Proteomic Analysis of Anti-Tumor Effects of 11-Dehydrosinulariolide on CAL-27 Cells
Abstract
:1. Introduction
2. Results and Discussion
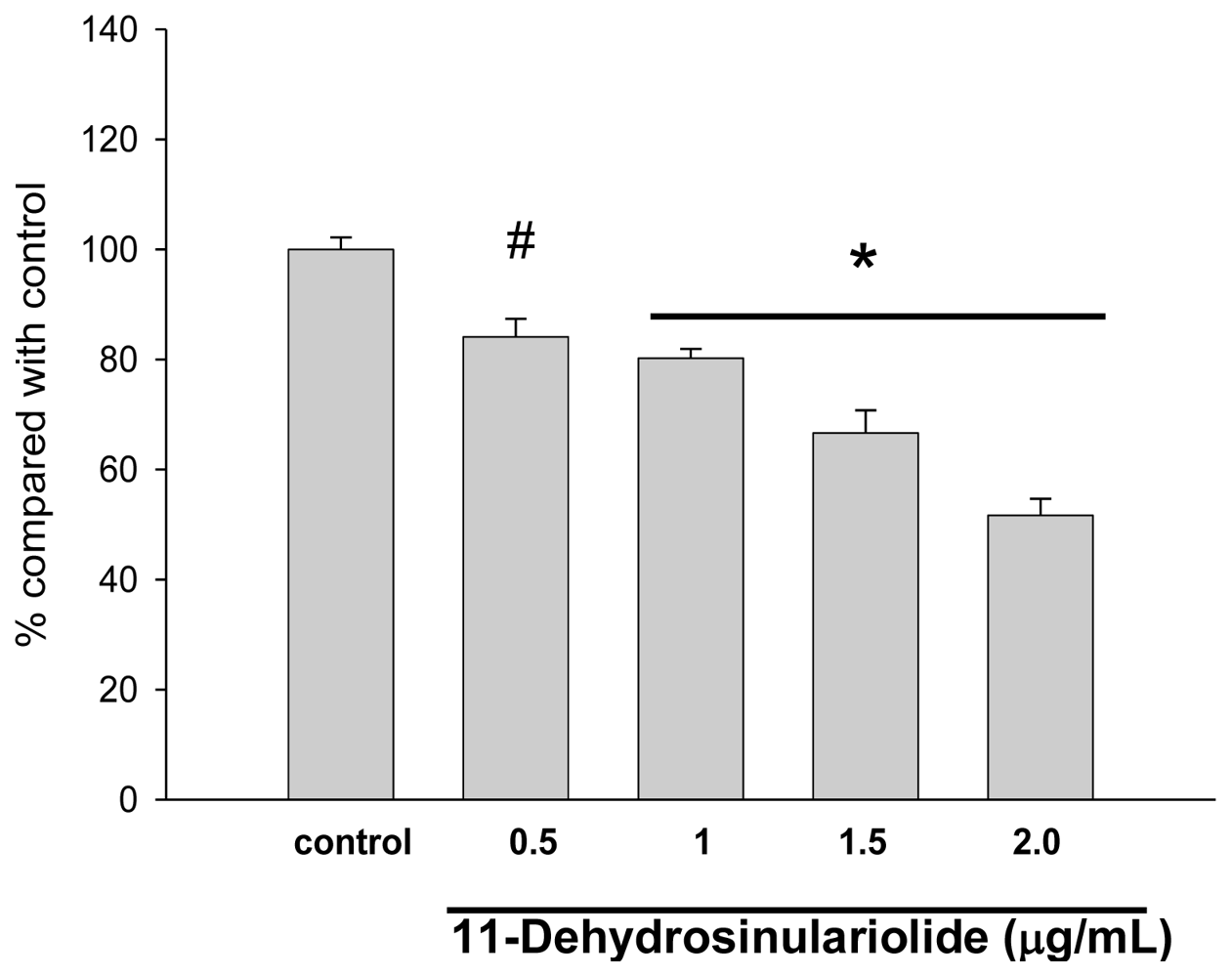
2.1. The Cytotoxic Effects of 11-Dehydrosinulariolide on CAL-27 Cells
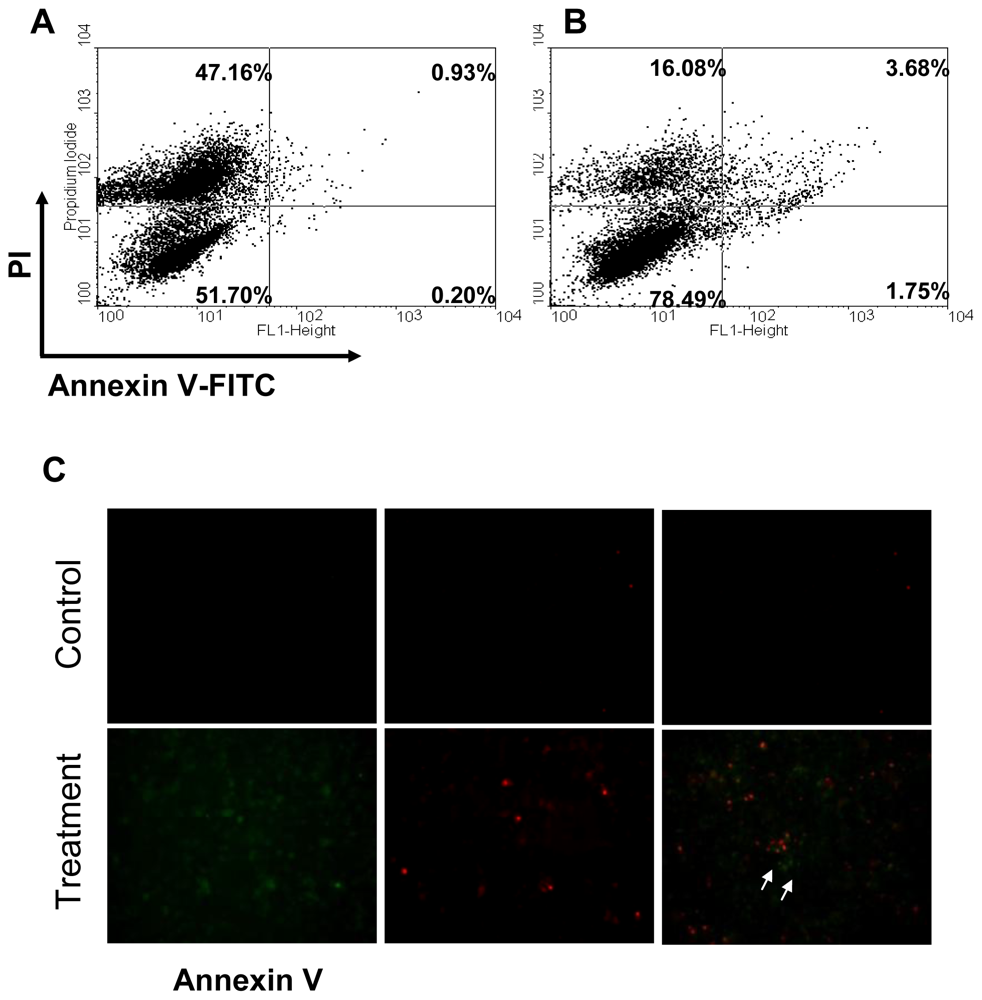
2.2. Apoptosis of CAL-27 Treated with 11-Dehydrosinulariolide
2.3. Proteomic Analysis of CAL-27 Cells Treated with 11-Dehydrosinulariolide
2.4. Validation by Western Blotting
2.5. Discussion
2.5.1. Protein Disulfide Isomerase A3 (PDIA3)
2.5.2. 60 kDa Heat Shock Protein (Hsp60)
2.5.3. 78 kDa Glucose-Regulated Protein (GRP78)
2.5.4. Heterogeneous Nuclear Ribonucleoprotein F (hnRNP F)
2.5.5. Prohibitin
2.5.6. Ubiquinol Cytochrome c Reductase Complex Core Protein 1 (UQCRC1)
3. Experimental Section
3.1. Cell Culture and Treatment with 11-Dehydrosinulariolide
3.2. MTT Assay
3.3. Apoptosis and Cell Cycle Analysis
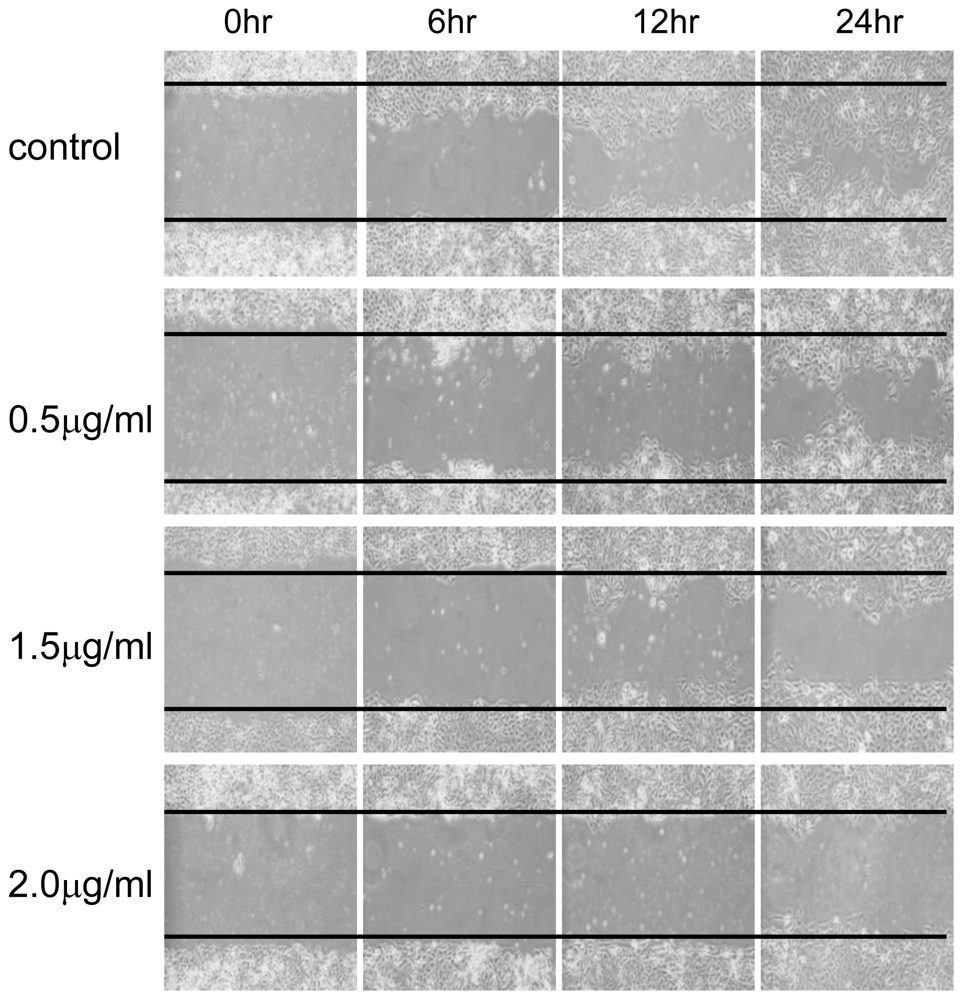
3.4. Wound Healing Assay
3.5. Protein Extraction and Estimation
3.6. Two-Dimensional Gel Electrophoresis
3.7. Protein Identification by LC-MS/MS
3.8. Western Blotting Analysis
4. Conclusion
Acknowledgements
- Samples Availability: Available from the authors.
Reference
- Chen, Y; Chang, J; Liao, C; Wang, H; Yen, T; Chiu, C; Lu, Y; Li, H; Cheng, A. Head and neck cancer in the betel quid chewing area: Recent advances in molecular carcinogenesis. Cancer Sci 2008, 99, 1507–1514. [Google Scholar]
- Liao, C; Chang, J; Wang, H; Ng, S; Hsueh, C; Lee, L; Lin, C; Chen, I; Huang, S; Cheng, A. Analysis of risk factors of predictive local tumor control in oral cavity cancer. Ann. Surg. Oncol 2008, 15, 915–922. [Google Scholar]
- Liao, C; Kang, C; Chang, J; Wang, H; Ng, S; Hsueh, C; Lee, L; Lin, C; Cheng, A. Survival of second and multiple primary tumors in patients with oral cavity squamous cell carcinoma in the betel quid chewing area. Oral Oncol 2007, 43, 811–819. [Google Scholar]
- Faulkner, DJ. Marine natural products. Nat. Prod. Rep 2002, 19, 1–48. [Google Scholar]
- Ruggieri, G. Drugs from the sea. Science 1976, 194, 491–497. [Google Scholar]
