Occurrence of Virulence Genes Associated with Diarrheagenic Escherichia coli Isolated from Raw Cow’s Milk from Two Commercial Dairy Farms in the Eastern Cape Province, South Africa
Abstract
:1. Introduction
2. Experimental Section
2.1. Study Sites
2.2. Isolation and Identification
2.3. DNA Extraction
2.4. Detection of Virulence Genes in E. coli Isolates
| Target Strain | Target Gene | Primer Sequence (5′–3′) | Conditions | Amplicon Size (bp) | References |
|---|---|---|---|---|---|
| E. coli | UidA | AAAACGGCAAGAAAAAGCAG ACGCGTGGTTAACAGTCTTGCG | An initial 2 min of denaturation at 94 °C followed by 25 cycles of 94 °C for 1 min, 58 °C for 1 min and 72 °C for 2 min. Amplified products were held at 4 °C after amplification. | 147 | [26] |
| EHEC | flicH7 | TACCATCGCAAAAGCAAC TCC GTCGGCAACGTTAGTGATACC | Initial denaturation at 95 °C for 15 min followed by 35 cycles of heat denaturation at 94 °C for 45 sec, primer annealing at 55 °C for 45 sec and DNA extension at 68 °C for 2 min. After the last cycle the samples were kept at 72 °C for 5 min to complete synthesis of all strands. | 230 | [27] |
| EIEC | Ial | CTGGATGGTATGGTGAGG GGAGGCCAATTATTTCC | 1 cycle for 2 min at 50 °C, 1 cycle for 5 min at 95 °C, 40 cycles for 45 sec at 95 °C, 45 sec at 55 °C and 45 sec at 72 °C and a final extension step for 10 min at 72 °C to complete synthesis of all strands. | 700 | [28] |
| EAEC | EagR | AGACTCTGGCGAAAGACTGTATC ATGGCTGTCTGTAATAGATGAGAAC | Initial denaturation at 95 °C for 15 min followed by 35 cycles of heat denaturation at 94 °C for 45 sec, primer annealing at 55 °C for 45 sec and DNA extension at 68 °C for 2 min. A final elongation step at 72 °C for 5 min. | 194 | [29] |
| EPEC | EaeA | TCAATGCAGTTCCGTTATCAGT GTAAAGTCCGTTACCCCAACC TG | An initial denaturation step at 94 °C for 5 min, followed by 36 cycles of 94 °C for 35 sec, annealing at 62 °C for 30 sec and elongation at 72 °C for 1 min. A final elongation step at 72 °C for 5 min. | 482 | [28] |
| ETEC | Lt | GCACACGGA GCTCCTCAGTC TCCTTCATCCTTTCAATGGCTT | An initial denaturation step at 94 °C for 5 min, followed by 36 cycles of 94 °C for 35 sec, annealing at 62 °C for 30 sec and elongation at 72 °C for 1 min. A final elongation step at 72 °C for 5 min. | 218 | [28] |
| UPEC | PapC | GACGGCTGTACTGCAGGGTGTGGCG ATATCCTTTCTGCAGGGATGCAATA | An initial denaturation step at 94 °C for 5 min, followed by 36 cycles of 94 °C for 35 sec, annealing at 62 °C for 30 sec and elongation at 72 °C for 1 min. A final elongation step at 72 °C for 5 min. | 328 | [30] |
2.5. Gel Electrophoresis
3. Results and Discussion
3.1. Isolation and Identification
3.1.1. Biochemical Confirmation
| Farms | Number of Samples | G (-ve) Rods | Cat (pos) | Ox (neg) | API 20E | uidA Gene | |
|---|---|---|---|---|---|---|---|
| Clinical Mastitis | Sub-Clinical Mastitis | ||||||
| A | 100 | 100 | 100 | 100 | 100 | 72 | 50 |
| B | 100 | 100 | 88 | 88 | 88 | 68 | 37 |
3.1.2. Molecular Characterization of E. coli
| Location | Amplified Genes | ||||||
