Process Analysis of Anaerobic Fermentation Exposure to Metal Mixtures
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Anaerobic Fermentation Experiment
2.3. Chemical and Microbial Analyses
2.4. Data Analysis
3. Results and Discussion
3.1. Biogas Properties of Cu-Treated Fermenters Combining with Fe, Ni and Zn
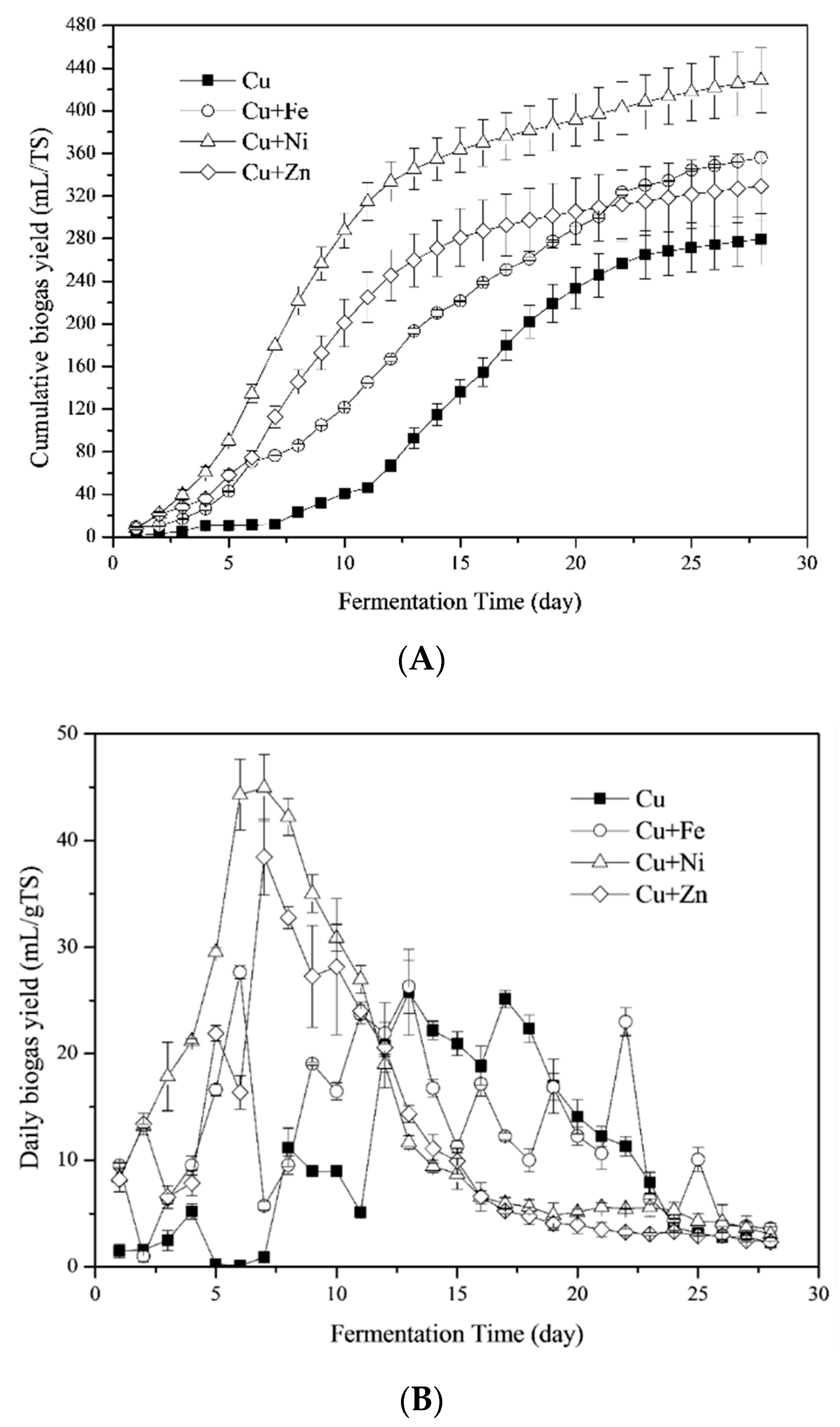
3.1.1. Biogas Yields
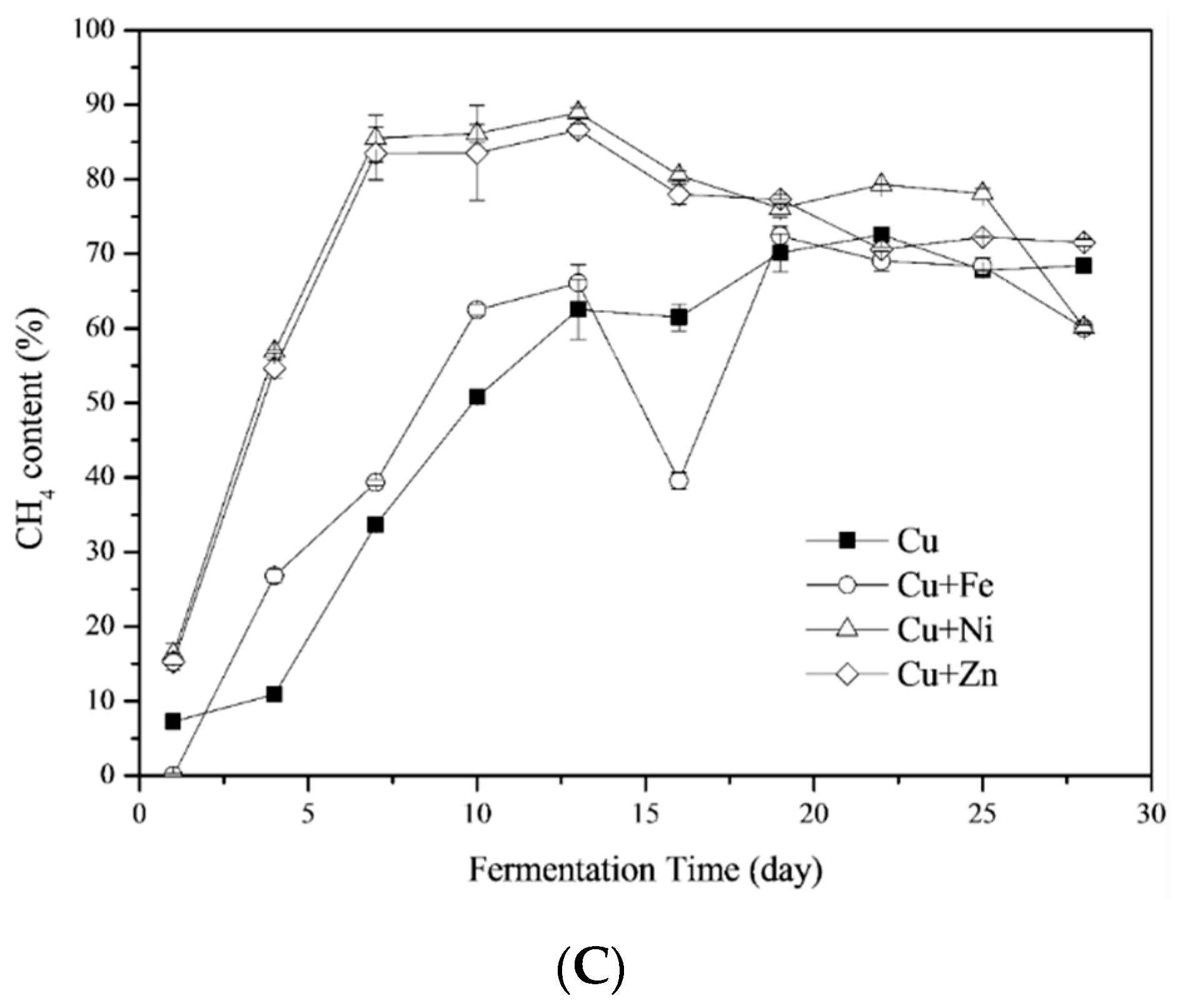
3.1.2. CH4 Yields
3.2. Process Stability as Indicated by the Variation of pH Values
3.3. Substrate Biodegradation
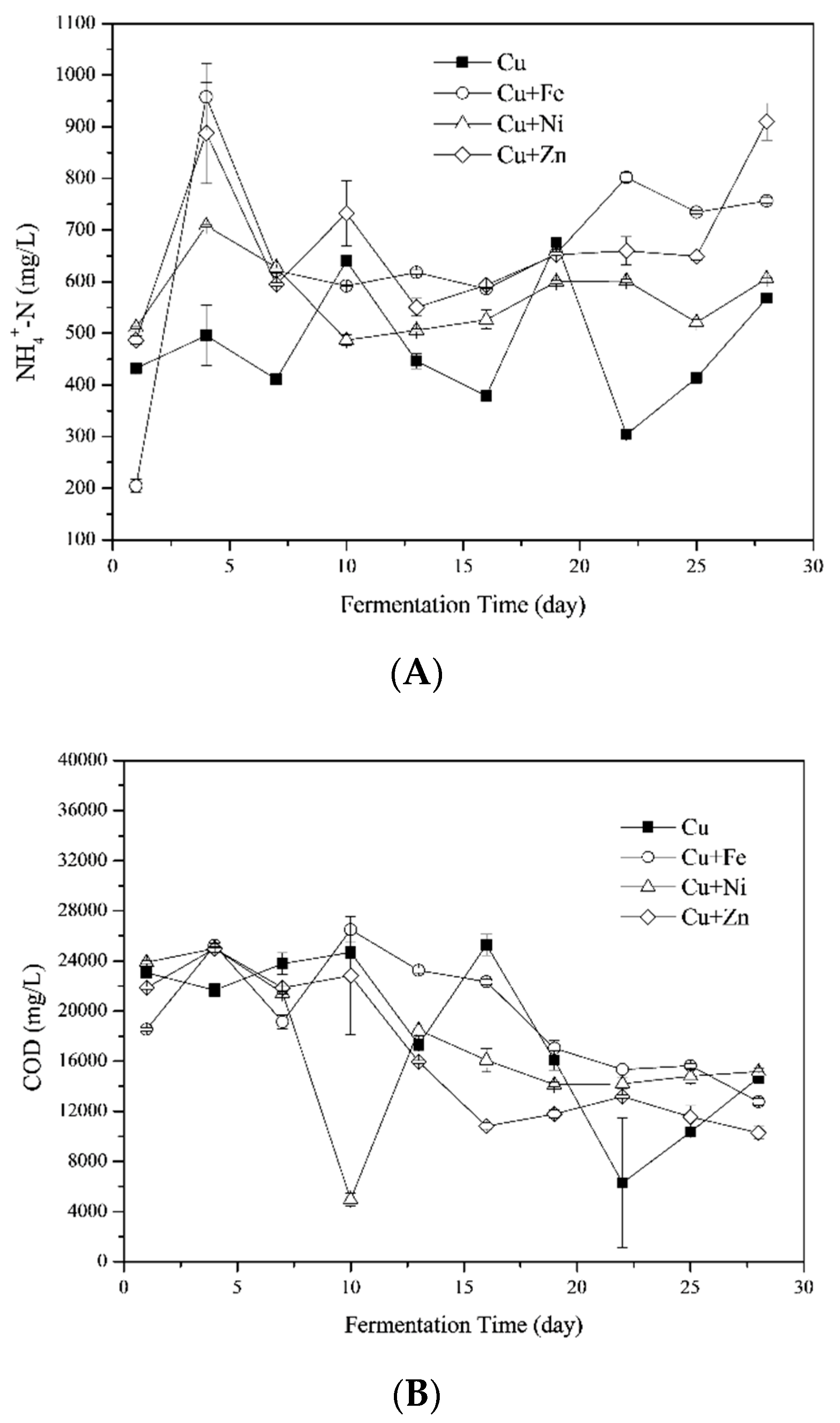
3.3.1. Variations of Ammonia Nitrogen (NH4+-N) Concentrations
3.3.2. Chemical Oxygen Demands (COD)
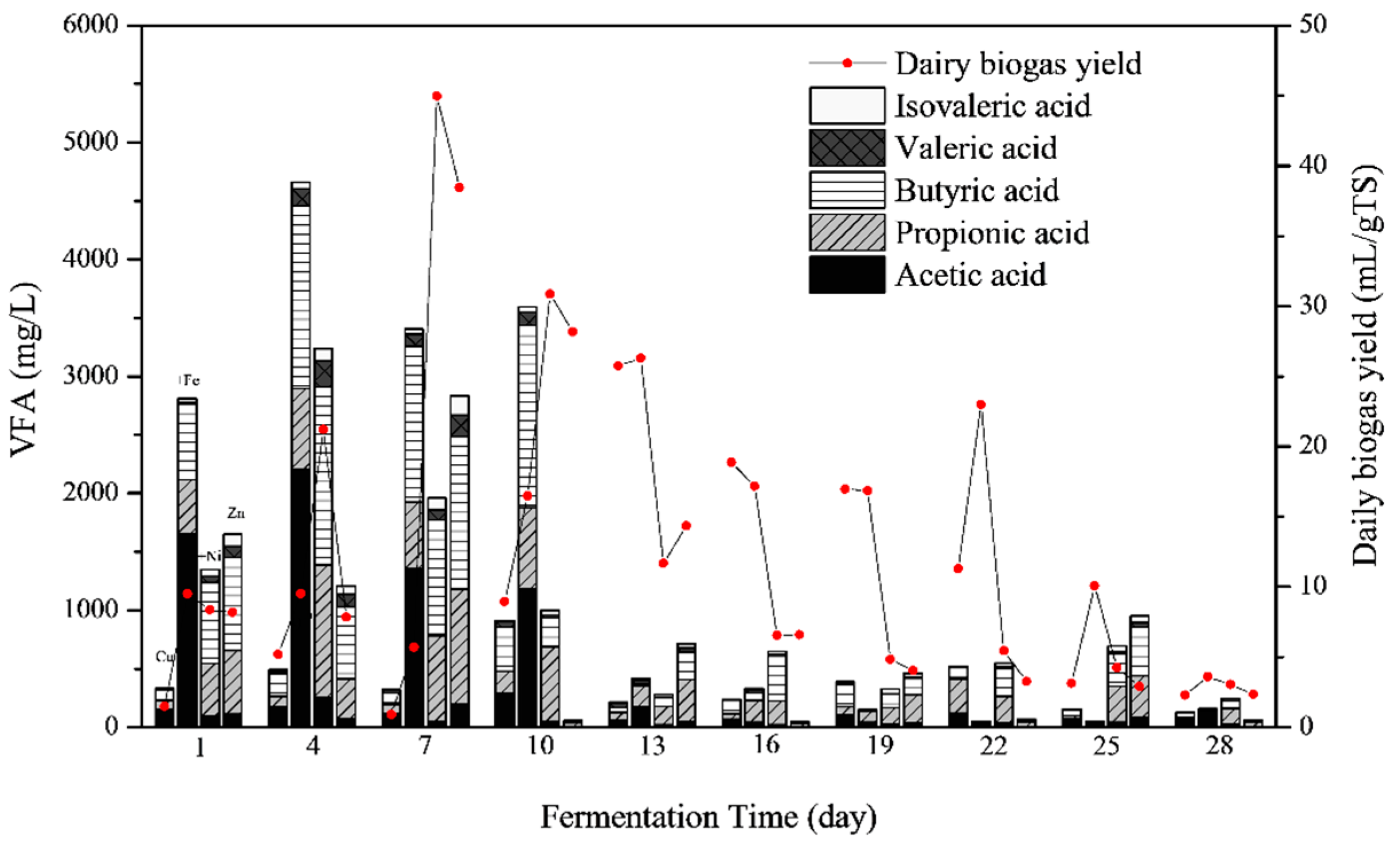
3.3.3. Responses of Volatile Fatty Acids (VFAs)
3.3.4. Degradation of Lignocelluloses
3.4. Responses of Enzyme Activity
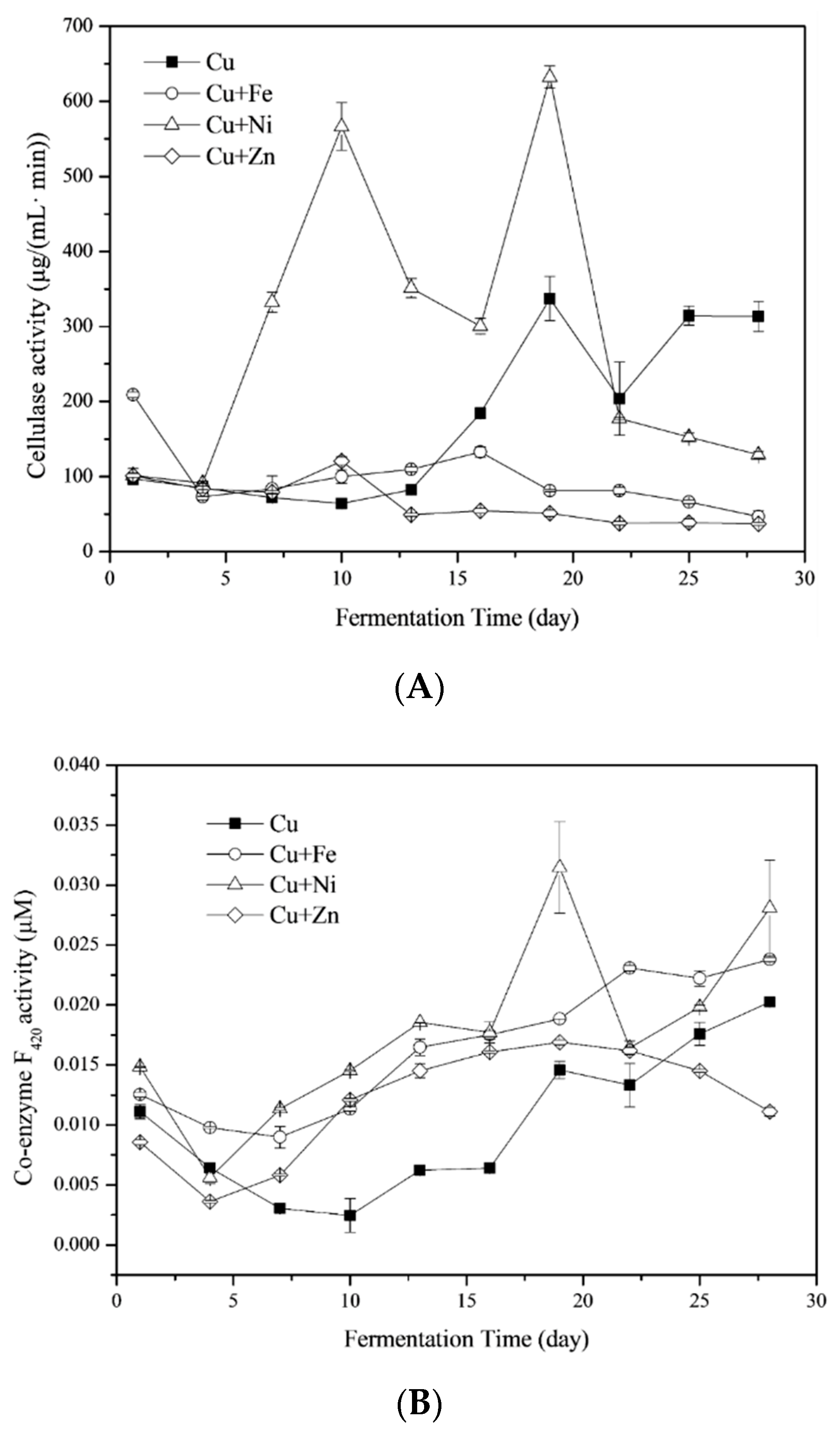
3.4.1. Cellulase
3.4.2. Coenzyme F420
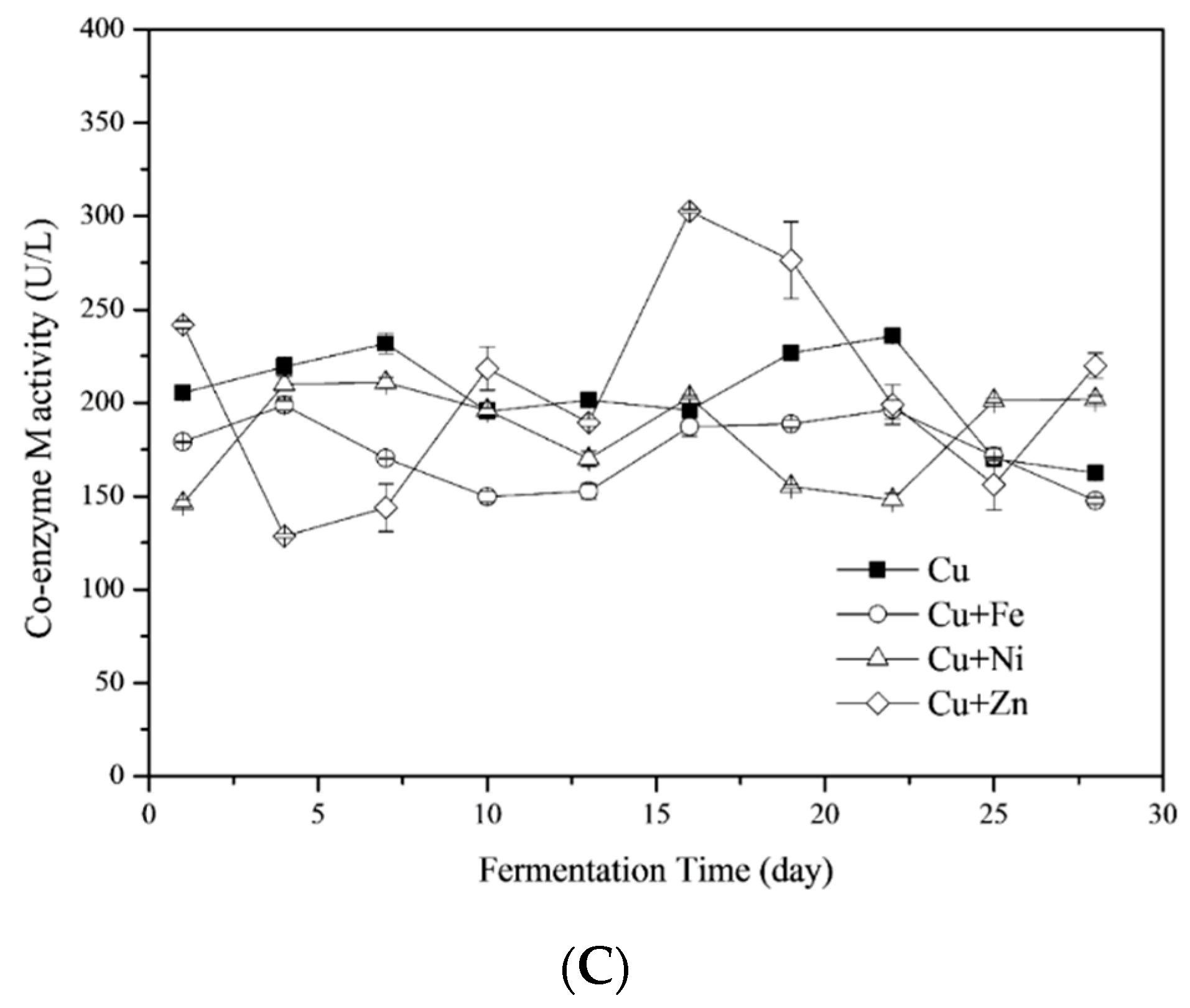
3.4.3. Coenzyme M
3.5. Impacts of Metal Mixtures on the Microbial Communities
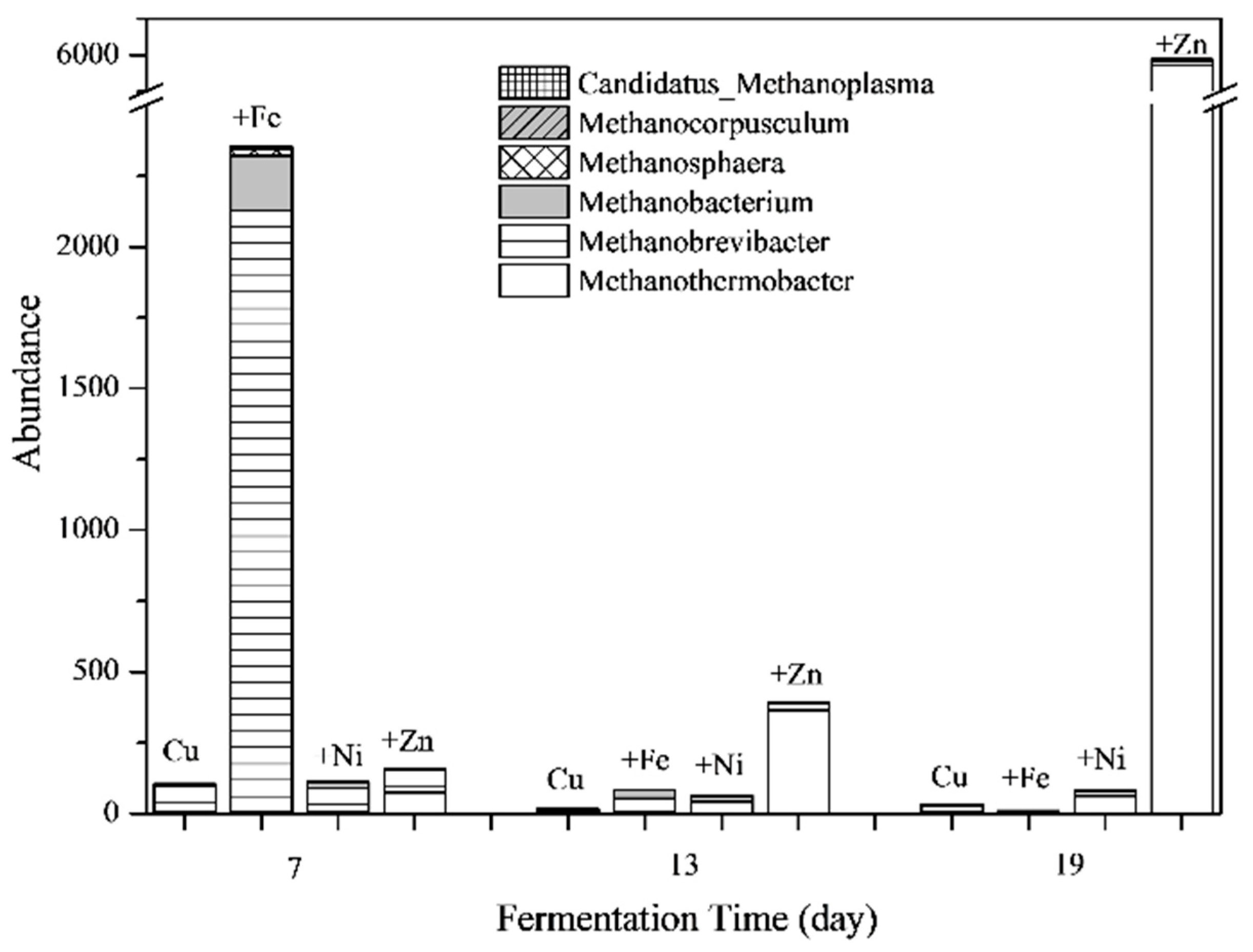
3.5.1. Structure of Microbial Communities
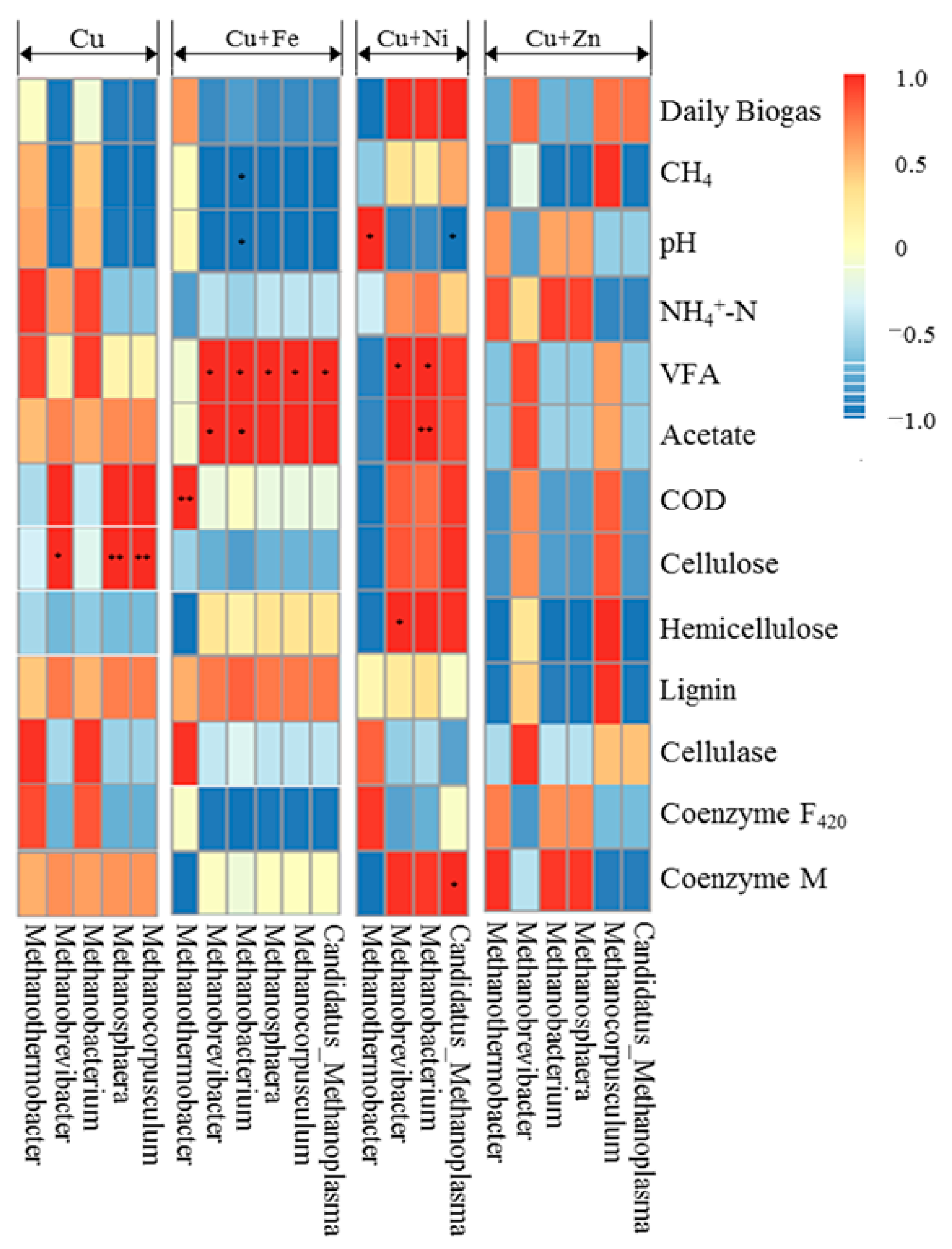
3.5.2. Methanogens and Their Relationships with Fermentation Parameters
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rezania, S.; Ponraj, M.; Talaiekhozani, A.; Sabbagh, F.; Sairan, F. Perspectives of phytoremediation using water hyacinth for removal of heavy metals, organic and inorganic pollutants in wastewater. J. Environ. Manag. 2015, 163, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Tian, Y.; Wang, L.; Zhang, L.; Dai, L. Ecophysiological characteristics and biogas production of cadmium-contaminated crops. Bioresour. Technol. 2013, 146, 628–636. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Zhang, H. Producing biogas from agricultural residues generated during phytoremediation process: Possibility, threshold, and challenges. Int. J. Green Energy 2016, 13, 1556–1563. [Google Scholar] [CrossRef]
- Fernandes, K.D.; Cañote, S.J.B.; Ribeiro, E.M.; Thiago Filho, G.L.; Fonseca, A.L. Can we use Cd-contaminated macrophytes for biogas production? Environ. Sci. Pollut. Res. 2018, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Han, X.; Tian, Y.; Li, Y.; Yang, K.; Hao, H.; Chai, Y.; Xu, X. Process analysis of anaerobic fermentation of Phragmites australis straw and cow dung exposing to elevated chromium (VI) concentrations. J. Environ. Manag. 2018, 224, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Manyiloh, C.E.; Mamphweli, S.N.; Meyer, E.L.; Okoh, A.I.; Makaka, G.; Simon, M. Microbial anaerobic digestion (Bio-Digesters) as an approach to the decontamination of animal wastes in pollution control and the generation of renewable energy. Int. J. Environ. Res. Public Health 2013, 10, 4390–4417. [Google Scholar] [CrossRef]
