Detection of Emerging and Re-Emerging Pathogens in Surface Waters Close to an Urban Area
Abstract
:1. Introduction
2. Experimental Section

2.1. Sample Collection
2.2. Isolation of Bacteria
2.2.1. Staphylococcus spp.
2.2.2. Salmonella spp.
2.2.3. Campylobacter spp.
2.2.4. Clostridium spp.
2.3. Virological Analysis
2.4. Nucleic Acid Extraction
2.5. One Step Quantitative PCR of Viruses (q-PCR)
| Viruses | Primer/Probe | Sequences (5′–3′) |
|---|---|---|
| Hepatitis A Virus [33] | F | HAV P3 (F): TCA CCG CCG TTT GCC TAG-5' |
| R | HAV P4 (R): GGA GAG CCC TGG AAG AAA G | |
| P | HAV P5 (-) (P): CCT GAA CCT GCA GGA ATT AA. FAM-3'BHQ1 | |
| Hepatitis E Virus [34] | F | HEV P3(F): GGT GGT TTC TGG GGT GAC AGG GT |
| R | HEV P4 (R): AGG GGT TGG TTG GAT GAA | |
| P | HEV P5 (P): TGA TTC TCA GCC CTT CGC. MGB-6-FAM | |
| Human Enteroviruses [35] | F | PanE P3(F): GGC CCC TGA ATG CGG CTA ATCC |
| R | PanE P4(R): GCG ATT GTC ACC ATW AGC AGY CA | |
| P | PanE P5 (P): CCG ACT ACT TTG GGW GTC CGT GT5. FAM-3'BHQ1 | |
| Human Norovirus GI [36] | F | Noro GI P3 (F): CGC TGG ATG CGN TTC CAT |
| R | Noro GI P4 (R): CCT TAG ACG CCA TCA TCA TTT AC | |
| P | Noro GI P5 (P): TGG ACA GGA GAY CGC RAT CT. TEXAS RED-BBQ 650 | |
| Human Norovirus GII [36] | F | Noro GII P3 (F): ATG TTC AGR TGG ATG AGR TTC TCW GA |
| R | Noro GII P4 (R): TCG ACG CCA TCT TCA TTC ACA | |
| P | Noro GII P5 (P): AGC ACG TGG GAG GGC GAT CG. HEX-BBQ 650 | |
| Human Adenovirus 41 | F | ADV41P3(F): GTACTTCAGCCTGGGGAACA |
| R | ADV41 P4 (R): GGTCGACTGGCACGAATC | |
| P | ADV41 P5 (P): AGACAGGTCACAGCGACTGA. FAM-BHQ1 |
| Viruses | Cq | A | E | R2 |
|---|---|---|---|---|
| HAV | = −3.3 log10·(q) + 40.81 | 2.01 | 100.92% | 0.997 |
| HEV | = −3.59 log10·(q) + 41.1 | 1.9 | 89.91% | 0.9932 |
| HE | = −3.34 log10·(q) + 38.44 | 1.991 | 99.25%; | 0.9926 |
| NoGGI | = −3.49 log10·(q) + 47.68 | 1.933 | 93.43% | 0.9928 |
| NoGGII | = −3.39 log10·(q) + 47 | 1.971 | 97.24% | 0.9979 |
| ADV41 | = −3.33 log10·(q) + 41.29 | 2.0 | 99.66%; | 0.9975 |