|---|---|---|---|---|---|---|---|
| uidA | fliCH7 | eae | lt | papC | Ial | eagR | |
| Dairy farm A | 50 (57.5%) | 27 (54%) | 9 (18%) | 2 (4%) | 3 (6%) | 4 (8%) | 5 (10%) |
| Dairy farm B | 37 (42.5%) | 16 (43.2%) | 9 (24.3%) | 5 (13.5%) | 1 (2.7%) | 2 (5.4%) | 4 (10.8%) |
| Total | 87 (100%) | 43 (49%) | 18 (21%) | 7 (8%) | 4 (4.6%) | 6 (7%) | 9 (10.3%) |
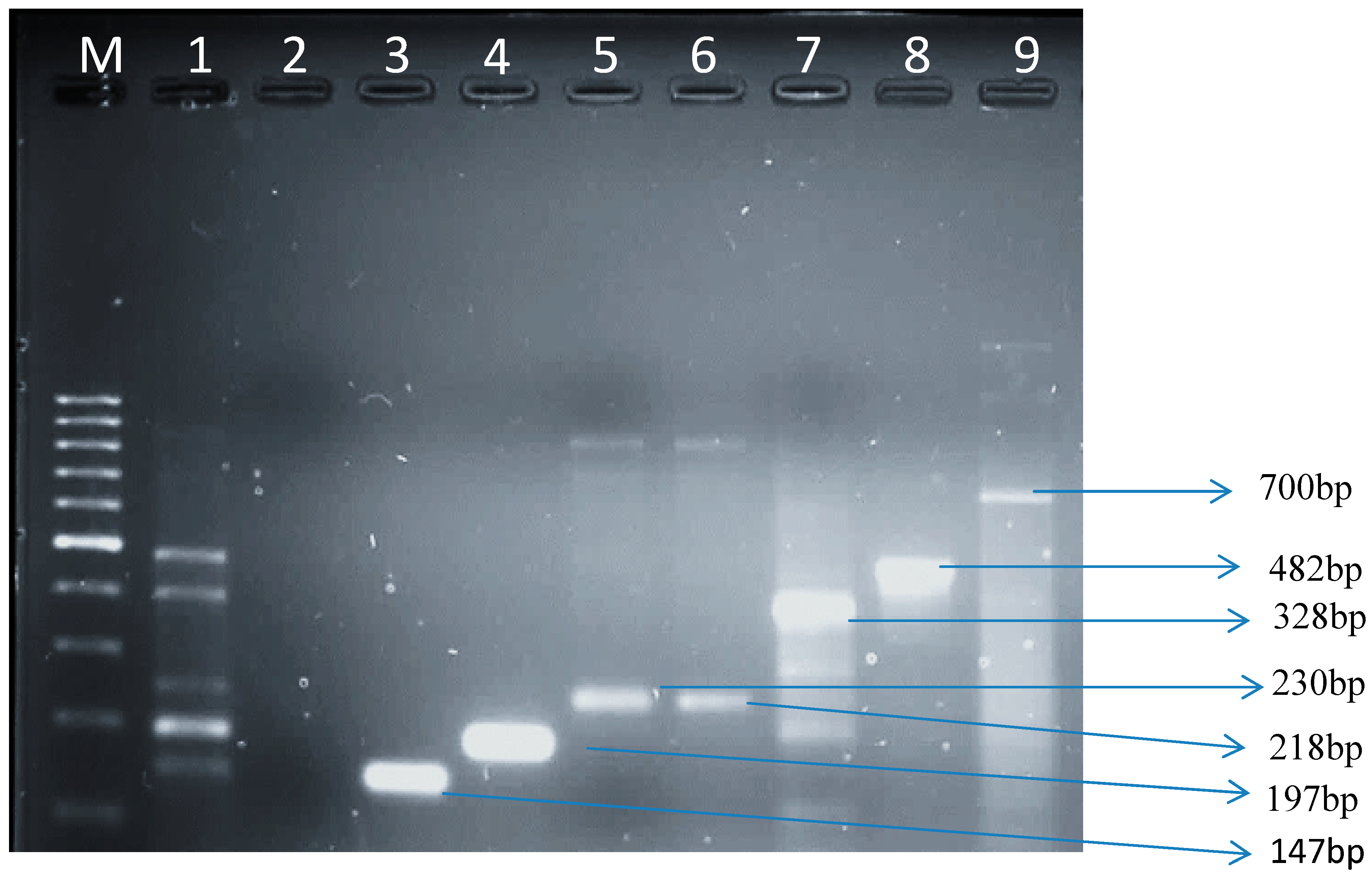
3.1.3. Virulence Genes from E. coli Isolates in Raw Cow’s Milk
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bélanger, L.A.; Garenaux, J.; Harel, M.; Boulianne, E.; Dozois, C. Escherichia coli from animal reservoirs as a potential source of human extraintestinal pathogenic Escherichia coli. Fems Immunol. Med. Microbiol. 2011, 62, 1–10. [Google Scholar] [CrossRef]
- Croxall, G.; Hale, J.; Weston, V.; Manning, G.; Cheetham, P.; Achtman, M.; McNally, A. Molecular epidemiology of extraintestinal pathogenic Escherichia coli isolates from a regional cohort of elderly patients highlights the prevalence of ST131 strains with increased antimicrobial resistance in both community and hospital care settings. J. Antimicrob. Chemother. 2011, 66, 2501–2508. [Google Scholar] [CrossRef] [PubMed]
- Croxen, M.A.; Finlay, B.B. Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. 2010, 26–38. [Google Scholar]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L.T. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Russo, T.A.; Johnson, J.R. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis 2002, 181, 1753–1754. [Google Scholar] [CrossRef]
- Bekal, S.; Brousseau, R.; Masson, L.; Préfontaine, G.; Fairbrother, J.M.; Harel, J. Rapid identification of Escherichia coli pathotypes by virulence gene detection with DNA microarray. J. Clin. Microbiol. 2003, 41, 2113–2125. [Google Scholar] [CrossRef]
- Huang, D.B.; Mohanty, A.; DuPont, H.L.; Okhuysen, P.C.; Chiang, T. A review of emerging enteric pathogen: Enteroaggregative Escherichia coli. J. Med. Microbiol. 2006, 1303–1311. [Google Scholar]
- Trabulsi, L.R.; Keller, R.; Gomes, T.A.T. Typical and atypical enteropathogenic Escherichia coli. Emerg. Infect. Dis. 2002, 8, 508–513. [Google Scholar] [CrossRef] [PubMed]
- Campellone, K.G. Cytoskeleton-modulating effectors of enteropathogenic and enterohaemorrhagic Escherichia coli: Tir, EspFU and actin pedestal assembly. Febs J. 2010, 277, 2390–2402. [Google Scholar] [CrossRef] [PubMed]
- Parsot, C. Shigella spp. and enteroinvasive Escherichia coli pathogenicity factors. Fems Microbiol. Lett. 2005, 252, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Piekrard, D.; Stevens, D.; Moriau, L.; Lior, H.; Lauwers, S. Isolation and virulence factors of Verocytotoxin producing Escherichia coli in human stool samples. Clin. Microbiol. Infec. 1997, 3, 531–540. [Google Scholar] [CrossRef]