- Zhang, H.; Tian, Y.; Wang, L.; Mi, X.; Chai, Y. Effect of ferrous chloride on biogas production and enzymatic activities during anaerobic fermentation of cow dung and Phragmites straw. Biodegradation 2016, 27, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.; Tian, Y.; Zhang, H.; Chai, Y. Copper stressed anaerobic fermentation: biogas properties, process stability, biodegradation and enzyme responses. Biodegradation 2017, 28, 369–381. [Google Scholar] [CrossRef]
- Tian, Y.; Zhang, H.; Chai, Y.; Wang, L.; Mi, X.; Zhang, L.; Ware, M.A. Biogas properties and enzymatic analysis during anaerobic fermentation of Phragmites australis straw and cow dung: influence of nickel chloride supplement. Biodegradation 2017, 28, 15–25. [Google Scholar] [CrossRef]
- Jain, S.K.; Gujral, G.S.; Jha, N.K.; Vasudevan, P. Production of biogas from Azolla pinnata R.Br and Lemna minor L.: Effect of heavy metal contamination. Bioresour. Technol. 1992, 41, 273–277. [Google Scholar] [CrossRef]
- Lin, C.Y.; Chen, C.C. Effect of heavy metals on the methanogenic UASB granule. Water Res. 1999, 33, 409–416. [Google Scholar] [CrossRef]
- Elbana, T.A.; Magdi Selim, H.; Akrami, N.; Newman, A.; Shaheen, S.M.; Rinklebe, J. Freundlich sorption parameters for cadmium, copper, nickel, lead, and zinc for different soils: Influence of kinetics. Geoderma 2018, 324, 80–88. [Google Scholar] [CrossRef]
- Jeke, N.N.; Zvomuya, F.; Badiou, P.; Ross, L.; Cicek, N. Biomass, nutrient, and trace element accumulation and partitioning in cattail (L.) during wetland phytoremediation of municipal biosolids. J. Environ. Qual. 2015, 44, 1541–1549. [Google Scholar] [CrossRef] [PubMed]
- Salem, Z.B.; Laffray, X.; Ashoour, A.; Ayadi, H.; Aleya, L. Metal accumulation and distribution in the organs of Reeds and Cattails in a constructed treatment wetland (Etueffont, France). Ecol. Eng. 2014, 64, 1–17. [Google Scholar] [CrossRef]
- Chan, P.C.; de Toledo, R.A.; Iu, H.I.; Shim, H. Effect of zinc supplementation on biogas production and short/long chain fatty acids accumulation during anaerobic co-digestion of food waste and domestic wastewater. Waste Biomass Valor. 2018, 1–11. [Google Scholar] [CrossRef]
- Scherer, P.; Lippert, H.; Wolff, G. Composition of the major elements and trace elements of 10 methanogenic bacteria determined by inductively coupled plasma emission spectrometry. Biol. Trace Elem. Res. 1983, 5, 149–163. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Z.; Suzuki, K.; Maekawa, T. Uptake and mass balance of trace metals for methane producing bacteria. Biomass Bioenergy 2003, 25, 427–433. [Google Scholar] [CrossRef]
