Bacterial Community Diversity of Oil-Contaminated Soils Assessed by High Throughput Sequencing of 16S rRNA Genes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site and Sample Treatment
2.2. DNA Extraction and PCR Amplification of Bacterial 16S rRNA Gene
2.3. High Throughput Sequencing of Bacterial 16S rRNA Gene
2.4. Statistical and Bioinformatics Analysis
3. Results
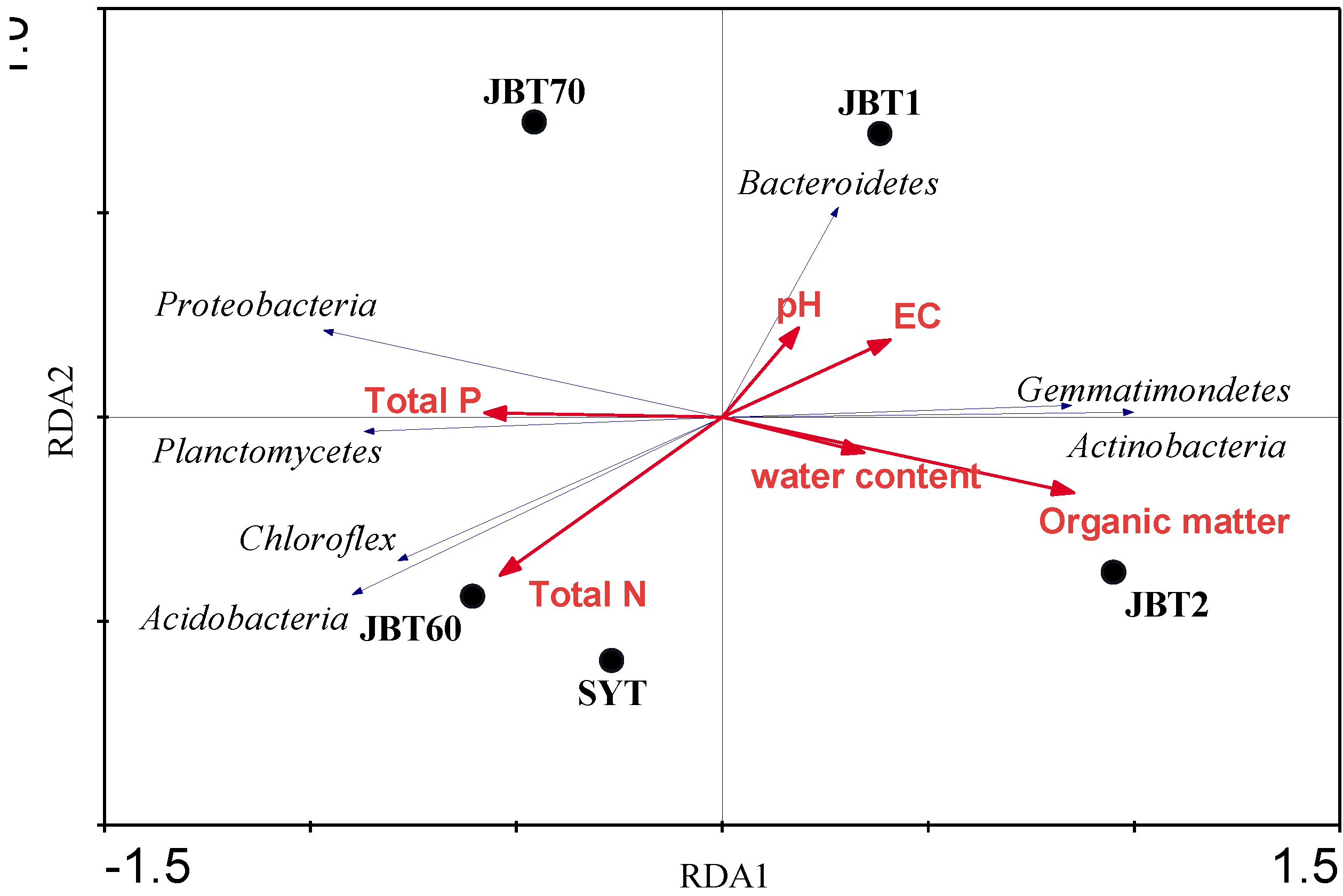
3.1. Physicochemical Properties of Soil
3.2. Microbial Richness and Diversity
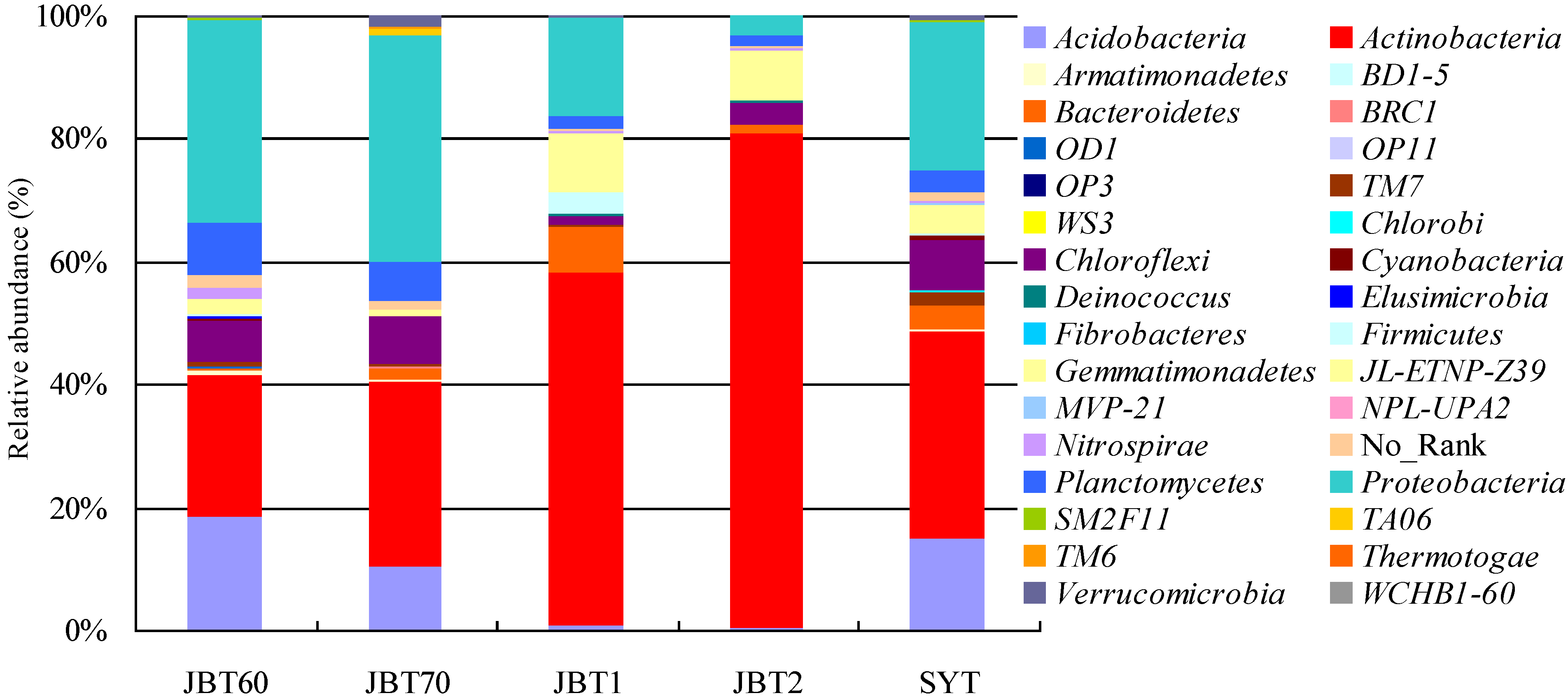
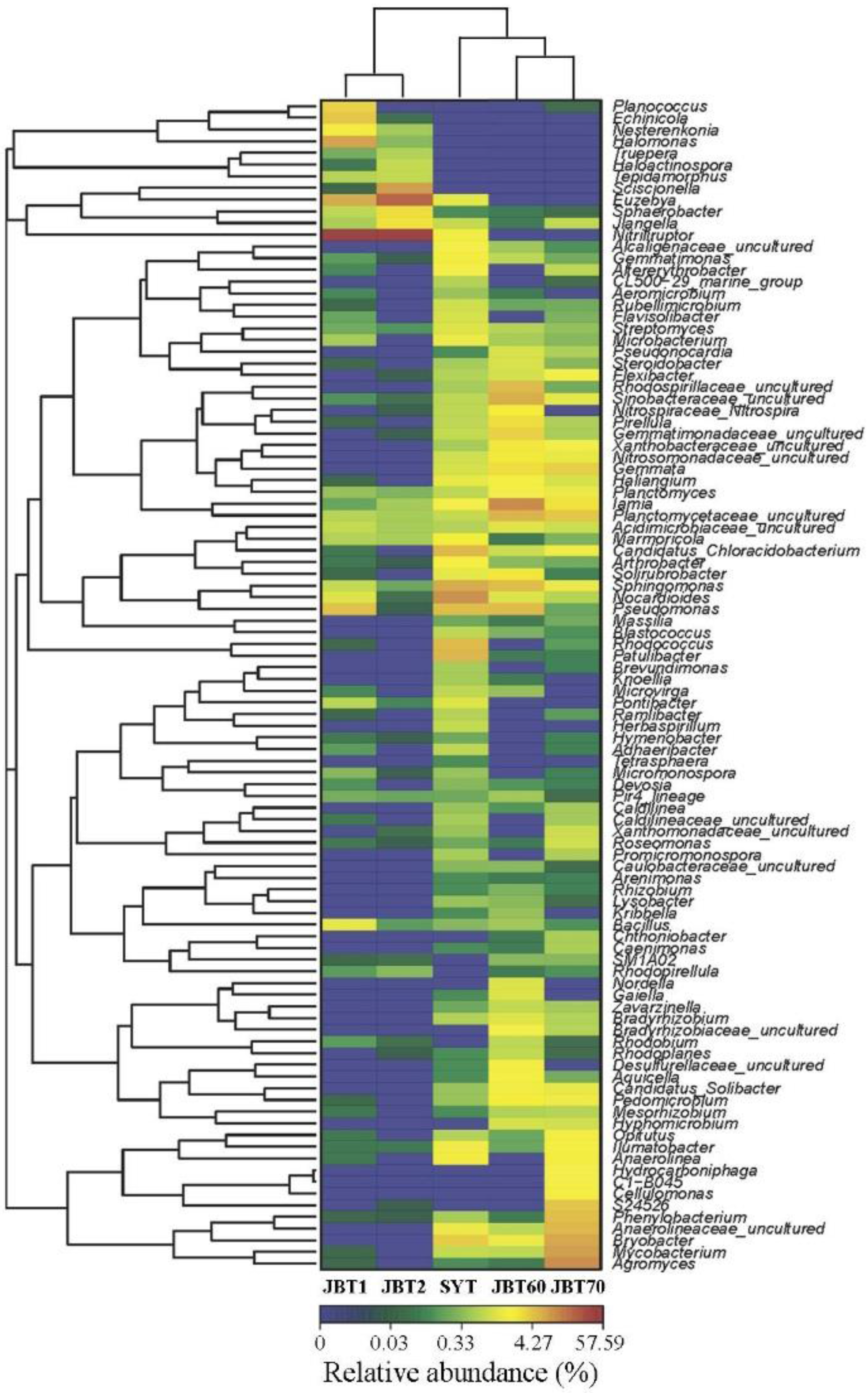
3.3. Taxonomic Composition
| Parameters | JBT60 | JBT70 | JBT1 | SYT | JBT2 |
|---|---|---|---|---|---|
| Soil characteristics | The oil contaminated soil from abandoned well mined in 1960s | The oil contaminated soil from abandoned well mined in 1970 | The Control closest to JBT60/70 | The oil contaminated soil from recent (one year) oil spilled site | The Control closest to SYT |
| pH | 7.36 ± 0.11 dD | 8.05 ± 0.04 bB | 9.40 ± 0.03 aA | 7.90 ± 0.05 cC | 9.35 ± 0.01 aA |
| Soil water content (%) | 7.37 ± 0.39 cC | 5.65 ± 0.30 dD | 14.17 ± 0.41 aA | 9.09 ± 0.29 bB | 13.52 ± 0.46 aA |
| EC (uS/cm) | 75.60 ± 3.06 eE | 192.30 ± 2.10 dD | 1716.43 ± 16.82 aA | 578.36 ± 6.15 cC | 1225.17 ± 10.41 bB |
| Total N (mg/g) | 1.30 ± 0.02 aA | 0.55 ± 0.03 cC | 0.70 ± 0.09 bB | 0.55 ± 0.01 cC | 0.70 ± 0.04 bB |
| Total P (mg/g) | 0.22 ± 0.01 aA | 0.32 ± 0.01 bB | 0.19 ± 0.01 bB | 0.21 ± 0.02 bB | 0.31 ± 0.05 aA |
| Organic content (%) | 1.86 ± 0.31 dC | 6.01 ± 0.31 bB | 5.24 ± 0.13 cB | 18.69 ± 0.69 aA | 5.34 ± 0.01 cB |
| Sample ID | Valid Reads | Cluster Distance (0.03) | |||||
|---|---|---|---|---|---|---|---|
| OTU | ACE | Chao1 | Coverage | Shannon | Simpson | ||