- Faulkner, DJ. Marine natural products. Nat. Prod. Rep 2001, 18, 1–49. [Google Scholar]
- Ojika, M; Islam, M; Shintani, T; Zhang, Y; Okamoto, T; Sakagami, Y. Three new cytotoxic acylspermidines from the soft coral, Sinularia sp. Biosci. Biotechnol. Biochem 2003, 67, 1410–1412. [Google Scholar]
- Ahmed, A; Shiue, R; Wang, G; Dai, C; Kuo, Y; Sheu, J. Five novel norcembranoids from Sinularia leptoclados and S. parva. Tetrahedron 2003, 59, 7337–7344. [Google Scholar]
- He, Q; Chen, J; Kung, H; Yuen, A; Chiu, J. Identification of tumor associated proteins in oral tongue squamous cell carcinoma by proteomics. Proteomics 2004, 4, 271–278. [Google Scholar]
- Herrmann, PC; Liotta, L; Petricoin, EF, III. Cancer proteomics: The state of the art. Dis Markers 2001, 17, 49–57. [Google Scholar]
- Hsieh, PW; Chang, FR; McPhail, AT; Lee, KH; Wu, YC. New cembranolide analogues from the formosan soft coral Sinularia flexibilis and their cytotoxicity. Nat. Prod. Res 2003, 17, 409–418. [Google Scholar]
- Aceret, TL; Coll, JC; Uchio, Y; Sammarco, PW. Antimicrobial activity of the diterpenes flexibilide and sinulariolide derived from Sinularia flexibilis Quoy and Gaimard 1833 (Coelenterata: Alcyonacea, Octocorallia). Comp. Biochem. Physiol. C Pharmacol. Toxicol. Endocrinol 1998, 120, 121–126. [Google Scholar]
- Rashid, MA; Gustafson, KR; Boyd, MR. HIV-inhibitory cembrane derivatives from a Philippines collection of the soft coral Lobophytum species. J. Nat. Prod 2000, 63, 531–533. [Google Scholar]
- Ahmed, AF; Hsieh, YT; Wen, ZH; Wu, YC; Sheu, JH. Polyoxygenated sterols from the Formosan soft coral Sinularia gibberosa. J. Nat. Prod 2006, 69, 1275–1279. [Google Scholar]
- Su, JH; Tseng, YJ; Huang, HH; Ahmed, AF; Lu, CK; Wu, YC; Sheu, JH. 9,11-Secosterols from the soft corals Sinularia lochmodes and Sinularia leptoclados. J. Nat. Prod 2006, 69, 850–852. [Google Scholar]
- Ahmed, AF; Tai, SH; Wu, YC; Sheu, JH. Sinugrandisterols AD, trihydroxysteroids from the soft coral Sinularia grandilobata. Steroids 2007, 72, 368–374. [Google Scholar]