- Lenártová, V.; Holovská, K.; Javorský, P. The influence of mercury on the antioxidant enzyme activity of rumen bacteria Streptococcus bovis and Selenomonas ruminantium. Fems Microbiol. Ecol. 1998, 27, 319–325. [Google Scholar] [CrossRef]
- Yue, Z.B.; Yu, H.Q.; Wang, Z.L. Anaerobic digestion of cattail with rumen culture in the presence of heavy metals. Bioresour. Technol. 2007, 98, 781–786. [Google Scholar] [CrossRef]
- Jackson-Moss, C.A.; Duncan, J.R. The effect of iron on anaerobic digestion. Biotechnol. Lett. 1990, 12, 149–154. [Google Scholar] [CrossRef]
- Glass, J.B.; Orphan, V.J. Trace metal requirements for microbial enzymes involved in the production and consumption of methane and nitrous oxide. Front. Microbiol 2012, 3, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Mudhoo, A.; Kumar, S. Effects of heavy metals as stress factors on anaerobic digestion processes and biogas production from biomass. Int. J. Environ. Sci. Technol. 2013, 10, 1383–1398. [Google Scholar] [CrossRef] [Green Version]
- Amen, T.W.M.; Eljamal, O.; Khalil, A.M.E.; Sugihara, Y.; Matsunaga, N. Methane yield enhancement by the addition of new novel of iron and copper-iron bimetallic nanoparticles. Chem. Eng. Process. Intensif. 2018, 130, 253–261. [Google Scholar] [CrossRef]
- Ragsdale, S.W. Nickel-based enzyme systems. J. Biol. Chem. 2009, 284, 18571–18575. [Google Scholar] [CrossRef] [PubMed]
- Warkentin, E.; Thauer, R.K.; Ermler, U. Structure and function of enzymes involved in the methanogenic pathway utilizing carbon dioxide and molecular hydrogen. J. Biosci. Bioeng. 2002, 93, 519–530. [Google Scholar]
- Zandvoort, B.M.H.; Van Hullebusch, E.D.; Fermoso, F.G.; Lens, P.N.L. Trace metals in anaerobic granular sludge reactors: Bioavailability and dosing strategies. Eng. Life Sci. 2006, 6, 293–301. [Google Scholar] [CrossRef]
- Thauer, R.K.; Kaster, A.; Goenrich, M.; Schick, M.; Hiromoto, T.; Shima, S. Hydrogenases from methanogenic archaea, mickel, a novel cofactor, and H2 storage. Annu. Rev. Biochem. 2010, 79, 507–536. [Google Scholar] [CrossRef]
- Sawers, R.G. Nickel in bacteria and archaea. In Encyclopedia of Metalloproteins; Kretsinger, R.H., Uversky, V.N., Permyakov, E.A., Eds.; Springer: New York, NY, USA, 2013; pp. 1490–1496. [Google Scholar]
- Chen, Y.; Cheng, J.J.; Creamer, K.S. Inhibition of anaerobic digestion process: A review. Bioresour. Technol. 2008, 99, 4044–4064. [Google Scholar] [CrossRef]
- Cao, Z.; Wang, S.; Wang, T.; Chang, Z.; Shen, Z.; Chen, Y. Using contaminated plants involved in phytoremediation for anaerobic digestion. Int. J. Phytoremed. 2015, 17, 201–207. [Google Scholar] [CrossRef]
- Clark, P.B.; Hillman, P.F. Enhancement of anaerobic digestion using duckweed (Lemna minor) enriched with iron. Water Environ. J. 1996, 10, 92–95. [Google Scholar] [CrossRef]