| JBT60 | 4097 | 1917 | 8338 | 4832 | 0.68 | 6.98 | 0.002 |
| JBT70 | 5777 | 1320 | 3582 | 2602 | 0.88 | 5.92 | 0.012 |
| JBT1 | 4760 | 852 | 2437 | 1663 | 0.90 | 5.11 | 0.026 |
| SYT | 2132 | 1161 | 4217 | 2838 | 0.63 | 6.70 | 0.0017 |
| JBT2 | 8187 | 996 | 2437 | 1799 | 0.94 | 5.37 | 0.012 |
| Total | 24953 | 6246 | -- | -- | -- | -- | -- |
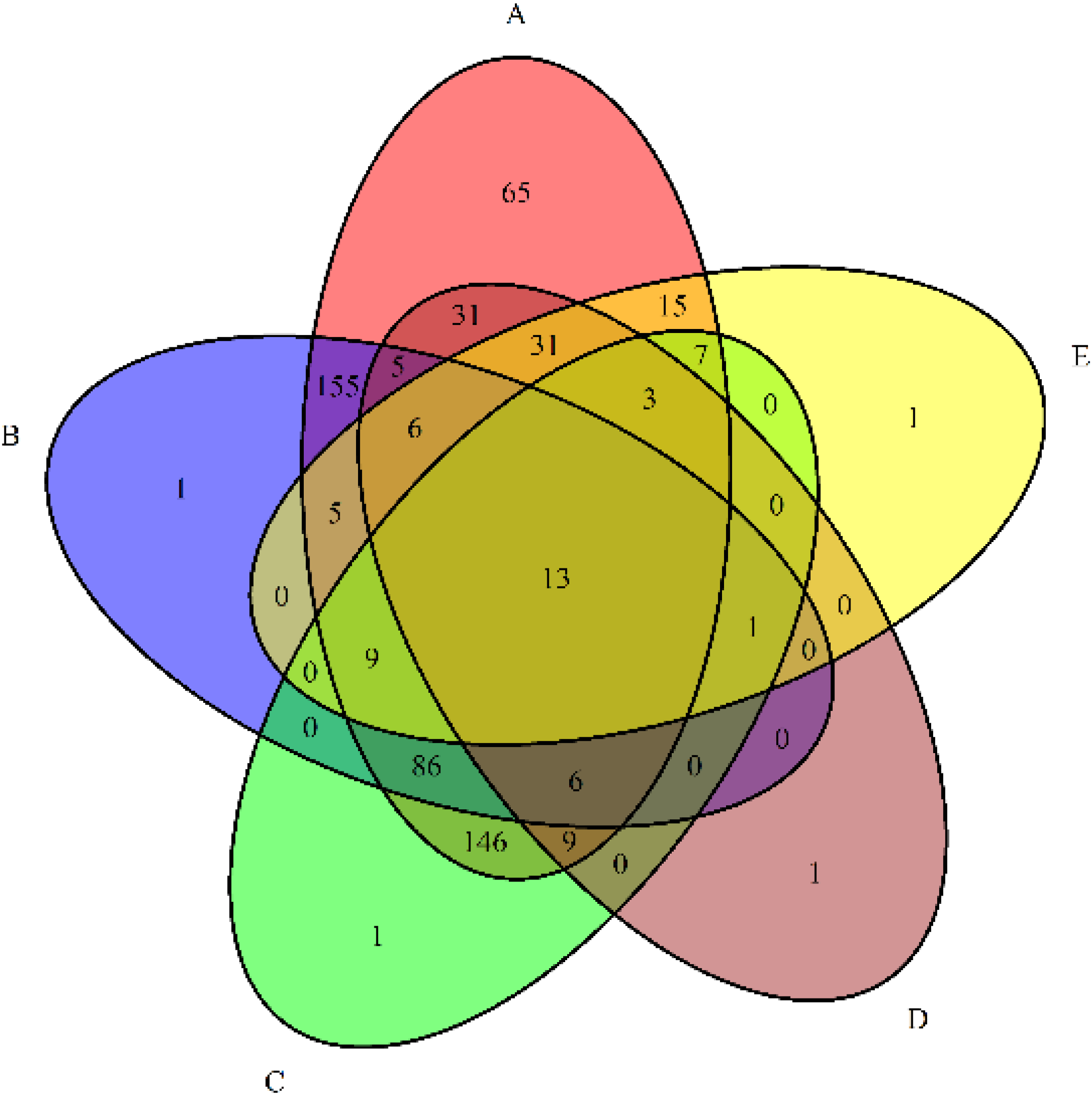
3.4. Relationships between Bacterial Communities among the Five Samples

4. Discussions
Supplementary Files
Supplementary File 1Acknowledgements
Author Contributions
Conflicts of Interest
References
- Zhang, Z.; Gai, L.; Hou, Z.; Yang, C.; Ma, C.; Wang, Z.; Sun, B.; He, X.; Tang, H.; Xu, P. Characterization and biotechnological potential of petroleum-degrading bacteria isolated from oil-contaminated soils. Bioresour. Technol. 2010, 101, 8452–8456. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Haruki, M.; Imanaka, T.; Morikawa, M.; Kanaya, S. Isolation and characterization of long-chain-alkane degrading Bacillus thermoleovorans from deep subterranean petroleum reservoirs. J. Biosci. Bioeng. 2001, 91, 64–70. [Google Scholar] [CrossRef]
- Atlas, R.M. Microbial degradation of petroleum hydrocarbons: An environmental perspective. Microbiol. Rev. 1981, 45, 180–209. [Google Scholar] [PubMed]
- Tamames, J.; Abellán, J.J.; Pignatelli, M.; Camacho, A.; Moya, A. Environmental distribution of prokaryotic taxa. BMC Microbiol. 2010, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- David, L.; Luc de, V. Microbial Species Diversity, Community Dynamics, and Metabolite Kinetics of Water Kefir Fermentation. Appl. Environ. Microbiol. 2014, 80, 2564–2572. [Google Scholar]