2.6. Microarray Design and Description
2.7. RNA Extraction, Labeling and Fragmentation
2.8. Hybridization and Washing
2.9. Microarray Scanning
2.10. Data Analysis
3. Results
3.1. Microbiological Results

3.2. Virological Results
| Viruses | Castel Giubileo Viral Copies/L | Mezzocammino Viral Copies/L |
|---|---|---|
| HAV | neg | neg |
| NoGGI | neg | 103 |
| NoGGII | neg | 103 |
| HE | neg | neg |
| HEV | neg | 102 |
| ADV41 | 102 | 105 |
3.3. Microarrays




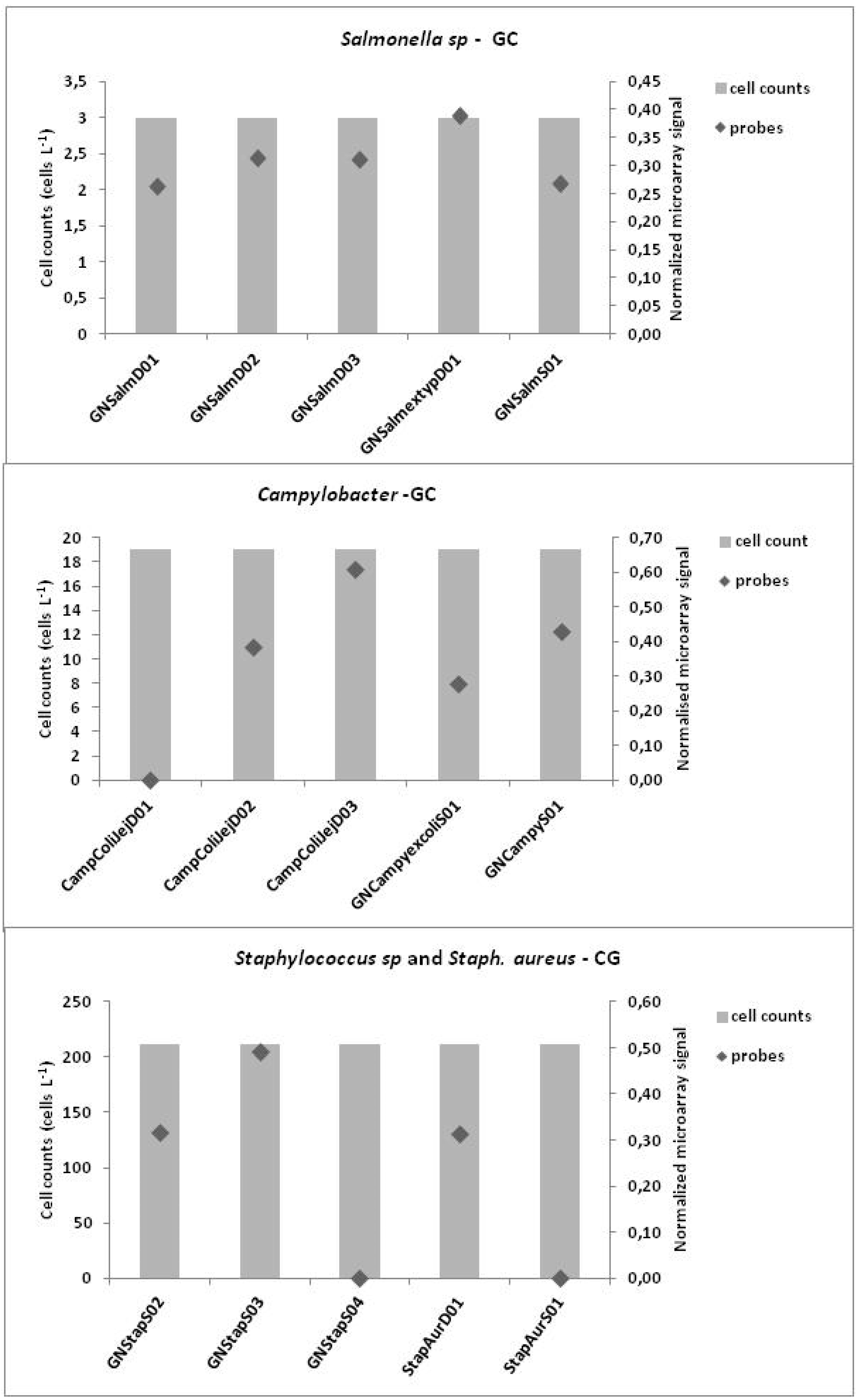

4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Brunkard, J.M.; Ailes, E.; Roberts, V.A.; Hill, V.; Hilborn, E.D.; Craun, G.F.; Rajasingham, A.; Kahler, A.; Garrison, L.; Hicks, L.; et al. Surveillance for waterborne disease outbreaks associated with drinking water—United States, 2007–2008. MMWR Surveill. Summ. 2011, 60, 38–68. [Google Scholar] [PubMed]
- Dwight, R.H.; Baker, D.B.; Semenza, J.C.; Olson, B.H. Health effects associated with recreational coastal water use: Urban versus rural California. Am. J. Public Health 2004, 94, 565–567. [Google Scholar] [CrossRef] [PubMed]
- Semenza, J.C.; Caplan, J.S.; Buescher, G.; Das, T.; Brinks, M.V.; Gershunov, A. Climate change and microbiological water quality at California beaches. Ecohealth 2012, 9, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Marcheggiani, S.; Carere, M.; Della Bella, V.; Grifoni, R.; Funari, E.; Mancini, L. Epidemie veicolate dall’acqua. In Cambiamenti Climatici e Salute: Criticità e Proposte Progettuali per una Strategia D’adattamento Ambientale; Sinisi, L., Ed.; ISPRA: Rome, Italy, 2010; pp. 25–35. (In Italian) [Google Scholar]
- Semenza, J.C. Climate change and human health. Int. J. Environ. Res. Public Health 2014, 11, 7347–7353. [Google Scholar] [CrossRef] [PubMed]
- Funari, E.; Blasi, M.F.; Carere, M.; Della Bella, V.; Mancini, L.; Marcheggiani, S.; Mattera, F.; Stefanelli, M. Flooding and health. In Environment and Health Risks from Climate Change and Variability in Italy; Wolf, T., Menne, B., Eds.; IGER Istituto Grafico Editoriale Romano: Rome, Italy, 2007; pp. 44–45. [Google Scholar]