- Pobeheim, H.; Munk, B.; Johansson, J.; Guebitz, G.M. Influence of trace elements on methane formation from a synthetic model substrate for maize silage. Bioresour. Technol. 2010, 101, 836–839. [Google Scholar] [CrossRef] [PubMed]
- Pobeheim, H.; Munk, B.; Lindorfer, H.; Guebitz, G.M. Impact of nickel and cobalt on biogas production and process stability during semi-continuous anaerobic fermentation of a model substrate for maize silage. Water Res. 2011, 45, 781–787. [Google Scholar] [CrossRef] [PubMed]
- Nkemka, V.N.; Murto, M. Evaluation of biogas production from seaweed in batch tests and in UASB reactors combined with the removal of heavy metals. J. Environ. Manag. 2010, 91, 1573–1579. [Google Scholar] [CrossRef] [PubMed]
- Willscher, S.; Mirgorodsky, D.; Jablonski, L.; Ollivier, D.; Merten, D.; Büchel, G.; Wittig, J.; Werner, P.H. Field scale phytoremediation experiments on a heavy metal and uranium contaminated site, and further utilization of the plant residues. Hydrometallurgy 2013, 131–132, 46–53. [Google Scholar] [CrossRef]
- Verma, V.K.; Singh, Y.P.; Rai, J.P.N. Biogas production from plant biomass used for phytoremediation of industrial wastes. Bioresour. Technol. 2007, 98, 1664–1669. [Google Scholar] [CrossRef] [PubMed]
- Lao, J.; Chen, X.; Qi, M.; Ji, B. Soil Agrochemical Analysis Manual; Lao, J., Ed.; Agriculture Press: Beijing, China, 1988. [Google Scholar]
- Wei, Y.; Van Houten, R.T.; Borger, A.R.; Eikelboom, D.H.; Fan, Y. Minimization of excess sludge production for biological wastewater treatment. Water Res. 2003, 37, 4453–4467. [Google Scholar] [CrossRef]
- Su, Y. Biogas Fermentation Detection Technology; Metallurgical Industry Press: Beijing, China, 2011. [Google Scholar]
- Allen, G.C.; Flores-Vergara, M.A.; Krasynanski, S.; Kumar, S.; Thompson, W.F. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 2006, 1, 2320–2325. [Google Scholar] [CrossRef]
- Zhang, X.; Tian, X.; Ma, L.; Feng, B.; Liu, Q.; Yuan, L.; Fan, C.; Huang, H.; Huang, H.; Yang, Q. Biodiversity of the symbiotic bacteria associated with toxic marine dinoflagellate Alexandrium tamarense. J. Biosci. Med. 2015, 3, 23–28. [Google Scholar] [CrossRef]
- Bitton, G. Wastewater Microbiology, 3rd ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2005. [Google Scholar]
- Rajagopal, R.; Massé, D.I.; Singh, G. A critical review on inhibition of anaerobic digestion process by excess ammonia. Bioresour. Technol. 2013, 143, 632–641. [Google Scholar] [CrossRef]
- Liu, T.; Sung, S. Ammonia inhibition on thermophilic aceticlastic methanogens. Water Sci. Technol. 2002, 45, 113–120. [Google Scholar] [CrossRef] [Green Version]
- Math-Alvarez, J.; Mtz.-Viturtia, A.; Llabrés-Luengo, P.; Cecchi, F. Kinetic and performance study of a batch two-phase anaerobic digestion of fruit and vegetable wastes. Biomass Bioenergy 1993, 5, 481–488. [Google Scholar] [CrossRef]