- Julia, G.; Lukas, Y.W.; Hauke, H.; Antonis, C. Evaluating T-RFLP protocols to sensitively analyze the genetic diversity and community changes of soil alkane degrading bacteria. Eur. J. Soil Biol. 2014, 65, 107–113. [Google Scholar]
- Thorsten, K.; José, L.S.; Sávia, G.; Lourdinha, F. Analysis of microbial community structure and composition in leachates from a young landfill by 454 pyrosequencing. Appl. Microbiol. Biotechnol. 2015, 99, 5657–5668. [Google Scholar]
- Trivedi, P.; He, Z.; van Nostrand, J.D.; Albrigo, G.; Zhou, J.; Wang, N. Huanglongbing alters the structure and functional diversity of microbial communities associated with citrus rhizosphere. ISME J. 2012, 6, 363–383. [Google Scholar] [CrossRef] [PubMed]
- Theron, J.; Cloete, T.E. Molecular techniques for determining microbial diversity and community structure in natural environments. Crit. Rev. Microbiol. 2000, 26, 37–57. [Google Scholar] [PubMed]
- Ye, L.; Zhang, T. Bacterial communities in different sections of a municipal wastewater treatment plant revealed by 16S rDNA 454 pyrosequencing. Appl. Microbiol. Biotechnol. 2013, 97, 2681–2690. [Google Scholar] [PubMed]
- Claesson, M.; O’Sullivan, O.; Wang, Q.; Nikkila, J.; Marchesi, J.; Smidt, H.; de Vos, W.; Ross, R.; O’Toole, P. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS ONE 2009, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bibby, K.; Viau, E.; Peccia, J. Pyrosequencing of the 16S rRNA gene to reveal bacterial pathogen diversity in biosolids. Water Res. 2010, 44, 4252–4260. [Google Scholar] [CrossRef] [PubMed]