- Freedman, RB; Hirst, TR; Tuite, MF. Protein disulphide isomerase: Building bridges in protein folding. Trends Biochem. Sci 1994, 19, 331–336. [Google Scholar]
- Hendershot, L. The ER function BiP is a master regulator of ER function. Mt. Sinai J. Med 2004, 71, 289–297. [Google Scholar]
- So, AYL; de la Fuente, E; Walter, P; Shuman, M; Bernales, S. The unfolded protein response during prostate cancer development. Cancer Metastasis Rev 2009, 28, 219–223. [Google Scholar]
- Hartl, F; Hlodan, R; Langer, T. Molecular chaperones in protein folding: The art of avoiding sticky situations. Trends Biochem. Sci 1994, 19, 20–25. [Google Scholar]
- Guimaraes, AJ; Frases, S; Gomez, FJ; Zancope-Oliveira, RM; Nosanchuk, JD. Monoclonal antibodies to heat shock protein 60 alter the pathogenesis of Histoplasma capsulatum. Infect. Immun 2009, 77, 1357–1367. [Google Scholar]
- He, L; Lemasters, JJ. Heat shock suppresses the permeability transition in rat liver mitochondria. J. Biol. Chem 2003, 278, 16755–16760. [Google Scholar]
- Ghosh, J; Dohi, T; Kang, B; Altieri, D. Hsp60 regulation of tumor cell apoptosis. J. Biol. Chem 2008, 283, 5188–5194. [Google Scholar]
- Haas, I. BiP (GRP78), an essential hsp70 resident protein in the endoplasmic reticulum. Cell. Mol. Life Sci 1994, 50, 1012–1020. [Google Scholar]
- Hughes, C; Shen, J; Subjeck, J. Resistance to etoposide induced by three glucose-regulated stresses in Chinese hamster ovary cells. Cancer Res 1989, 49, 4452–4454. [Google Scholar]
- Jamora, C; Dennert, G; Lee, A. Inhibition of tumor progression by suppression of stress protein GRP78/BiP induction in fibrosarcoma B/C10ME. Proc. Natl. Acad. Sci. USA 1996, 93, 7690–7694. [Google Scholar]
- Song, M; Park, Y; Lee, J; Park, K. Induction of glucose-regulated protein 78 by chronic hypoxia in human gastric tumor cells through a protein kinase C-ɛ/ERK/AP-1 signaling cascade. Cancer Res 2001, 61, 8322–8330. [Google Scholar]
- Pyrko, P; Schonthal, AH; Hofman, FM; Chen, TC; Lee, AS. The unfolded protein response regulator GRP78/BiP as a novel target for increasing chemosensitivity in malignant gliomas. Cancer Res 2007, 67, 9809–9816. [Google Scholar]