- Tian, H.; Karachalios, P.; Angelidaki, I.; Fotidis, I.A. A proposed mechanism for the ammonia-LCFA synergetic co-inhibition effect on anaerobic digestion process. Chem. Eng. J. 2018, 349, 574–580. [Google Scholar] [CrossRef]
- Tian, Y.; Yang, K.; Li, S.; Zhang, H.; Han, X.; Hao, H.; Chai, Y.; Xu, X. How does copper stress influence the relationships among the anaerobic co-fermentation parameters? J. Renew. Sustain. Energy 2018, 10, 053107. [Google Scholar] [CrossRef]
- Tabatabaei, M.; Sulaiman, A.; Nikbakht, A.M. Influential parameters on biomethane generation in anaerobic wastewater treatment plants. In Alternative Fuel; Manzanera, M., Ed.; InTech: London, UK, 2011; pp. 227–262. ISBN 978-953-30-372-9. [Google Scholar]
- Taherzadeh, M.J.; Karimi, K. Pretreatment of lignocellulosic wastes to improve ethanol and biogas production: A review. Int. J. Mol. Sci. 2008, 9, 1621–1651. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Chang, Z.; Ye, X.; Xu, R.; Du, J.; Chen, G. Methane production from wheat straw with anaerobic sludge by heme supplementation. Bioresour. Technol. 2014, 172, 91–96. [Google Scholar] [CrossRef]
- Miao, H.; Lu, M.; Zhao, M.; Huang, Z.; Ren, H.; Yan, Q.; Ruan, W. Enhancement of Taihu blue algae anaerobic digestion efficiency by natural storage. Bioresour. Technol. 2013, 149, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Harper, S.R.; Pohland, F.G. Recent developments in hydrogen management during anaerobic biological wastewater treatment. Biotechnol. Bioeng. 1986, 28, 585–602. [Google Scholar] [CrossRef]
- Nettmann, E.; Bergmann, I.; Pramschu, S.; Mundt, K.; Plogsties, V.; Herrmann, C.; Klocke, M. Polyphasic analyses of methanogenic archaeal communities in agricultural biogas plants. Appl. Environ. Microbiol. 2010, 76, 2540–2548. [Google Scholar] [CrossRef]
- Tahuer, R.K. Biochemistry of methanogenesis: A tribute to Marjory Stephenson. Microbiology 1998, 144, 2377–2406. [Google Scholar] [CrossRef]
- Omar, B.; Abou-shanab, R.; El-gammal, M.; Fotidis, I.A.; Kougias, P.G.; Zhang, Y.; Angelidaki, I. Simultaneous biogas upgrading and biochemicals production using anaerobic bacterial mixed cultures. Water Res. 2018, 142, 86–95. [Google Scholar] [CrossRef]
- Pramanik, P.; Kim, P.J. Effect of limited nickel availability on methane emission from EDTA treated soils: Coenzyme M an alternative biomarker for methanogens. Chemosphere 2013, 90, 873–876. [Google Scholar] [CrossRef] [PubMed]