- Bielaszewska, M.; Middendorf, B.; Kock, R.; Friedrich, A.W.; Fruth, A.; Karch, H.; Schmidt, A.; Mellmann, A. Shiga toxin-negative attaching and effacing Escherichia coli: Distinct clinical associations with bacterial phylogeny and virulence traits and inferred in-host pathogen evolution. Clin. Infect. Dis. 2008, 47, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Stephan, R.; Schumacher, S.; Corti, S.; Krause, G.; Danuser, J.; Beutin, L. Prevalence and characteristics of shiga toxin-producing Esherichia coli in Swiss raw milk cheeses collected at producer level. J. Dairy Sci. 2008, 91, 2561–2565. [Google Scholar] [CrossRef] [PubMed]
- Arisoy, M.; Ayasev, D.; Ekim, M. Detection of virulence factors of Escherichia coli from children by multiplex polymerase chain reaction. Int. J. Clin Pract. 2006, 60, 170–173. [Google Scholar] [CrossRef]
- Yamamoto, S. Molecular epidemiology of uropathogenic Escherichia coli. J. Infect. Chemother. 2007, 13, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Hagan, E.C.; Mobley, H.L. Uropathogenic Escherichia coli outer membrane antigens expressed during urinary tract infection. Infect. Immun. 2007, 75, 3941–3949. [Google Scholar] [CrossRef] [PubMed]
- Belanger, S.D.; Boissinot, M.; Menard, C.; Picard, F.J.; Bergeron, M.G. Rapid detection of Shiga toxin-producing bacteria in feces by multiplex PCR with molecular beacons on the smart cycler. J. Clin. Microbiol. 2002, 40, 1436–1440. [Google Scholar] [CrossRef] [PubMed]
- Gilgen, M.; Hubner, P.; Hofelen, C.; Luthy, J.; Candrian, U. PCR-based detection of verotoxin producing Esherichia. coli (VTEC) in ground beef. Res. Microbiol. 1998, 149, 145–154. [Google Scholar] [CrossRef]
- Fagan, P.K.; Hornitzky, M.A.; Bettelheim, K.A.; Djordjevic, S.P. Detection of Shiga-like toxin (stx1 and stx2), intimin (eaeA), and enterohemorrhagic Escherichia coli (EHEC) hemolysin (EHEC hlyA) genes in animal feces by multiplex PCR. Appl. Environ. Microbiol. 1999, 65, 868–872. [Google Scholar] [PubMed]
- Rappelli, P.; Maddau, G.; Mannu, F.; Colombo, M.; Fiori, P.L.; Cappuccinelli, P. Development of a set of multiplex PCR assays for the simultaneous identification of enterotoxigenic, enteropathogenic, enterohemorrhagic and enteroinvasive Escherichia coli. New Microbiol. 2001, 24, 77–83. [Google Scholar] [PubMed]
- Chapman, P.A.; Cerdan, A.; Malo, A.T.; Ellin, M.; Ashton, R.; Harkin, M.A. Escherichia coli O157 in cattle and sheep at slaughter, on beef and lamb carcasses and in raw beef and lamb products in south Yorkshire, UK. Int. J. Food Microbiol. 2001, 64, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Chirsten, G.L.; Davidson, P.M.; McAllister, J.S.; Roth, L.A. Coliform and other indicator bacteria. In Standard Methods for the Examination of Dairy Products, 16th ed.; Marshall, R.T., Ed.; American Public Health Association: Washington, DC, USA, 1993; Chapter 7; pp. 247–269. [Google Scholar]
- Houghtby, G.A.; Maturin, L.J.; Koenig, E.K. Microbiological count methods. In Standard Methods for the Examination of Dairy Products, 16th ed.; Marshall, R.T., Ed.; American Public Health Association: Washington, DC, USA, 1993; Chapter 6; pp. 213–246. [Google Scholar]