- Rao, RV; Hermel, E; Castro-Obregon, S; del Rio, G; Ellerby, LM; Ellerby, HM; Bredesen, DE. Coupling endoplasmic reticulum stress to the cell death program. J. Biol. Chem 2001, 276, 33869–33874. [Google Scholar]
- Honore, B; Baandrup, U; Vorum, H. Heterogeneous nuclear ribonucleoproteins F and H/H′ show differential expression in normal and selected cancer tissues. Exp. Cell Res 2004, 294, 199–209. [Google Scholar]
- Fogel, BL; McNally, MT. A cellular protein, hnRNP H, binds to the negative regulator of splicing element from Rous sarcoma virus. J. Biol. Chem 2000, 275, 32371–32378. [Google Scholar]
- Jacquenet, S; Mereau, A; Bilodeau, PS; Damier, L; Stoltzfus, CM; Branlant, C. A second exon splicing silencer within human immunodeficiency virus type 1 tat exon 2 represses splicing of Tat mRNA and binds protein hnRNP H. J. Biol. Chem 2001, 276, 40464–40475. [Google Scholar]
- Oberg, D; Fay, J; Lambkin, H; Schwartz, S. A downstream polyadenylation element in human papillomavirus type 16 L2 encodes multiple GGG motifs and interacts with hnRNP H. J. Virol 2005, 79, 9254–9269. [Google Scholar]
- McNally, LM; Yee, L; McNally, MT. Heterogeneous nuclear ribonucleoprotein H is required for optimal U11 small nuclear ribonucleoprotein binding to a retroviral RNA-processing control element. J. Biol. Chem 2006, 281, 2478–2488. [Google Scholar]
- Garneau, D; Revil, T; Fisette, JF; Chabot, B. Heterogeneous nuclear ribonucleoprotein F/H proteins modulate the alternative splicing of the apoptotic mediator Bcl-x. J. Biol. Chem 2005, 280, 22641–22650. [Google Scholar]
- Crawford, JB; Patton, JG. Activation of α-tropomyosin exon 2 is regulated by the SR protein 9G8 and heterogeneous nuclear ribonucleoproteins H and F. Mol. Cell. Biol 2006, 26, 8791–8802. [Google Scholar]
- Yoshida, T; Kokura, K; Makino, Y; Ossipow, V; Tamura, T. Heterogeneous nuclear RNA-ribonucleoprotein F binds to DNA via an oligo (dG)-motif and is associated with RNA polymerase II. Genes Cells 1999, 4, 707–719. [Google Scholar]