- Lira-Silva, E.; Santiago-Martínez, M.G.; García-Contreras, R.; Zepeda-Rodríguez, A.; Marín-Hernández, A.; Moreno-Sánchez, R.; Jasso-Chávez, R. Cd2+ resistance mechanisms in Methanosarcina acetivorans involve the increase in the coenzyme M content and induction of biofilm synthesis. Environ. Microbiol. Rep. 2013, 5, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.; Shen, L.; Zou, Y.; Qi, Z.; Yin, J.; Jiang, J.; Guo, L.; He, L.; Chen, Z.; Tang, Z.; et al. Comparative genome analysis of Prevotella intermedia strain isolated from infected root canal reveals features related to pathogenicity and adaptation. BMC Genom. 2015, 16, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Petersen, L.M.; Bautista, E.J.; Nguyen, H.; Hanson, B.M.; Chen, L.; Lek, S.H.; Sodergren, E.; Weinstock, G.M. Community characteristics of the gut microbiomes of competitive cyclists. Microbiome 2017, 5, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Su, X.-L.; Shi, X.-S.; Guo, R.-B.; Qiu, Y.-L.; Tian, Q.; Yuan, X.-Z.; Zhang, J. Acetobacteroides hydrogenigenes gen. nov., sp. nov., an anaerobic hydrogen-producing bacterium in the family Rikenellaceae isolated from a reed swamp. Int. J. Syst. Evol. Microbiol. 2014, 64, 2986–2991. [Google Scholar] [CrossRef] [PubMed]
- Fosses, A.; Maté, M.; Franche, N.; Liu, N.; Denis, Y.; Borne, R.; De Philip, P.; Fierobe, H.P.; Perret, S. A seven-gene cluster in Ruminiclostridium cellulolyticum is essential for signalization, uptake and catabolism of the degradation products of cellulose hydrolysis. Biotechnol. Biofuels 2017, 10, 1–14. [Google Scholar] [CrossRef]
- Liu, T.; Ahn, H.; Sun, W.; McGuinness, L.R.; Kerkhof, L.J.; Häggblom, M.M. Identification of a Ruminococcaceae species as the Methyl tert-Butyl Ether (MTBE) degrading bacterium in a Methanogenic consortium. Environ. Sci. Technol. 2016, 50, 1455–1464. [Google Scholar] [CrossRef]
- Weiss, G.A.; Hennet, T. The role of milk sialyllactose in Sialyllactose in Intestinal Bacterial Colonization. Adv. Nutr. 2012, 3, 483–488. [Google Scholar] [CrossRef]
- Slobodkin, A. Tepidimicrobium. In Bergey’s Manual of Systematics of Archaea and Bacteria; Whitman, W.B., Ed.; Wiley: Hoboken, NJ, USA, 2017; pp. 1–6. [Google Scholar]
- Xie, J.; Fan, J.; Wang, Z.; He, T. The stress concentration effect around the hole in flat plate. Adv. Mater. Res. 2012, 538–541, 1573–1578. [Google Scholar] [CrossRef]
- Cibis, K.G.; Wibberg, D.; Maus, I.; Schlüter, A.; Pühler, A.; Winkler, A.; König, H.; Stolze, Y. Complete genome sequence of the strain Defluviitoga tunisiensis L3, isolated from a thermophilic, production-scale biogas plant. J. Biotechnol. 2015, 203, 17–18. [Google Scholar]
- Matulova, M.; Delort, A.M.; Nouaille, R.; Gaudet, G.; Forano, E. Concurrent maltodextrin and cellodextrin synthesis by Fibrobacter succinogenes S85 as identified by 2D NMR spectroscopy. Eur. J. Biochem. 2001, 268, 3907–3915. [Google Scholar] [CrossRef] [PubMed]