- Maugeri, T.L.; Carbone, M.; Fera, M.T.; Irrera, G.P.; Gugliandolo, C. Distribution of potentially pathogenic bacteria as free-living and plankton-associated in a marine coastal zone. J. Appl. Microbiol. 2004, 97, 354–361. [Google Scholar] [CrossRef] [PubMed]
- López-Saucedo, C.; Cerna, J.F.; Villegas-Sepulveda, N.; Thompson, R.; Velazquez, F.R.; Toress, J.; Tarr, P.I.; Estrada-Garda, T. Single multiplex polymerase chain reaction to detect diverse loci associated with diarrhoeagenic Escherichia coli. Emerg. Infect. Dis. 2003, 9, 127–131. [Google Scholar] [CrossRef]
- Tsai, L.Y.; Palmer, C.L.; Sangeermano, L.R. Detection of Escherichia coli in sludge by polymerase chain reaction. Appl. Environ. Microbiol. 1993, 59, 353–357. [Google Scholar]
- Wang, G.; Clifford, G.C.; Frank, G.R. Detection in Escherichia coli of the genes encoding the major virulence factors, the genes defining the O157:H7 serotype, and components of the type 2 shiga toxin family by multiplex PCR. J. Clin. Microbiol. 2002, 40, 3613–3619. [Google Scholar] [CrossRef]
- Stacy-Phipps, S.J.; Mecca, J.; Weiss, J.B. Multiplex PCR assay and simple preparation method for stool specimens detect enterotoxigenic Escherichia coli DNA during the course of infection. J. Clin. Microbiol. 1995, 33, 1054–1059. [Google Scholar]
- Kong, R.Y.C.; Lee, S.K.Y.; Law, T.W.F.; Law, S.H.W.; Wu, R.S.S. Rapid detection of six types of bacterial pathogens in marine waters by multiplex PCR. Water Res. 2002, 36, 2802–2812. [Google Scholar] [CrossRef] [PubMed]
- Vidal, R.; Vidal, M.; Lagos, R.; Levine, M.; Prado, V. Multiplex PCR for diagnosis of enteric infections associated with diarrheagenic Escherichia coli. J. Clin. Microbiol. 2004, 42, 1787–1789. [Google Scholar] [CrossRef]
- Cagney, C.H.; Crowley, G.; Duffy, J.J.; Sheridan, S.; O’Brien, E.; Carney, W.M.; Anderson, D.A.; Dowell, I.; Blair, S.; Bisho, R.H. Prevalence and numbers of Escherichia coli O157:H7 in minced beef and beef burgers from butcher shops and supermarkets in the Republic of Ireland. J. Food Microbiol. 2004, 21, 203–212. [Google Scholar] [CrossRef]
- Dozois, C.M.; Curtiss, R., 3rd. Pathogenic diversity of Escherichia coli and the emergence of exotic’ islands in the gene stream. Vet. Res. 1999, 30, 157–179. [Google Scholar]
- Odonkor, S.T.; Ampofi, J.K. Escherichia coli as an indicator of bacteriological quality of water: An overview. Microbiol. Res. 2013, 4, 1–11. [Google Scholar] [CrossRef]
- Farzan, A.; Friendship, R.M.; Cook, A.; Pollari, F. Occurrence of Salmonella, Campylobacter, Yersinia enterocolitica, Escherichia coli O157 and Listeria monocytogenes in swine. Zoonoses Public Health 2010, 57, 388–396. [Google Scholar] [CrossRef]
- Oliver, S.P.; Jayarao, B.M.; Almedia, R.A. Food borne pathogens in milk and the dairy environment food safety and public health implications. Foodborne Pathog. Dis. 2005, 2, 1115–1129. [Google Scholar]
- Collins, C.H.; Lyne, P.M.; Grange, J. Microbiological Methods; Collins, C.H., Lyne, P.M., Grange, J., Falkinham III, J. O., Eds.; Butterworth-Heinemann: London, UK, 1995. [Google Scholar]