- Olopade, OI; Adeyanju, MO; Safa, AR; Hagos, F; Mick, R; Thompson, CB; Recant, WM. Overexpression of BCL-x protein in primary breast cancer is associated with high tumor grade and nodal metastases. Cancer J. Sci. Am 1997, 3, 230–237. [Google Scholar]
- Clarke, MF; Apel, IJ; Benedict, MA; Eipers, PG; Sumantran, V; Gonzalez-Garcia, M; Doedens, M; Fukunaga, N; Davidson, B; Dick, JE. A recombinant bcl-x s adenovirus selectively induces apoptosis in cancer cells but not in normal bone marrow cells. Proc. Natl. Acad. Sci. USA 1995, 92, 11024–11028. [Google Scholar]
- Montano, MM; Ekena, K; Delage-Mourroux, R; Chang, W; Martini, P; Katzenellenbogen, BS. An estrogen receptor-selective coregulator that potentiates the effectiveness of antiestrogens and represses the activity of estrogens. Proc. Natl. Acad. Sci. USA 1999, 96, 6947–6952. [Google Scholar]
- Sun, L; Liu, L; Yang, XJ; Wu, Z. Akt binds prohibitin 2 and relieves its repression of MyoD and muscle differentiation. J. Cell Sci 2004, 117, 3021–3029. [Google Scholar]
- Vander Heiden, MG; Choy, JS; VanderWeele, DJ; Brace, JL; Harris, MH; Bauer, DE; Prange, B; Kron, SJ; Thompson, CB; Rudin, CM. Bcl-x L complements Saccharomyces cerevisiae genes that facilitate the switch from glycolytic to oxidative metabolism. J. Biol. Chem 2002, 277, 44870–44876. [Google Scholar]
- Fusaro, G; Dasgupta, P; Rastogi, S; Joshi, B; Chellappan, S. Prohibitin induces the transcriptional activity of p53 and is exported from the nucleus upon apoptotic signaling. J. Biol. Chem 2003, 278, 47853–47861. [Google Scholar]
- Rajalingam, K; Wunder, C; Brinkmann, V; Churin, Y; Hekman, M; Sievers, C; Rapp, UR; Rudel, T. Prohibitin is required for Ras-induced Raf-VMEK-VERK activation and epithelial cell migration. Nat. Cell Biol 2005, 7, 837–843. [Google Scholar]
- Nijtmans, LGJ; de Jong, L; Sanz, MA; Coates, PJ; Berden, JA; Back, JW; Muijsers, AO; van der Spek, H; Grivell, LA. Prohibitins act as a membrane-bound chaperone for the stabilization of mitochondrial proteins. EMBO J 2000, 19, 2444–2451. [Google Scholar]