- Leahy, S.C.; Kelly, W.J.; Ronimus, R.S.; Wedlock, N.; Altermann, E.; Attwood, G.T. Genome sequencing of rumen bacteria and archaea and its application to methane mitigation strategies. Animal 2013, 7, 235–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schnürer, A.; Müller, B.; Dicksved, J.; Gonda, H.; Danielsson, R.; Bertilsson, J.; Sun, L. Methane production in dairy cows correlates with rumen methanogenic and bacterial community structure. Front. Microbiol. 2017, 8, 1–15. [Google Scholar]
- Pfister, P.; Nolling, J.; Wasserfallen, A.; Conway de Macario, E.; Reeve, J. Phylogenetic analysis of 18 thermophilic Methanobacterium isolates supports the proposals to create a new genus, Methanothermobacter gen. nov., and to reclassify several isolates in three species, Methanothermobacter thermautotrophicus comb. nov., Methano. Int. J. Syst. Evol. Microbiol. 2011, 50, 43–53. [Google Scholar]
- Hammel, K.E.; Cornwell, K.L.; Diekert, G.B.; Thauer, R.K. Evidence for a nickel-containing carbon monoxide dehydrogenase in Methanobrevibacter arboriphilicus. J. Bacteriol. 1984, 157, 975–978. [Google Scholar]



| Feedstocks | Metals | Reactor Type and Volume | Temp (°C) | HRT (d) | Biogas Yield | CH4 Yield | References |
|---|---|---|---|---|---|---|---|
| Phragmites straw + Cow dung | Cu | Pilot (30 L) | 37 ± 1 | 33 | 110.59 mL/gTS | Maximum 86.72% | [8] |
| Phytolacca americana L. | Cu | Batch (0.7 L) | 37 | 50 | 0.12 L/gVS | 52% | [30] |
| Zea mays L. | Cu | Batch (0.7 L) | 37 | 50 | 0.45 L/gVS | ~70% | [30] |
| Brassica napus L. | Cu | Batch (0.7 L) | 37 | 50 | 0.40 L/gVS | ~70% | [30] |
| Elsholtzia splendens | Cu | Batch (0.7 L) | 37 | 50 | 0.27 L/gVS | 56% | [30] |
| Oenothera biennis L. | Cu | Batch (0.7 L) | 37 | 50 | 0.39 L/gVS | 56% | [30] |
| Phragmites straw + Cow dung | Fe | Batch (0.25 L) | 35 ± 1 | 26 | 26.20–32.46 mL/gTS | Maximum 67.9% | [7] |
| Synthetic waste | Fe | UASB (9 L) | 35 | 9 | 0.362 L/g COD removed | 80.5–93.0% | [20] |
| Lemnaceae + Poultry manure | Fe | Batch (1 L) | 32 ± 2 | 50–80 | 0.281 L/g VS | 65–80% | [31] |
| Lemnaceae + Poultry manure | Fe | Semi-cont. (25 L) | 32 ± 2 | 8.3–16.6 | 22.76 L/d | NR | [31] |
| Synthetic model substrate for maize silage | Ni | Batch (1 L) | 35 | 30 | NR | 188–404 LN/kg organic dry matter | [32] |
| Model substrate for maize | Ni | Semi-cont. (5 L) | 35 | ~60 | 0.6–8.0 LN/d | 36–55% | [33] |
| Phragmites straw + Cow dung | Ni | Batch (0.25 L) | 35 ± 1 | 26 | 27.49–32.70 mL/gTS | Maximum 70.41% | [9] |
| Azolla pinnata R.Br | Fe, Cu, Cd, Ni, Pb, Zn, Mn and Co | Batch (2 L) | 37 | 36–42 | 132–189 L/kg | 45–83% | [10] |
| Lemna minor L. | Fe, Cu, Cd, Ni, Pb, Zn, Mn and Co | Batch (2 L) | 37 | 36–42 | 132–176 L/kg | 43–85% | [10] |
| Seaweed | Cd, Cu, Ni, Zn | Batch (0.5 L) | 37 | 30 | NR | 0.09–0.12 LN CH4/g VSa (44.4–49.7%) | [34] |