- Guh, A.; Phan, Q.; Nelson, R.; Purviance, K.; Milardo, E.; Kinney, S.; Mshar, P.; Kasacek, W.; Cartter, M. Outbreak of Escherichia coli O157 associated with raw milk, Connecticut, 2008. Clin. Infect. Dis. 2010, 51, 1411–1417. [Google Scholar] [CrossRef] [PubMed]
- Okoh, A.I.; Osode, A.N. Enterotoxigenic Escherichia coli (ETEC): A recurring decimal in infants’ and travelers’ diarrhea. Rev. Environ. Health 2008, 23, 135–148. [Google Scholar]
- Blum, S.; Sela, N.; Heller, E.D.; Sela, S.; Leitner, G. Identification of a bovine mastitis Escherichia coli subset. Vet. Microbiol. 2008, 132, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Wenz, J.R.; Barrington, G.M.; Garry, F.B.; Ellis, R.P.; Magnuson, R.J. Escherichia coli isolates serotypes, genotypes, and virulence genes and clinical coliform mastitis severity. J. Dairy Sci. 2006, 89, 3408–3412. [Google Scholar] [CrossRef] [PubMed]
- Altalhi, A.D.; Hassan, S.A. Bacterial quality of raw milk investigated by Escherichia coli and isolates analysis for specific virulence-gene markers. Food Control 2009, 20, 913–917. [Google Scholar]
- Ghanbarpour, R.; Oswald, E. Phylogenetic distribution of virulence genes in Escherichia coli isolated from bovine mastitis in Iran. Res. Vet. Sci. 2010, 88, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Bean, A.; Williamson, J.; Cursons, R.T. Virulence genes of Escherichia coli strains isolated from mastitic milk. J. Vet. Med. B-Infect. Dis. Vet. Public Health 2004, 51, 285–287. [Google Scholar] [CrossRef] [PubMed]
- Dogan, B.; Klaessig, S.; Rishniw, M.; Almeida, R.A.; Oliver, S.P.; Simpson, K.; Schukken, Y.H. Adherent and invasive Escherichia coli are associated with persistent bovine mastitis. Vet. Microbiol. 2006, 116, 270–282. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Grover, S.; Batish, B.K. Application of multiplex PCR assay based on uidR and fliCH7 genes for detection of Escherichia coli O157:H7 in milk. J. General Appl. Microbiol. 2013, 59, 11–19. [Google Scholar] [CrossRef]
- Osode, A.N.; Okoh, A.I. Survival of free-living and plankton-associated Escherichia coli in the final effluents of a waste water treatment facility in a peri-urban community of the Eastern Cape Province of South Africa. Afr. J. Micobiol. Res. 2010, 4, 1424–1432. [Google Scholar]
- Bettelheim, K.A. Role of non-O157 VTEC. J. Appl. Microbiol. 2000, 88, S38–S50. [Google Scholar] [CrossRef] [PubMed]
- Elliott, E.J.; Robins-Browne, R.M.; O’Loughlin, E.V.; Bennett-Wood, V.; Bourke, J.; Henning, P.; Hogg, G.G.; Knight, J.; Pouch, H.; Redmond, D. Nationwide study of haemolytic uraemic syndrome: Clinical, microbiological, and epidemiological features. Arch. Dis Child. 2001, 85, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Murinda, S.E.; Nguyen, L.T.; Man, H.M.; Almedia, R.A. Detection of sorbitol negative and sorbitol-positive shiga toxin-producing Escherichia coli, Listeria monocytogenes, Campylobacter jejuni and Salmonella species in dairy farm environments. Foodborne Pathog. Dis. 2004, 1, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Ruan, X.; Crupper, S.S.; Schultz, B.D.; Robertson, D.C.; Zhang, W. Escherichia coli expressing EAST1 toxin did not cause an increase of cAMP or cGMP levels in cells, and no diarrhea in 5-day old gnotobiotic pigs. PLoS One 2012. [Google Scholar] [CrossRef]