- Kirchman, D.L.; Cottrell, M.T.; Lovejoy, C. The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environ. Microbiol. 2010, 12, 1132–1143. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, H.F.; Cury, J.C.; do Carmo, F.L.; dos Santos, A.L.; Tiedje, J.; van Elsas, J.D.; Soares Rosado, A.; Peixoto, R.S. Mangrove bacterial diversity and the impact of oil contamination revealed by pyrosequencing: Bacterial proxies for oil pollution. PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Wang, L.; Zhang, S. Investigating landscape pattern and its dynamics in Daqing, China. Int. J. Remote Sens. 2005, 26, 2259–2280. [Google Scholar] [CrossRef]
- Blakemore, L.C.; Searle, P.L.; Daly, B.K. Methods for chemical analysis of soils. New Zealand Soil Bureau Sci. Rep. 1987, 80. [Google Scholar]
- Marín, M.; Garcia-Lechuz, J.M.; Alonso, P.; Villanueva, M.; Alcala, L.; Gimeno, M.; Cercenado, E.; Sánchez-Somolinos, M.; Radice, C.; Bouza, E. The role of universal 16S rRNA gene PCR and sequencing in the diagnosis of prosthetic joint infection. J. Clin. Microbiol. 2011. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [PubMed]
- Krishna, P.; Babu, A.G.; Reddy, M.S. Bacterial diversity of extremely alkaline bauxite residue site of alumina industrial plant using culturable bacteria and residue 16S rRNA gene clones. Extremophiles 2014, 18, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Gentleman, R.; Ihaka, R. The R Language. J. East Chin. Geol. Institute 1996, 15, 693–704. [Google Scholar]
- Wu, S.; Wang, G.; Angert, E.R.; Wang, W.; Li, W.; Zou, H. Composition, diversity, and origin of the bacterial community in grass carp intestine. PLoS One 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Schreiter, S.; Ding, G.-C.; Grosch, R.; Kropf, S.; Antweiler, K.; Smalla, K. Soil type dependent effects of a potential biocontrol inoculant on indigenous bacterial communities in the rhizosphere of field-grown lettuce. FEMS Microbiol. Ecol. 2014, 90, 718–730. [Google Scholar] [CrossRef] [PubMed]