- United Nations. The Human Right to Water and Sanitation. Resolution Adopted by the General Assembly on 28 July 2010. Available online: http://contrattoacqua.it/public/upload/1/2/tab_elms_docs/1404320102n0947935.pdf (accessed on 25 February 2015).
- World Health Organization. Microbial Fact Sheet. In Guidelines for Drinking-Water Quality; WHO Library Cataloguing-in-Publication Data: Geneva, Switzerland, 2011; pp. 231–306. Available online: http://www.who.int/water_sanitation_health/publications/2011/9789241548151_ch11.pdf (accessed on 25 February 2015).
- World Health Organization. Emerging Issues in Water and Infectious Disease; WHO Library Cataloguing-in-Publication Data: Geneva, Switzerland, 2003; Available online: http://www.who.int/water_sanitation_health/emerging/emerging.pdf (accessed on 25 February 2015).
- European Centre for Disease Prevention and Control. Annual Epidemiological Report 2014—Food and Waterborne Diseases and Zoonoses; ECDC: Stockholm, Sweden, 2014; Available online: http://ecdc.europa.eu/en/publications/Publications/food-waterborne-diseases-annual-epidemiological-report-2014.pdf (accessed on 29 April 2015).
- Copeland, C.; Beers, B.; Thompson, M.; Pinkerton, R.; Barrett, L.; Sevilleja, J.E.; Alencar, S.; Lima, A.; Guerrant, R. Faecal contamination of drinking water in a Brazilian shanty town: Importance of household storage and new human faecal marker testing. J. Water Health 2009, 7, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Birkbecka, T.H.; RingØ, E. Pathogenesis and the gastrointestinal tract of growing fish. In Microbial Ecology of Growing Animals; Holzapfel, W.H., Naughton, P.J., Eds.; Elsevier: London, UK, 2005; pp. 208–234. [Google Scholar]
- USGS. Science for a Changing World. The Effects of Urbanization on Water Quality: Waterborne Pathogens. Available online: http://water.usgs.gov/edu/urbanpath.html (accessed on 25 February 2015).