- Frank, V.; Hahn, G.; Tolle, A. Incidence and identification of enterotoxigenic Escherichia coli strains in milk and milk products. Zentral. Bakteriol. Microbiol. Hyg. 1984, 257, 51–59. [Google Scholar]
- Ericsson, C.D. Travelers’ diarrhea. Int. J. Antimicrobial Agents 2003, 21, 116–124. [Google Scholar] [CrossRef]
- Foodborne Disease Outbreaks: Guidelines for Investigation and Control; World Health Organization: Geneva, Switzerland, 2007; p. 21.
- Evans, D.J.; Evans, D.G., Jr. Colonization factor antigens of human pathogens. Current topics—Escherichia coli in diarrheal disease. Microbiol. Immun. 1990, 151, 129–145. [Google Scholar]
- Gordillo, M.E.; Reeve, G.R.; Pappas, J.; Mathewson, J.J.; DuPont, H.L.; Murray, B.E. Molecular characterization of strains of enteroinvasive Escherichia coli O143, including isolates from a large outbreak in Houston, Texas. J. Clin. Microbiol. 1992, 30, 889–893. [Google Scholar] [PubMed]
- Marier, R.; Wells, J.G.; Swanson, R.C.; Dalthan, W.; Mehlman, I.J. An outbreak of enteropathogenic Escherichia coli food borne disease traced to imported French cheese. Lancet 1973, 302, 1376–1378. [Google Scholar] [CrossRef]
- Harris, J.R.; Mariano, J.; Wells, J.G.; Payne, B.J.; Donnell, H.D.; Cohen, M.L. Person-to-person transmission in an outbreak of enteroinvasive Escherichia coli. Amer. J. Epidemol. 1985, 122, 245–252. [Google Scholar]
- Todar, K. Pathogenic Escherichia coli. In Todar’s Online Textbook on Bacteriology; Department of Bacteriology, University of Wisconsin: Madison, WI, USA, 2008. [Google Scholar]
- Lan, R.; Alles, M.C.; Donohoe, K.; Martinez, M.B.; Reeves, P.R. Molecular evolutionary relationships of enteroinvasive Escherichia coli and Shigella spp. Infect. Immun. 2004, 72, 5080–5088. [Google Scholar] [CrossRef] [PubMed]
- Kahali, S.; Sarkar, B.; Rajendran, K.; Khanam, J.; Nandy, S.R.K.; Bhattacharya, S.K.; Ramamurthy, T. Virulence characteristics and molecular epidemiology of enteroaggregative Escherichia coli isolates from hospitalized diarrheal patients in Kolkata, India. J. Clin. Microbiol. 2004, 42, 4111–4120. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Nakazawa, M. Detection and sequences of the Enteroaggregative Escherichia coli heat-stable enterotoxin 1 gene in enterotoxigenic E. coli strains isolated from piglets and calves with diarrhea. J. Clin. Microbiol. 1997, 35, 223–227. [Google Scholar] [PubMed]
- Vallance, B.A.; Finlay, B.B. Exploitation of host cells by enteropathogenic Escherichia coli. Proc. Nat. Acad. Sci. USA 2000, 97, 8799–8806. [Google Scholar] [CrossRef] [PubMed]