- Abed, R.M.; Al-Kindi, S.; Al-Kharusi, S. Diversity of Bacterial Communities along a Petroleum Contamination Gradient in Desert Soils. Microb. Ecol. 2014, 69, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Sutton, N.B.; Maphosa, F.; Morillo, J.A.; Al-Soud, W.A.; Langenhoff, A.A.; Grotenhuis, T.; Rijnaarts, H.H.M.; Smidt, H. Impact of long-term diesel contamination on soil microbial community structure. Appl. Environ. Microbiol. 2013, 79, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Evans, F.F.; Seldin, L.; Sebastian, G.V.; Kjelleberg, S.; Holmström, C.; Rosado, A.S. Influence of petroleum contamination and biostimulation treatment on the diversity of Pseudomonas spp. in soil microcosms as evaluated by 16S rRNA based—PCR and DGGE. Lett. Appl. Microbiol. 2004, 38, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Ruberto, L.; Vazquez, S.C.; Mac Cormack, W.P. Effectiveness of the natural bacterial flora, biostimulation and bioaugmentation on the bioremediation of a hydrocarbon contaminated Antarctic soil. Int. Biodeter. Biodegradation 2003, 52, 115–125. [Google Scholar] [CrossRef]
- Zhang, D.; Mörtelmaier, C.; Margesin, R. Characterization of the bacterial archaeal diversity in hydrocarbon-contaminated soil. Sci. Total Environ. 2012, 421–422, 184–196. [Google Scholar] [CrossRef] [PubMed]
- Chan, H. Biodegradation of petroleum oil achieved by bacteria and nematodes in contaminated water. Sep. Purif. Technol. 2011, 80, 459–466. [Google Scholar] [CrossRef]
- Baek, K.H.; Kim, H.S.; Oh, H.M.; Yoon, B.D.; Kim, J.; Lee, I.S. Effects of crude oil, oil components, and bioremediation on plant growth. J. Environ. Sci. Health 2004, 39, 2465–2472. [Google Scholar] [CrossRef]
