Prevalence and Characterization of Integrons in Multidrug Resistant Acinetobacter baumannii in Eastern China: A Multiple-Hospital Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. A. baumannii Isolates
| Province | City | Hospital Code | Number of A. baumannii Strains Isolated |
|---|---|---|---|
| Fujian | Fuzhou | A | 254 |
| B | 2 | ||
| C | 20 | ||
| D | 15 | ||
| Xiamen | E | 25 | |
| F | 1 | ||
| G | 3 | ||
| Quanzhou | H | 14 | |
| Longyan | I | 5 | |
| Nanping | J | 56 | |
| Jiangsu | Nanjing | K | 3 |
| Zhejiang | Haining | L | 3 |
| Hangzhou | M | 17 | |
| Taizhou | N | 2 | |
| Wenzhou | O | 2 | |
| Shandong | Yantai | P | 3 |
| Total | 425 | ||
2.2. Antimicrobial Susceptibility Test
2.3. Characterization of Integrons and Their Gene Cassettes
| Integrase Gene | Primer Sequences | PCR Product Size (bp) | Reference |
|---|---|---|---|
| intl1 | F: 5’-CAG TGG ACA TAA GCC TGT TC-3’; | 160 | 20 |
| R: 5’-CCC GAG GCA TAG ACT GTA-3’ | |||
| intl2 | F: 5’-TTG CGA GTA TCC ATA ACC TG-3’; | 288 | 20 |
| R: 5’-TTA CCT GCA CTG GAT TAA GC-3’ | |||
| intl3 | F: 5’-GCCTCCGGCAGCGACTTTCAG-3’; | 1041 | 20 |
| R: 5’-ACGGATCTGCCAAACCTGACT-3’ | |||
| intl CS | F: 5’-GGC ATC CAA GCA GCA AG-3’; | Unknown | 20 |
| R: 5’-AAGCAG ACT TGA CCT GA-3’ |
2.4. Strain Typing of Integron-Positive A. baumannii Isolates
2.5. Ethical Statement
2.6. Statistical Analysis
3. Results
3.1. Prevalence of intI1, intI2 and intI3 Genes in A. baumannii
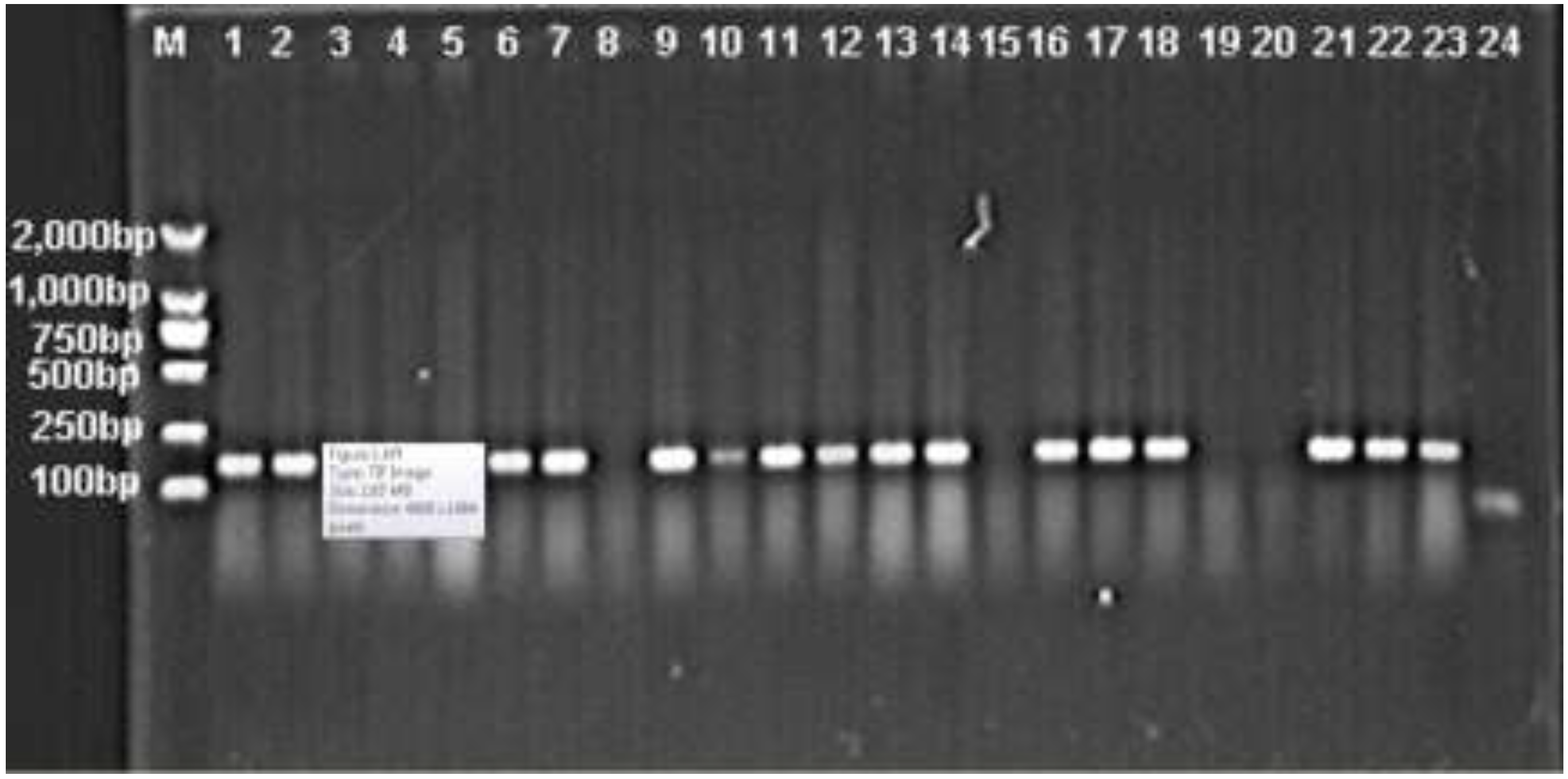
3.2. Comparison of the Prevalence of Antimicrobial Resistance between Integron-Positive and -Negative A. baumannii Isolates
| Antimicrobial | Integron-Positive A. baumannii Isolates (n = 296) | Integron-Negative A. baumannii Isolates (n = 129) | p Value |
|---|---|---|---|
| Ampicillin/sulbactam | 89.9% | 62.4% | <0.05 |
| Piperacillin/tazobactam | 92.7% | 65.2% | <0.05 |
| Ceftazidime | 98.5% | 75.4% | <0.05 |
| Ceftriaxone | 100% | 98% | <0.05 |
| Cefepime | 94.3% | 74.6% | <0.05 |
| Aztreonam | 99.6% | 93.1% | <0.05 |
| Imipenem | 86.6% | 65.8% | <0.05 |
| Meropenem | 89.9% | 57.6% | <0.05 |
| Amikacin | 87.8% | 42.7% | <0.05 |
| Gentamicin | 98.1% | 71.7% | <0.05 |
| Tobramycin | 93.3% | 62.7% | <0.05 |
| Ciprofloxacin | 98.7% | 77.7% | <0.05 |
| Levofloxacin | 86.2% | 55.4% | <0.05 |