| Seaweed | Cd, Cu, Ni, Zn | UASB (0.8 L) | 37 ± 1 | 8.8–0.5 | 0.22–3.04 LN CH4/L·d | 0.16–0.23 LN CH4/g CODa (62.9–73.7%) | [34] |
| Triticale | Al, Ni, Zn, Co, U, La | Stirring reactor (5 L) | 38 ± 1 | NR | ~780 LN/kgVS | 440 LN/kgVS | [35] |
| Brassica juncea | Al, Ni, Zn, Co, U, La | Stirring reactor (5 L) | 38 ± 1 | NR | ~640 LN/kgVS | 425 LN/kgVS | [35] |
| Helianthus annuus | Al, Ni, Zn, Co, U, La | Stirring reactor (5 L) | 38 ± 1 | NR | ~360 LN/kgVS | 163 LN/kgVS | [35] |
| Eichhornia crassipes | Cu- and Cr-rich brass and electroplating industry effluent | Batch (NR) | 35 ± 1 | 20 | 11.10–27.80 L/kg dw | 29.80–63.82% | [36] |
| Trapa bispinnosa | Cu- and Cr-rich brass and electroplating industry effluent | Batch (>0.15 L) | 35 ± 1 | 20 | 10.45–20.90 L/kg dw | 27.00–57.04% | [36] |
| Characteristics | Corn Stover | Fresh Cow Dung |
|---|---|---|
| TS (%dry weight) | 95.59 ± 0.23 | 16.49 ± 0.16 |
| VS (% TS) | 90.72 ± 0.24 | 84.00 ± 0.48 |
| TN (% TS) | 1.21 ± 0.03 | 3.22 ± 0.11 |
| TOC (% TS) | 13.94 ± 0.64 | 14.81 ± 0.37 |
| Ratio of C/N | 11.52 ± 0.05 | 4.45 ± 0.30 |
| Cellulose (% TS) | 20.19 ± 1.24 | 23.56 ± 1.47 |
| Hemicellulose (% TS) | 14.05 ± 2.25 | 16.41 ± 0.48 |
| Lignin (% TS) | 13.55 ± 0.07 | 15.41 ± 1.11 |
| Cu (μg/g) | 8.57 ± 0.20 | 38.63 ± 0.30 |
| Ni (μg/g) | 1.71 ± 0.37 | 1.78 ± 0.10 |
| Cd (μg/g) | Negligible | Negligible |
| Zn (μg/g) | 14.89 ± 1.61 | 152.44 ± 2.06 |
| Fe (μg/g) | 520.80 ± 67.03 | 610.80 ± 12.87 |
| Co (μg/g) | 0.34 ± 0.24 | 0.61 ± 0.03 |
| Cr (μg/g) | 8.33 ± 1.13 | 3.01 ± 0.63 |
| Metals | Lignin (% TS) | Hemicellulose (% TS) | Cellulose (% TS) | Total Lignocellulose (% TS) |
|---|---|---|---|---|
| Cu | 19.63 ± 0.85 | 13.23 ± 0.75 | 19.34 ± 1.46 | 52.21 ± 3.06 |
| Cu + Fe | 16.92 ± 0.90 | 11.44 ± 0.61 | 16.46 ± 0.83 | 44.83 ± 2.34 ** |
| Cu + Ni | 19.95 ± 1.15 | 12.05 ± 0.69 | 20.21 ± 0.74 | 52.21 ± 2.58 |
| Cu + Zn | 14.56 ± 1.03 ** | 12.34 ± 0.61 | 14.43 ± 1.18 ** | 41.34 ± 2.82 ** |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, Y.; Zhang, H.; Zheng, L.; Li, S.; Hao, H.; Yin, M.; Cao, Y.; Huang, H. Process Analysis of Anaerobic Fermentation Exposure to Metal Mixtures. Int. J. Environ. Res. Public Health 2019, 16, 2458. https://doi.org/10.3390/ijerph16142458
Tian Y, Zhang H, Zheng L, Li S, Hao H, Yin M, Cao Y, Huang H. Process Analysis of Anaerobic Fermentation Exposure to Metal Mixtures. International Journal of Environmental Research and Public Health. 2019; 16(14):2458. https://doi.org/10.3390/ijerph16142458
Chicago/Turabian StyleTian, Yonglan, Huayong Zhang, Lei Zheng, Shusen Li, He Hao, Meixiao Yin, Yudong Cao, and Hai Huang. 2019. "Process Analysis of Anaerobic Fermentation Exposure to Metal Mixtures" International Journal of Environmental Research and Public Health 16, no. 14: 2458. https://doi.org/10.3390/ijerph16142458