- Cho, O.; Choi, K.Y.; Zylstra, G.J.; Kim, Y.S.; Kim, S.K.; Lee, J.H.; Sohn, Y.; Kwon, G.S.; Kim, Y.M.; Kim, E. Catabolic role of a three-component salicylate oxygenase from Sphingomonas yanoikuyae B1 in polycyclic aromatic hydrocarbon degradation. Biochem. Biophys. Res. Commun. 2005, 327, 656–662. [Google Scholar] [CrossRef] [PubMed]
- Habe, H.; Omori, T. Genetics of polycyclic aromatic hydrocarbon metabolism in diverse aerobic bacteria. Biosci. Biotechnol. Biochem. 2003, 67, 225–243. [Google Scholar] [CrossRef] [PubMed]
- Singer, A.C.; Wong, C.S.; Crowley, D.E. Differential enantioselective transformation of atropisomeric polychlorinated biphenyls by multiple bacterial strains with different inducing compounds. Appl. Environ. Microbiol. 2002, 68, 5756–5759. [Google Scholar] [CrossRef] [PubMed]
- Bell, T.H.; Yergeau, E.; Martineau, C.; Juck, D.; Whyte, L.G.; Greer, C.W. Identification of nitrogen-incorporating bacteria in petroleum-contaminated arctic soils by using [15N] DNA-based stable isotope probing and pyrosequencing. Appl. Environ. Microbiol. 2011, 77, 4163–4171. [Google Scholar] [CrossRef] [PubMed]
- Kindaichi, T.; Yuri, S.; Ozaki, N.; Ohashi, A. Ecophysiological role and function of uncultured Chloroflexi in an anammox reactor. Water Sci. Technol. 2012, 66, 2556–2561. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, D.Y.; van Pelt, S.; Tourova, T.P.; Evtushenko, L.I. Nitriliruptor alkaliphilus gen. nov., sp. nov., a deep-lineage haloalkaliphilic actinobacterium from soda lakes capable of growth on aliphatic nitriles, and proposal of Nitriliruptoraceae fam. nov. and Nitriliruptorales ord. nov. Int. J. Syst. Evol. Microbiol. 2009, 59, 248–253. [Google Scholar] [CrossRef] [PubMed]