| Sulfamethoxazole/trimenthoprim | 98.3% | 85.3% | <0.05 |
| Minocycline | 39.3% | 28.8% | 0.119 |
3.3. PFGE Analysis of intI1 Gene-Positive A. baumannii Isolates

3.4. Distribution of intI1 Gene-Positive A. baumannii Isolates
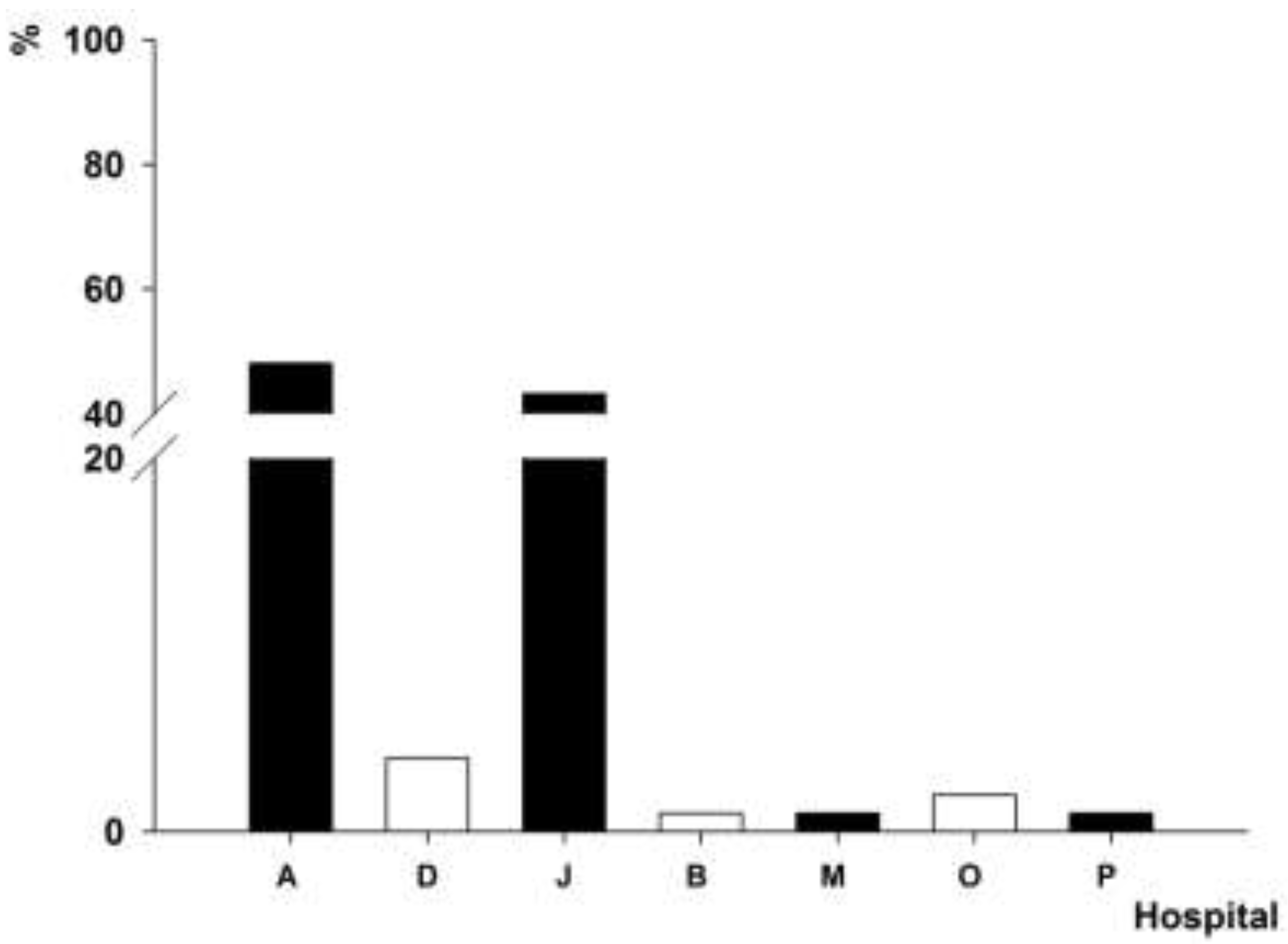


3.5. Antimicrobial Resistance Pattern of intI1 Gene-Positive A. baumannii Isolates with PFGE Genotypes P1 and P4
3.6. Sequencing of int I Gene Cassette
4. Discussion
5. Conclusions
Acknowledgements
Author Contributions
Conflicts of Interest
References
- Zhang, H.Z.; Zhang, J.S.; Qiao, L. The Acinetobacter baumannii group: A systemic review. World J. Emerg. Med. 2013, 4, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Howard, A.; O’Donoghue, M.; Feeney, A.; Sleator, R.D. Acinetobacter baumannii: An emerging opportunistic pathogen. Virulence 2012, 3, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Peleg, A.Y.; Seifert, H.; Paterson, D.L. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 2008, 21, 538–582. [Google Scholar] [CrossRef] [PubMed]
- Antunes, L.C.; Visca, P.; Towner, K.J. Acinetobacter baumannii: Evolution of a global pathogen. Pathog. Dis. 2014, 71, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Durante-Mangoni, E.; Zarrilli, R. Global spread of drug-resistant Acinetobacter baumannii: Molecular epidemiology and management of antimicrobial resistance. Future Microbiol. 2011, 6, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Giamarellou, H.; Antoniadou, A.; Kanellakopoulou, K. Acinetobacter baumannii: A universal threat to public health? Int. J. Antimicrob. Agents 2008, 32, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Gootz, T.D.; Marra, A. Acinetobacter baumannii: An emerging multidrug-resistant threat. Expert. Rev. Anti Infect. Ther. 2008, 6, 309–325. [Google Scholar] [CrossRef] [PubMed]
- Chang-Tai, Z.; Yang, L.; Zhong-Yi, H.; Chang-Song, Z.; Yin-Ze, K.; Yong-Ping, L.; Chun-Lei, D. High frequency of integrons related to drug-resistance in clinical isolates of Acinetobacter baumannii. Indian J. Med. Microbiol. 2011, 29, 118–123. [Google Scholar] [PubMed]
- Lee, Y.T.; Huang, L.Y.; Chen, T.L.; Siu, L.K.; Fung, C.P.; Cho, W.L.; Yu, K.W.; Liu, C.Y. Gene cassette arrays, antibiotic susceptibilities, and clinical characteristics of Acinetobacter baumannii bacteremic strains harboring class 1 integrons. J. Microbiol. Immunol. Infect. 2009, 42, 210–219. [Google Scholar] [PubMed]