- International Joint Commission. The Impact of Urban Development on Water Quality. Available online: http://www.ijc.org/php/publications/html/12br/english/report/physical/iudwq.html (accessed on 25 February 2015).
- Sherchand, J. Future emerging issues in waterborne diseases and microbial agents. J. Inst. Med. 2012, 34, 1–3. [Google Scholar]
- Kot, M.; Castleden, H.; Gagnon, G.A. The human dimension of water safety plans: A critical review of literature and information gaps. Environ. Rev. 2015, 23, 24–29. [Google Scholar] [CrossRef]
- European Union. Directive No. 83 of 3 November 1998 on the Quality of Water Intended for Human Consumption. Official Journal 330/32. Available online: http://dwi.defra.gov.uk/stakeholders/legislation/eudir98_83_EC.pdf (accessed on 19 May 2015).
- European Union. Directive No. 7 of 15 February 2006 Concerning the Management of Bathing Water Quality and Repealing Directive 76/160/EEC. Official Journal L 64/37. Available online: http://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX:32006L0007 (accessed on 19 May 2015).
- Marcheggiani, S.; Mancini, L. Microbiological quality of river sediments and primary prevention. In Ecosystems Biodiversity; Intech Open: Rijeka, Croatia, 2011; pp. 117–138. [Google Scholar]
- Tanchou, V. Review of Methods for the Rapid Identification of Pathogens in Water Samples—ERNCIP Thematic A = πr2 Chemical & Biological Risks in the Water Sector—Task 7, Deliverable 1. Institute for the Protection and Security of the Citizen. Publications Office of the European Union. 2014. Available online: http://publications.jrc.ec.europa.eu/repository/bitstream/JRC92395/lbna26881enn.pdf (accessed on 28 April 2015).
- Mitchell, R.; Dong-Gu, J. Environmental Microbiology, 2nd ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2010. [Google Scholar]
- Cecchi, G.; Fabiani, C.; Mancini, L.; Munafò, M. Pressures of human activity on water status. In Tevere Pilot River Basin Article 5 Report; Gangemi Editore: Rome, Italy, 2005; pp. 79–87. [Google Scholar]
- Meyer, V.K.; Kober, C.; Niessner, R.; Seidel, M. Regeneration of recombinant antigen microarrays for the automated monitoring of antibodies against zoonotic pathogens in Swine Sera. Sensors 2015, 15, 2614–2628. [Google Scholar] [CrossRef] [PubMed]
- Kegel, J.U.; del Amo, Y.; Costes, L.; Medlin, L.K. Testing a microarray to detect and monitor toxic microalgae in Arcachon Bay in France. Microarrays 2013, 2, 1–23. [Google Scholar] [CrossRef]
- Berzano, M.; Marcheggiani, S.; Rombini, S.; Spurio, R. The application of oligonucleotide probes and microarrays for the identification of freshwater diatoms. Hydrobiologia 2012, 695, 52–72. [Google Scholar] [CrossRef]
- Andreani, P.; Bernabei, S.; Cecchi, G.; Charavgis, F.; D’Angelo, A.M.; Formichetti, P.; Mancini, L.; Martinelli, A.; Notargiacomo, T.; Pace, G.; et al. Perennial water courses types. In Tevere Pilot River Basin Article 5 Report; Gangemi Editore: Rome, Italy, 2005; pp. 40–54. [Google Scholar]
- American Public Health Association. Standard Methods for the Examination of Water and Wastewater, 21st ed.; APHA: Washington, DC, USA, 2008. [Google Scholar]
- United States Environmental Protection Agency EPA. Method 1600: Membrane Filter Test Method for Enterococci in Water; Office of Water, United States Environmental Protection Agency: Washington, DC, USA, 2002.
- Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for the Detection of Salmonella spp.; International Organization for Standardization: Geneva, Switzerland, 2007.