- Rousk, J.; Baath, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef] [PubMed]
- Nacke, H.; Thürmer, A.; Wollherr, A.; Will, C.; Hodac, L.; Herold, N.; Schöning, I.; Schrumpf, M.; Daniel, R. Pyrosequencing-Based Assessment of Bacterial Community Structure Along Different Management Types in German Forest and Grassland Soils. PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Ke, L.; Wong, T.W.; Wong, Y.S.; Tam, N.F. Fate of polycyclic aromatic hydrocarbon (PAH) contamination in a mangrove swamp in Hong Kong following an oil spill. Mar. Pollut. Bull. 2002, 45, 339–347. [Google Scholar] [CrossRef]
- Li, C.H.; Zhou, H.W.; Wong, Y.S.; Tam, N.F.Y. Vertical distribution and anaerobic biodegradation of polycyclic aromatic hydrocarbons in mangrove sediments in Hong Kong, South China. Sci. Total Environ. 2009, 407, 5772–5779. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, M.; Zi, X.; Wang, Q. Bacterial Community Diversity of Oil-Contaminated Soils Assessed by High Throughput Sequencing of 16S rRNA Genes. Int. J. Environ. Res. Public Health 2015, 12, 12002-12015. https://doi.org/10.3390/ijerph121012002
Peng M, Zi X, Wang Q. Bacterial Community Diversity of Oil-Contaminated Soils Assessed by High Throughput Sequencing of 16S rRNA Genes. International Journal of Environmental Research and Public Health. 2015; 12(10):12002-12015. https://doi.org/10.3390/ijerph121012002
Chicago/Turabian StylePeng, Mu, Xiaoxue Zi, and Qiuyu Wang. 2015. "Bacterial Community Diversity of Oil-Contaminated Soils Assessed by High Throughput Sequencing of 16S rRNA Genes" International Journal of Environmental Research and Public Health 12, no. 10: 12002-12015. https://doi.org/10.3390/ijerph121012002