- Ruiz, J.; Navia, M.M.; Casals, C.; Sierra, J.M.; Jiménez De Anta, M.T.; Vila, J. Integron-mediated antibiotic multiresistance in Acinetobacter baumannii clinical isolates from Spain. Clin. Microbiol. Infect. 2003, 9, 907–911. [Google Scholar] [CrossRef] [PubMed]
- Turton, J.F.; Kaufmann, M.E.; Glover, J.; Coelho, J.M.; Warner, M.; Pike, R.; Pitt, T.L. Detection and typing of integrons in epidemic strains of Acinetobacter baumannii found in the United Kingdom. J. Clin. Microbiol. 2005, 43, 3074–3082. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.L.; Ma, L.; Chang, J.C.; Su, L.H.; Chu, C.; Leu, H.S.; Siu, L.K. Variable resistance patterns of integron-associated multidrug-resistant Acinetobacter baumannii isolates in a surgical intensive care unit. Microb. Drug. Resist. 2004, 10, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.Y.; Kim, K.S.; Jeong, Y.W.; Cho, J.W.; Park, J.C.; Lee, J.C. Epidemiological typing and prevalence of integrons in multiresistant Acinetobacter strains. Acta. Pathol. Microbiol. Immunol. Scand. 2002, 110, 247–252. [Google Scholar] [CrossRef]
- Labbate, M.; Case, R.J.; Stokes, H.W. The integron/gene cassette system: An active player in bacterial adaptation. Methods Mol. Biol. 2009, 532, 103–125. [Google Scholar] [PubMed]
- Partridge, S.R.; Tsafnat, G.; Coiera, E.; Iredell, J.R. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS. Microbiol. Rev. 2009, 33, 757–784. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.M.; Collis, C.M. Mobile gene cassettes and integrons: Capture and spread of genes by site-specific recombination. Mol. Microbiol. 1995, 15, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.L.; Siu, L.K.; Wu, R.C.; Shaio, M.F.; Huang, L.Y.; Fung, C.P.; Lee, C.M.; Cho, W.L. Comparison of one-tube multiplex PCR, automated ribotyping and intergenic spacer (ITS) sequencing for rapid identification of Acinetobacter baumannii. Clin. Microbiol. Infect. 2007, 13, 801–806. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.X.; Rawte, P.; Brown, S.; Lo, S.; Siebert, H.; Pong-Porter, S.; Low, D.E.; Jamieson, F.B. Evaluation of CLSI agar dilution method and Trek Sensititre broth microdilution panel for determining antimicrobial susceptibility of Streptococcus pneumoniae. J. Clin. Microbiol. 2011, 49, 704–706. [Google Scholar] [CrossRef] [PubMed]