- Sanz, MA; Tsang, WY; Willems, EM; Grivell, LA; Lemire, BD; van der Spek, H; Nijtmans, LGJ. The mitochondrial prohibitin complex is essential for embryonic viability and germline function in Caenorhabditis elegans. J. Biol. Chem 2003, 278, 32091–32099. [Google Scholar]
- Lohrum, MAE; Woods, DB; Ludwig, RL; Balint, E; Vousden, KH. C-terminal ubiquitination of p53 contributes to nuclear export. Mol. Cell. Biol 2001, 21, 8521–8532. [Google Scholar]
- Mihara, M; Erster, S; Zaika, A; Petrenko, O; Chittenden, T; Pancoska, P; Moll, UM. p53 has a direct apoptogenic role at the mitochondria. Mol. Cell 2003, 11, 577–590. [Google Scholar]
- Chipuk, JE; Green, DR. Cytoplasmic p53: Bax and forward. Cell Cycle 2004, 3, 429–431. [Google Scholar]
- Mishra, S; Murphy, LC; Nyomba, B; Murphy, LJ. Prohibitin: A potential target for new therapeutics. Trends Mol. Med 2005, 11, 192–197. [Google Scholar]
- Trumpower, B; Edwards, CA. Purification of a reconstitutively active iron-sulfur protein (oxidation factor) from succinate. cytochrome c reductase complex of bovine heart mitochondria. J. Biol. Chem 1979, 254, 8697. [Google Scholar]
- Schapira, AHV; Cooper, JM; Morgan-Hughes, JA; Landon, DN; Clark, JB. Mitochondrial myopathy with a defect of mitochondrial-protein transport. N. Engl. J. Med 1990, 323, 37–42. [Google Scholar]
- Slipetz, D; Aprille, J; Goodyer, P; Rozen, R. Deficiency of complex III of the mitochondrial respiratory chain in a patient with facioscapulohumeral disease. Am. J. Hum. Genet 1991, 48, 502–510. [Google Scholar]
- Fiskum, G; Starkov, A; Polster, BM; Chinopoulos, C. Mitochondrial mechanisms of neural cell death and neuroprotective interventions in Parkinson’s disease. Ann. N. Y. Acad. Sci 2003, 991, 111–119. [Google Scholar]
- Gille, L; Nohl, H. The ubiquinol/bc1 redox couple regulates mitochondrial oxygen radical formation. Arch. Biochem. Biophys 2001, 388, 34–38. [Google Scholar]
- Nicolay, K; de Kruijff, B. Effects of adriamycin on respiratory chain activities in mitochondria from rat liver, rat heart and bovine heart. Evidence for a preferential inhibition of complex III and IV. Biochim. Biophys. Acta Bioenerg 1987, 892, 320–330. [Google Scholar]