- Bugarel, M.; Martin, A.; Fach, P.; Beutin, L. Virulence profiling of enterohemorrhagic (EHEC) and enteropathogenic (EPEC) Escherichia coli strains: A basis of molecular risk assessment of atypical and typical EPEC strains. BMC Micobiol. 2011, 11. [Google Scholar] [CrossRef]
- Aleixo, J.A.; Aver, G.P. Prevalence of enteropathogenic and enterotoxigenic Escherichia coli in foods of animal origin in southern Brazil. Ciência. Rural 1996, 26, 1121–1129. [Google Scholar] [CrossRef]
- Adesiyun, A.; Webb, L.; Romain, H.; Kaminjolo, J. Prevalence and characteristics of strains of Escherichia coli isolated from milk and feces of cows on dairy farms in Trinidad. J. Food Protect. 1997, 60, 174–181. [Google Scholar]
- Holko, I.; Bisova, T.; Holkova, Z.; Kmet, V. Virulence markers of Escherichia coli strains isolated from traditional cheeses made from unpasteurised sheep milk in Slovakia. Food Control 2006, 17, 393–396. [Google Scholar] [CrossRef]
- Zuber, M. E. coli: Serotypes other than O157:H7; MSPH DOH, Regional Epidemiologist: Tallahasse FL, USA, 1999. [Google Scholar]
- Tennant, S.M.; Tauschek, M.; Azzopardi, K.; Bigham, A.; Bennett-Wood, V.; Hartland, E.L.; Qi, W.; Whittam, T.S.; Robins-Browne, R.M. Characterization of atypical enteropathogenic E. coli strains of clinical origin. BMC Microbiol. 2009, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dogan, B.; Rishniw, M.; Bruant, G.; Harel, J.; Schukken, Y.H.; Simpson, K.W. Phylogroup and lpfA influence epithelial invasion by mastitis associated Escherichia coli. Vet. Microbiol. 2012, 159, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Alteri, C.J.; Hagan, E.C.; Sivick, K.E.; Smith, S.N.; Mobley, H.L.T. Mucosal immunization with iron receptor antigens protects against urinary tract infection. PLoS Pathog. 2009, 5. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caine, L.-A.; Nwodo, U.U.; Okoh, A.I.; Ndip, R.N.; Green, E. Occurrence of Virulence Genes Associated with Diarrheagenic Escherichia coli Isolated from Raw Cow’s Milk from Two Commercial Dairy Farms in the Eastern Cape Province, South Africa. Int. J. Environ. Res. Public Health 2014, 11, 11950-11963. https://doi.org/10.3390/ijerph111111950
Caine L-A, Nwodo UU, Okoh AI, Ndip RN, Green E. Occurrence of Virulence Genes Associated with Diarrheagenic Escherichia coli Isolated from Raw Cow’s Milk from Two Commercial Dairy Farms in the Eastern Cape Province, South Africa. International Journal of Environmental Research and Public Health. 2014; 11(11):11950-11963. https://doi.org/10.3390/ijerph111111950
Chicago/Turabian StyleCaine, Lesley-Anne, Uchechukwu U. Nwodo, Anthony I. Okoh, Roland N. Ndip, and Ezekiel Green. 2014. "Occurrence of Virulence Genes Associated with Diarrheagenic Escherichia coli Isolated from Raw Cow’s Milk from Two Commercial Dairy Farms in the Eastern Cape Province, South Africa" International Journal of Environmental Research and Public Health 11, no. 11: 11950-11963. https://doi.org/10.3390/ijerph111111950