- Kristo, I.; Pitiriga, V.; Poulou, A.; Zarkotou, O.; Kimouli, M.; Pournaras, S.; Tsakris, A. Susceptibility patterns to extended-spectrum cephalosporins among Enterobacteriaceae harbouring extended-spectrum β-lactamases using the updated Clinical and Laboratory Standards Institute interpretive criteria. Int. J. Antimicrob. Agents. 2013, 41, 383–387. [Google Scholar] [CrossRef] [PubMed]
- Khorsi, K.; Messai, Y.; Hamidi, M.; Ammari, H.; Bakour, R. High prevalence of multidrug-resistance in Acinetobacter baumannii and dissemination of carbapenemase-encoding genes blaOXA-23-like, blaOXA-24-like and blaNDM-1 in Algiers hospitals. Asian. Pac. J. Trop. Med. 2015, 8, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.F.; Peng, C.F.; Hsu, H.J.; Toh, H.S. Use of inverse PCR for analysis of class 1 integrons carrying an unusual 3’ conserved segment structure. Antimicrob. Agents Chemother. 2011, 55, 943–945. [Google Scholar] [CrossRef] [PubMed]
- Çıçek, A.Ç.; Düzgün, A.Ö.; Saral, A.; Kayman, T.; Çızmecı, Z.; Balcı, P.Ö.; Dal, T.; Fırat, M.; Tosun, İ.; Alıtntop, Y.A.; et al. Detection of class 1 integron in Acinetobacter baumannii isolates collected from nine hospitals in Turkey. Asian. Pac. J. Trop. Biomed. 2013, 3, 743–747. [Google Scholar] [CrossRef]
- Koeleman, J.G.; Stoof, J.; Van Der Bijl, M.W.; Vandenbroucke-Grauls, C.M.; Savelkoul, P.H. Identification of epidemic strains of Acinetobacter baumannii by integrase gene PCR. J. Clin. Microbiol. 2001, 39, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Tenover, F.C.; Arbeit, R.D.; Goering, R.V.; Mickelsen, P.A.; Murray, B.E.; Persing, D.H.; Swaminathan, B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing. J. Clin. Microbiol. 1995, 33, 2233–2239. [Google Scholar] [PubMed]
- Huovinen, P. Antibiotic usage and the incidence of resistance. Clin. Microbiol. Infect. 1999, 5, 4S12–4S16. [Google Scholar] [CrossRef]
- Tenover, F.C. Mechanisms of antimicrobial resistance in bacteria. Am. J. Infect. Control. 2006, 34, S3–S10. [Google Scholar] [CrossRef] [PubMed]
