a feasibility study quantifying the difference between benign and malignant masses. Am. J. Med. Sci. 1996, 311, 211–214. [Google Scholar] [CrossRef]
- Chevallet, J.P.; Maillot, N.; Lim, J.H. Concept Propagation Based on Visual Similarity Application to Medical Image Annotation. In Proc Third Asia Information Retrieval Symposium, Singapore, October 16-18, 2006; 4182, pp. 514–521.
- Soares, F.; Andruszkiewic, P.; Freire, M.M. Self-Similarity Analysis Applied to 2D Breast Cancer Imaging. In Proc International Conf. on Systems and Networks Communications, Cap Esterel, French Riviera, France, August 25-31, 2007; pp. 1–6.
- Nishikawa, R.M.; Giger, M.L.; Doi, K.; Metz, C.E.; Yin, F.; Vyborny, C.J.; Schmidt, R.A. Effect of case selection on the performance of computer-aided detection schemes. Med Phys 1994, 21, 265–269. [Google Scholar] [CrossRef]
- Kupinski, M.A.; Giger, M.L. Feature selection with limited database. Med Phys 1999, 26, 2176–2182. [Google Scholar]
- Zheng, B.; Chang, Y.H.; Good, W.F.; Gur, D. Adequacy testing of training set sample sizes in the development of a computer-assisted diagnosis scheme. Acad Radiol 1997, 4, 497–502. [Google Scholar] [CrossRef]
- Mitchell, T.M. Machine Learning; WCB/McGraw-Hill: Boston, MA, 1997; pp. 230–248. [Google Scholar]
- Park, S.C.; Sukthankar, R.; Mummert, L.; Satyanarayanan, M.; Zheng, B. Optimization of reference library used in content-based medical image retrieval scheme. Med Phys 2007, 34, 4331–4339. [Google Scholar] [CrossRef]
- Tourassi, G.D.; Harrawood, B.; Singh, S.; Lo, J.Y. Information-theoretic CAD system in mammography: entropy-based indexing for computational efficiency and robust performance. Med Phys 2007, 34, 3193–3204. [Google Scholar] [CrossRef]
- Mazurowski, M.A.; Zurada, J.M.; Tourassi, G.D. Selection of examples in case-based computer-aided decision systems. Phys Med Biol 2008, 53, 6079–6096. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Doi, K. Reduction of bias and variance for evaluation of computer-aided diagnostic schemes. Med Phys 2006, 33, 868–875. [Google Scholar] [CrossRef]
- Sinha, U.; Kangarloo, H. Principal component analysis for content-based image retrieval. RadioGraphics 2002, 22, 1271–1289. [Google Scholar]
- Brodley, C.; Kak, A.; Shyu, C.; Dy, J.G.; Broderick, L.S.; Aisen, A.M. Content-based retrieval from medical image database: A synergy of human interaction, machine learning and computer vision. In Proc 16th National Conference on Artificial Intelligence and 11th Conference on Innovative Applications of Artificial Intelligence, Orlando, FL, July 18-22, 1999; pp. 760–767.
- Yang, L.; Jin, R.; Sukthankar, R.; Zheng, B.; Mummert, L.; Satyanarayanan, M.; Chen, M.; Jukic, D. Learning distance metrics for interactive search-assisted diagnosis of mammograms. Proc SPIE 2007, 6514, 65141H. [Google Scholar]
- Deserno, T.M.; Guild, M.O.; Plodowski, B.; Spitzer, K.; Wein, B.B.; Schubert, H.; Ney, H.; Seidi, T. Extended query refinement for medical image retrieval. J Digit Imaging 2008, 21, 280–289. [Google Scholar] [CrossRef] [PubMed]
- Traina, C.; Traina, A.; Araujo, M.; Bueno, J.M.; Chino, F.; Razente, H.; Azevedo-Marques, P.M. Using an image-extended relational database to support content-based image retrieval in a PACS. Comput Methods Programs Biomed 2005, 80, S71–S83. [Google Scholar] [CrossRef]
- Diamond – interactive search of non-indexed data, http://diamond.cs.cmu.edu.
- Huston, L.; Sukthankar, R.; Wickremesinghe, R.; Satyanarayanan, M.; Ganger, G.; Riedel, E.; Ailamaki, A. Diamond: a storage architecture for early discard in interactive search. In Proc of the 3rd USENIX Conference on File and Storage Technologies, San Francisco, CA, March, 2004; Available online from: http://diamond.cs.cmu.edu/papers/fast2004-diamond.pdf.
- Zheng, B.; Abrams, G.; Leader, J.K.; Park, S.C.; Maitz, G.S.; Gur, D. Mass margins spiculation: agreement between ratings by observers and a computer scheme. Proc SPIE 2007, 6514, 65141P. [Google Scholar]
- Muramatsu, C.; Li, Q.; Suzuki, K.; Schmidt, R.A.; Shiraishi, J.; Newstead, G.M.; Doi, K. Investigation of psychophysical measure for evaluation of similar images for mammographic masses: preliminary results. Med Phys 2005, 32, 2295–2304. [Google Scholar] [CrossRef]
- Paquerault, S.; Yarusso, L.M.; Papaioannou, J.; Jiang, Y.; Nishikawa, R.M. Radial gradient-based segmentation of mammographic microcalcifications: observer evaluation and effect on CAD performance. Med Phys 2004, 31, 2648–2657. [Google Scholar] [CrossRef]
- Rosner, B. Fundamentals of biostatistics; Duxbury, Pacific Grove, CA, 2000. [Google Scholar]
- Muramatsu, C.; Li, Q.; Schmidt, R.A.; Shiraishi, J.; Suzuki, K.; Newstead, G.M.; Doi, K. Determination of subjective similarity for pairs of masses and pairs of clustered microcalcifications on mammograms: comparison of similarity ranking scores and absolute similarity ratings. Med Phys 2007, 34, 2890–2895. [Google Scholar] [CrossRef] [PubMed]
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Zheng, B. Computer-Aided Diagnosis in Mammography Using Content-Based Image Retrieval Approaches: Current Status and Future Perspectives. Algorithms 2009, 2, 828-849. https://doi.org/10.3390/a2020828
Zheng B. Computer-Aided Diagnosis in Mammography Using Content-Based Image Retrieval Approaches: Current Status and Future Perspectives. Algorithms. 2009; 2(2):828-849. https://doi.org/10.3390/a2020828
Chicago/Turabian StyleZheng, Bin. 2009. "Computer-Aided Diagnosis in Mammography Using Content-Based Image Retrieval Approaches: Current Status and Future Perspectives" Algorithms 2, no. 2: 828-849. https://doi.org/10.3390/a2020828