- Popoff, M.Y.; le Minor, L. Formules Antigeniques des Serovars de Salmonella, 8th ed.; WHO Collaborating Centre for Reference and Research on Salmonella: Paris, France, 2001. [Google Scholar]
- Zhou, P.M.; Hussain, S.K.; Liles, M.R.; Arias, C.R. A simplified and cost-effective enrichment protocol for the isolation of Campylobacter spp. from retail broiler meat without microaerobic incubation. BMC Microbiol. 2011, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcheggiani, S.; Iaconelli, M.; D’angelo, A.M.; Pierdominici, E.; la Rosa, G.; Muscillo, M.; Equestre, M.; Mancini, L. Microbiological and 16S rRNA analysis of sulphite-reducing clostridia from river sediments in central Italy. BMC Microbiol. 2008, 8. [Google Scholar] [CrossRef] [PubMed]
- Costafreda, M.I.; Bosch, A.; Pintó, R.M. Development, evaluation, and standardization of a real-time TaqMan reverse transcription-PCR assay for quantification of hepatitis A virus in clinical and shellfish samples. appl. environ. microbiol. 2006, 72, 3846–3855. [Google Scholar] [CrossRef] [PubMed]
- Jothikumara, N.; Cromeansa, T.L.; Robertson, B.H.; Mengd, X.J.; Vincent, R. A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J. Virol. Methods 2006, 131, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Nnamdigadi Chigor, V.; Ifeanyi Okoh, A. Quantitative RT-PCR detection of hepatitis A virus, rotaviruses and enteroviruses in the Buffalo River and source water dams in the eastern cape province of South Africa. Int. J. Environ. Res. Public Health 2012, 9, 4017–4032. [Google Scholar] [CrossRef] [PubMed]
- Loisya, F.; Atmarb, R.L.; Guillona, P.; le Canna, P.; Pommepuya, M.; le Guyader, F.S. Real time RT-PCR for norovirus screening in shellfish. J. Virol. Methods 2005, 123, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.; Medlin, L.K.; Raine, R. MIDTAL (Microarrays for the Detection of Toxic Algae): A Protocol for a Successful Microarray Hybridisation and Analysis; Lewis, J., Medlin, L.K., Raine, R., Eds.; Koeltz: Koenigstein, Germany, 2012; p. 16. [Google Scholar]
- Gescher, G.; Metfies, K.; Medlin, L.K. The ALEX Chip—Development of a DNA chip for identification and monitoring of Alexandrium. Harmful Algae 2008, 7, 485–494. [Google Scholar] [CrossRef]
- Kegel, J.U.; Amo, Y.D.; Medlin, L.K. Introduction to project MIDTAL: Its methods and samples from Arcachon Bay, France. Environ. Sci. Pollut. Res. 2012, 20, 6690–6704. [Google Scholar] [CrossRef]
- Dittami, S.M.; Edvardsen, B. GPR-Analyzer: A simple tool for quantitative analysis of hierarchical multispecies microarrays. Environ. Sci. Pollut. Res. Int. 2013, 20, 6808–6815. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, C.; Montacutelli, R.; del Vecchio, R.; Solimini, A.G.; Marinelli, L.; Lombardi, A.M.; de Giusti, M.; MariaFara, G.; Boccia, A. Microbiological quality of surface waters of Rome and it’s County from 1890 to 2010: A systematic review of Roman Hygiene School. Epidemiol. Biostat. Public Health 2014, 11, 1–10. [Google Scholar]
- Oberste, M.S.; Peñaranda, S.; Rogers, S.L.; Henderson, E.; Nix, W.A. Comparative evaluation of Taqman real-time PCR and semi-nested VP1 PCR for detection of enteroviruses in clinical specimens. J. Clin. Virol. 2010, 49, 73–74. [Google Scholar] [CrossRef] [PubMed]