- Abbo, A.; Navon-Venezia, S.; Hammer-Muntz, O.; Krichali, T.; Siegman-Igra, Y.; Carmeli, Y. Multidrug-resistant Acinetobacter baumannii. Emerg. Infect. Dis. 2005, 11, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Huddleston, J.R. Horizontal gene transfer in the human gastrointestinal tract: Potential spread of antibiotic resistance genes. Infect. Drug. Resist. 2014, 7, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Leverstein-van Hall, M.A.; Box, A.T.; Blok, H.E.; Paauw, A.; Fluit, A.C.; Verhoef, J. Evidence of extensive interspecies transfer of integron-mediated antimicrobial resistance genes among multidrug-resistant Enterobacteriaceae in a clinical setting. J. Infect. Dis. 2002, 186, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Krauland, M.G.; Marsh, J.W.; Paterson, D.L.; Harrison, L.H. Integron-mediated multidrug resistance in a global collection of nontyphoidal Salmonella enterica isolates. Emerg. Infect. Dis. 2009, 15, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.M.; Stokes, H.W. Integrons: Novel DNA elements which capture genes by site-specific recombination. Genetica. 1993, 90, 115–132. [Google Scholar] [CrossRef] [PubMed]
- Fluit, A.C.; Schmitz, F.J. Resistance integrons and super-integrons. Clin. Microbiol. Infect. 2004, 10, 272–288. [Google Scholar] [CrossRef] [PubMed]
- Taherikalani, M.; Maleki, A.; Sadeghifard, N.; Mohammadzadeh, D.; Soroush, S.; Asadollahi, P.; Asadollahi, K.; Emaneini, M. Dissemination of class 1, 2 and 3 integrons among different multidrug resistant isolates of Acinetobacter baumannii in Tehran hospitals, Iran. Pol. J. Microbiol. 2011, 60, 169–174. [Google Scholar] [PubMed]
- Gu, B.; Tong, M.; Zhao, W.; Liu, G.; Ning, M.; Pan, S.; Zhao, W. Prevalence and characterization of class I integrons among Pseudomonas aeruginosa and Acinetobacter baumannii isolates from patients in Nanjing, China. J. Clin. Microbiol. 2007, 45, 241–243. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.M.; Li, Z.D.; Pan, Y.P.; Wei, H. Association of integrons and inserted gene with antibiotics resistance of 100 strains of Acinetobacter baumannii isolates. Chin. J. Microecol. 2013, 25, 261–263. (in Chinese). [Google Scholar]