- Goormaghtigh, E; Huart, P; Brasseur, R; Ruysschaert, JM. Mechanism of inhibition of mitochondrial enzymatic complex I–III by adriamycin derivatives. Biochim. Biophys. Acta Biomembr 1986, 861, 83–94. [Google Scholar]
- Tuquet, C; Dupont, J; Mesneau, A; Roussaux, J. Effects of tamoxifen on the electron transport chain of isolated rat liver mitochondria. Cell Biol. Toxicol 2000, 16, 207–219. [Google Scholar]
- Huang, H; Brennan, T; Muir, M; Mason, R. Functional α1 and β2 adrenergic receptors in human osteoblasts. J. Cell. Physiol 2009, 220, 267–275. [Google Scholar]
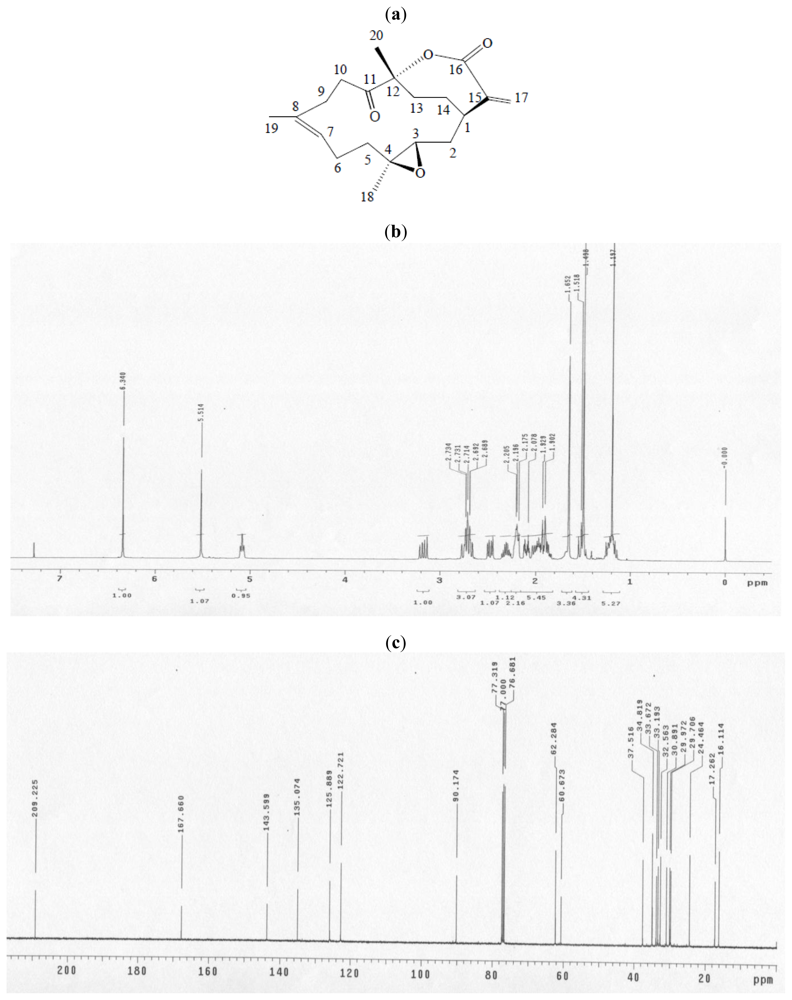


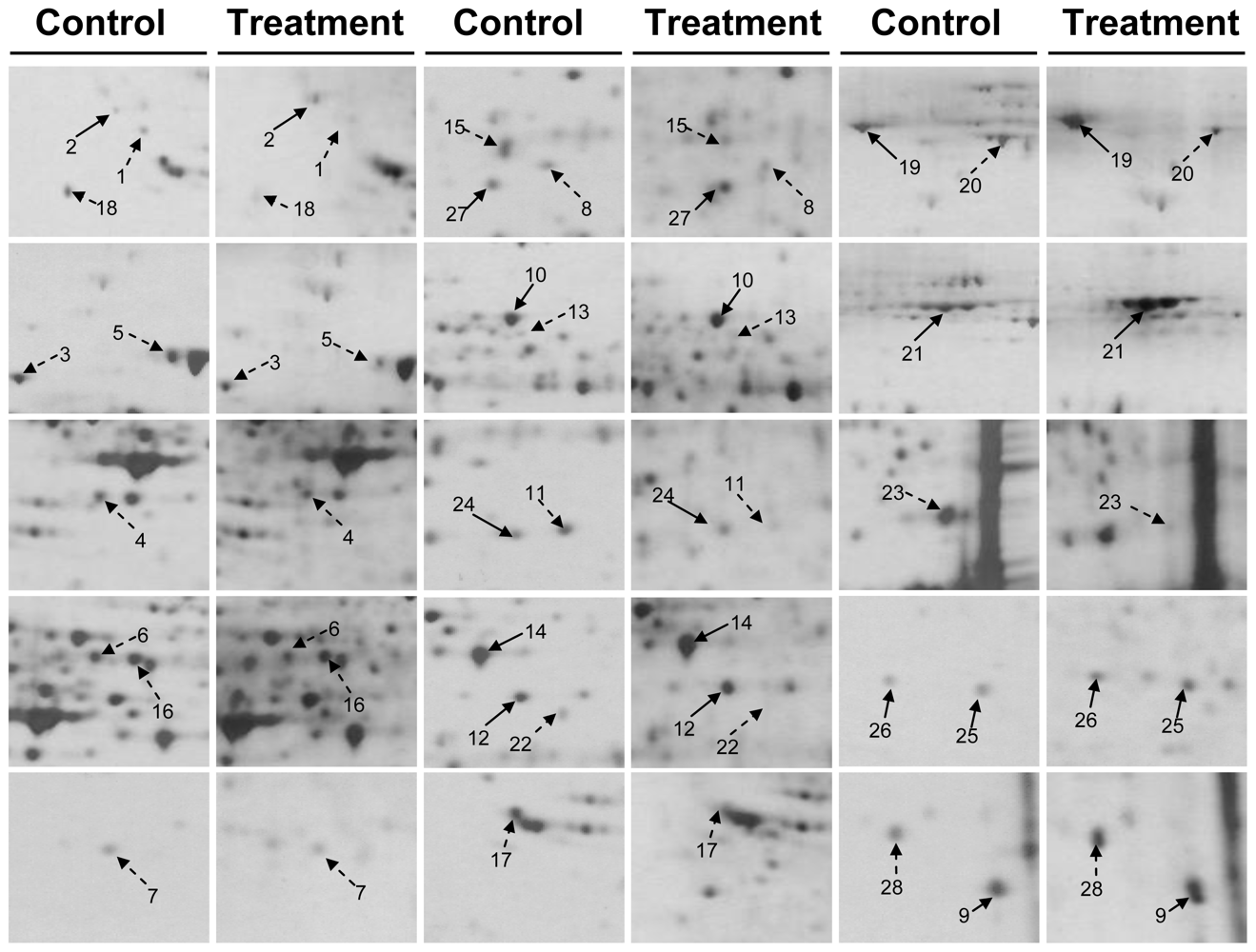

| Spot no. | Protein name | Accessio n no. | Calculate Mr/pI | Peptide matched | Sequence covered % | MASCOT score | Regulation (fold-change) * |
|---|---|---|---|---|---|---|---|
| 1 | Reticulocalbin-1 | Q15293 | 38.86/4.86 | 7 | 28 | 148 | −2.7 |
| 2 | Protein SET (Phosphatase 2A inhibitor I2PP2A) | Q01105 | 33.47/4.23 | 5 | 20 | 87 | +2.2 |
| 3 | 78 kDa glucose-regulated protein precursor | P11021 | 72.28/5.07 | 8 | 8 | 152 | −2.0 |
| 4 | Heterogeneous nuclear ribonucleoprotein F | P52597 | 45.64/5.38 | 29 | 35 | 345 | −2.1 |
| 5 | 60 kDa heat shock protein | P10809 | 61.02/5.70 | 43 | 46 | 753 | −3.7 |
| 6 | Actin-like protein 6A | O96019 | 47.43/5.39 | 19 | 23 | 83 | −2.2 |
| 7 | Proteasome subunit beta type 4 precursor | P28070 | 29.18/5.72 | 6 | 35 | 118 | −2.1 |
| 8 | F-actin capping protein subunit beta | P47756 | 31.33/5.36 | 22 | 31 | 242 | −2.8 |
| 9 | Peptidyl-prolyl cis-trans isomerase A | P62937 | 18.0/7.68 | 2 | 9 | 49 | +2.3 |
| 10 | Protein disulfide-isomerase A3 precursor | P14136 | 56.75/5.98 | 84 | 57 | 1168 | +2.2 |
| 11 | Purine nucleoside phosphorylase | P00491 | 32.09/6.45 | 9 | 23 | 208 | −6.4 |
| 12 | UPF0160 protein MYG1 | Q9HB07 | 42.41/6.25 | 2 | 5 | 73 | +3.8 |
| 13 | Xaa-Pro dipeptidase | P12955 | 54.51/5.64 | 11 | 14 | 199 | −2.0 |
| 14 | SPFH domain-containing protein 2 precursor | O94905 | 37.81/5.47 | 17 | 6 | 104 | +2.2 |