- Van Dam, H.; Mertens, A.; Sinkeldam, J. A coded checklist and ecological indicator values of freshwater diatoms from the Netherlands. Neth. J. Aquat. Ecol. 1994, 28, 117–133. [Google Scholar] [CrossRef]
- Coste, M. Étude des Méthodes Biologiques D’appreciation Quantitative de la Qualité des Eaux; Rapport, Q.E., Lyon, A.F., Eds.; Bassin Rhône- Mediterranée Corse, CEMAGREF: Lyon, France, 1982; p. 218. (In France) [Google Scholar]
- Rott, E.; Pfister, P.; van Dam, H.; Pipp, E.; Pall, K.; Binder, N.; Ortler, K. Indikationslisten für Aufwuchsalgen in Österreichischen Fliessgewässern, Teil 2: Trophieindikation und autökologische Anmerkungen Bundesministerium für Land- und Forstwirtschaf; Wasserwirtschaftskataster: Wien, Austria, 1999. (In German) [Google Scholar]
- Rapport, D.J.; Whittford, W.G. How ecosystems respond to stress. BioScience 1999, 49, 193–203. [Google Scholar] [CrossRef]
- Tancioni, L.; Caprioli, R.; Dawood, H.; Al-Khafaji, A.; Mancini, L.; Boglione, C.; Ciccotti, E.; Cataudella, S. Gonadal disorder in the Thinlip grey mullet (Liza Ramada, Risso 1827) as a biomarker of environmental stress in surface waters. Int. J. Environ. Res. Public Health 2015, 12, 1817–1833. [Google Scholar] [CrossRef] [PubMed]
- Davies, G.I.; McIver, L.; Kim, Y.; Hashizume, M.; Iddings, S.; Chan, V. Water-borne diseases and extreme weather events in Cambodia: Review of impacts and implications of climate change. Int. J. Environ. Res. Public Health 2015, 12, 191–213. [Google Scholar] [CrossRef]
- CEC—Council of European Communities. Directive 2000/60/EEC of 23 October 2000 Establishing a Framework for Community Action in the Field of Water Policy. Available online: http://faolex.fao.org/docs/pdf/eur23005.pdf (accessed on 19 May 2015).
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marcheggiani, S.; D'Ugo, E.; Puccinelli, C.; Giuseppetti, R.; D'Angelo, A.M.; Gualerzi, C.O.; Spurio, R.; Medlin, L.K.; Guillebault, D.; Weigel, W.; et al. Detection of Emerging and Re-Emerging Pathogens in Surface Waters Close to an Urban Area. Int. J. Environ. Res. Public Health 2015, 12, 5505-5527. https://doi.org/10.3390/ijerph120505505
Marcheggiani S, D'Ugo E, Puccinelli C, Giuseppetti R, D'Angelo AM, Gualerzi CO, Spurio R, Medlin LK, Guillebault D, Weigel W, et al. Detection of Emerging and Re-Emerging Pathogens in Surface Waters Close to an Urban Area. International Journal of Environmental Research and Public Health. 2015; 12(5):5505-5527. https://doi.org/10.3390/ijerph120505505
Chicago/Turabian StyleMarcheggiani, Stefania, Emilo D'Ugo, Camilla Puccinelli, Roberto Giuseppetti, Anna Maria D'Angelo, Claudio Orlando Gualerzi, Roberto Spurio, Linda K. Medlin, Delphine Guillebault, Wilfried Weigel, and et al. 2015. "Detection of Emerging and Re-Emerging Pathogens in Surface Waters Close to an Urban Area" International Journal of Environmental Research and Public Health 12, no. 5: 5505-5527. https://doi.org/10.3390/ijerph120505505
APA StyleMarcheggiani, S., D'Ugo, E., Puccinelli, C., Giuseppetti, R., D'Angelo, A. M., Gualerzi, C. O., Spurio, R., Medlin, L. K., Guillebault, D., Weigel, W., Helmi, K., & Mancini, L. (2015). Detection of Emerging and Re-Emerging Pathogens in Surface Waters Close to an Urban Area. International Journal of Environmental Research and Public Health, 12(5), 5505-5527. https://doi.org/10.3390/ijerph120505505