- Fouad, M.; Attia, A.S.; Tawakkol, W.M.; Hashem, A.M. Emergence of carbapenem-resistant Acinetobacter baumannii harboring the OXA-23 carbapenemase in intensive care units of Egyptian hospitals. Int. J. Infect. Dis. 2013, 17, e1252–e1254. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.F.; Liou, M.L.; Tu, C.C.; Yeh, H.W.; Lan, C.Y. Molecular epidemiology of integron-associated antimicrobial gene cassettes in the clinical isolates of Acinetobacter baumannii from northern Taiwan. Ann. Lab. Med. 2013, 33, 242–247. [Google Scholar] [CrossRef] [PubMed]
- Gombac, F.; Riccio, M.L.; Rossolini, G.M.; Lagatolla, C.; Tonin, E.; Monti-Bragadin, C.; Lavenia, A.; Dolzani, L. Molecular characterization of integrons in epidemiologically unrelated clinical isolates of Acinetobacter baumannii from Italian hospitals reveals a limited diversity of gene cassette arrays. Antimicrob. Agents Chemother. 2002, 46, 3665–3668. [Google Scholar] [CrossRef] [PubMed]
- Nemec, A.; Dolzani, L.; Brisse, S.; van den Broek, P.; Dijkshoorn, L. Diversity of aminoglycoside-resistance genes and their association with class 1 integrons among strains of pan-European Acinetobacter baumannii clones. J. Med. Microbiol. 2004, 53, 1233–1240. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.B.; Ou, J.M. Study on the integrons in Acinetobacter and the relationship with antibiotic resistance. Lab. Med. Clin. 2010, 7, 816–819. (in Chinese). [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Li, H.; Yang, J.; Zhan, R.; Chen, A.; Yan, Y. Prevalence and Characterization of Integrons in Multidrug Resistant Acinetobacter baumannii in Eastern China: A Multiple-Hospital Study. Int. J. Environ. Res. Public Health 2015, 12, 10093-10105. https://doi.org/10.3390/ijerph120810093
Chen J, Li H, Yang J, Zhan R, Chen A, Yan Y. Prevalence and Characterization of Integrons in Multidrug Resistant Acinetobacter baumannii in Eastern China: A Multiple-Hospital Study. International Journal of Environmental Research and Public Health. 2015; 12(8):10093-10105. https://doi.org/10.3390/ijerph120810093
Chicago/Turabian StyleChen, Jing, Hong Li, Jinsong Yang, Rong Zhan, Aiping Chen, and Yansheng Yan. 2015. "Prevalence and Characterization of Integrons in Multidrug Resistant Acinetobacter baumannii in Eastern China: A Multiple-Hospital Study" International Journal of Environmental Research and Public Health 12, no. 8: 10093-10105. https://doi.org/10.3390/ijerph120810093