| 15 | 60S acidic ribosomal protein P0 | P05388 | 34.25/5.71 | 88 | 44 | 933 | −2.7 |
| 16 | Ubiquinol cytochrome c reductase complex core protein 1 | P31930 | 52.61/5.94 | 23 | 28 | 296 | −2.6 |
| 17 | 40S ribosomal protein SA | P08865 | 32.83/4.79 | 72 | 51 | 930 | −1.8 |
| 18 | Nascent polypeptide-associated complex subunit alpha | Q13765 | 23.37/4.52 | 25 | 32 | 494 | −3.1 |
| 19 | Nucleolin | P19338 | 76.56/4.6 | 51 | 34 | 519 | +4.1 |
| 20 | Transitional endoplasmic reticulum ATPase | P55072 | 89.26/5.14 | 127 | 46 | 1117 | −1.6 |
| 21 | Neutral alpha-glucosidase AB precursor | Q14697 | 106.8/5.74 | 85 | 39 | 988 | +3.0 |
| 22 | Isocitrate dehydrogenase subunit alpha | P50213 | 39.56/6.47 | 35 | 28 | 413 | −3.8 |
| 23 | Fructose-bisphosphate aldolase A | P04075 | 39.39/8.0 | 103 | 60 | 1330 | −9.0 |
| 24 | Prohibitin | P35232 | 29.78/5.57 | 23 | 36 | 115 | +2.4 |
| 25 | Lactoylglutathione lyase | Q04760 | 20.7/5.24 | 52 | 56 | 344 | +1.5 |
| 26 | Translationally-controlled tumor protein (TCTP) | P13693 | 19.58/4.84 | 35 | 52 | 198 | +2.0 |
| 27 | Inorganic pyrophosphatase | Q15181 | 32.63/5.54 | 45 | 55 | 530 | +3.0 |
| 28 | Protein DJ-1 | Q99497 | 19.87/6.33 | 38 | 65 | 238 | +2.0 |
© 2011 by the authors; licensee MDPI, Basel, Switzerland This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Liu, C.-I.; Chen, C.-C.; Chen, J.-C.; Su, J.-H.; Huang, H.H.; Chen, J.Y.-F.; Wu, Y.-J. Proteomic Analysis of Anti-Tumor Effects of 11-Dehydrosinulariolide on CAL-27 Cells. Mar. Drugs 2011, 9, 1254-1272. https://doi.org/10.3390/md9071254
Liu C-I, Chen C-C, Chen J-C, Su J-H, Huang HH, Chen JY-F, Wu Y-J. Proteomic Analysis of Anti-Tumor Effects of 11-Dehydrosinulariolide on CAL-27 Cells. Marine Drugs. 2011; 9(7):1254-1272. https://doi.org/10.3390/md9071254
Chicago/Turabian StyleLiu, Chih-I, Cheng-Chi Chen, Jiing-Chuan Chen, Jui-Hsin Su, Han Hsiang Huang, Jeff Yi-Fu Chen, and Yu-Jen Wu. 2011. "Proteomic Analysis of Anti-Tumor Effects of 11-Dehydrosinulariolide on CAL-27 Cells" Marine Drugs 9, no. 7: 1254-1272. https://doi.org/10.3390/md9071254




