New Cytotoxic Oxygenated Sterols from the Marine Bryozoan Cryptosula pallasiana
Abstract
:1. Introduction
2. Results and Discussion
3. Experimental Section
3.1. General Experimental Procedures
3.2. Animal Material
3.3. Extraction and Isolation
3.3.1. Liebermann-Burchard Test
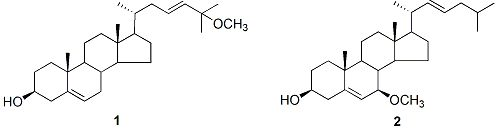
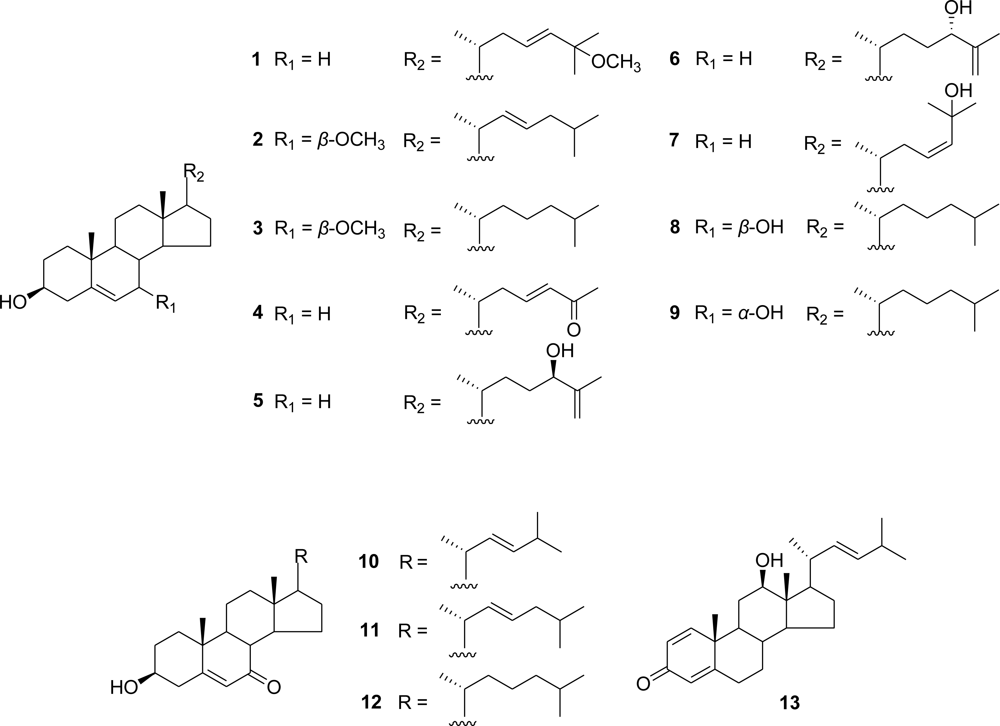
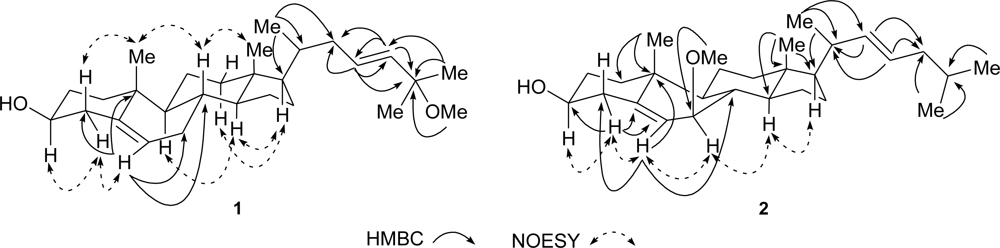
3.3.2. (23E)-25-Methoxy-cholesta-5,23-dien-3β-ol (1)
3.3.3. (22E)-7β-Methoxy-cholesta-5,22-dien-3β-ol (2)
3.3.4. 7β-Methoxy-cholest-5-en-3β-ol (3)
3.3.5. (23E)-3β-Hydroxy-27-norcholesta-5,23-dien-25-one (4)
3.3.6. 24(R)-Cholesta-5,25-diene-3β,24-diol (5)
3.3.7. 24(S)-Cholesta-5,25-diene-3β,24-diol (6)
3.4. MTT Cytotoxicity Assays
Supporting Information
- S1: HR-ESI-MS of compound 1
- S2: EI-MS of compound 1
- S3: 1H NMR spectrum (CDCl3, 500 MHz) of compound 1
- S4: 13C NMR spectrum (CDCl3, 125 MHz) of compound 1
- S5: DEPT spectrum of compound 1
- S6: HSQC spectrum of compound 1
- S7: COSY spectrum of compound 1
- S8: HMBC spectrum of compound 1
- S9: TOCSY spectrum of compound 1
- S10: NOESY spectrum of compound 1
- S11: HR-ESI-MS of compound 2
- S12: EI-MS of compound 2
- S13: 1H NMR spectrum (CDCl3, 500 MHz) of compound 2
- S14: 13C NMR spectrum (CDCl3, 125 MHz) of compound 2
- S15: DEPT spectrum of compound 2
- S16: HSQC spectrum of compound 2
- S17: HMBC spectrum of compound 2
- S18: COSY spectrum of compound 2
- S19: TOCSY spectrum of compound 2
- S20: NOESY spectrum of compound 2
Acknowledgments
References
- Faulkner, DJ. Marine natural products. Nat Prod Rep 1998, 15, 113–158. [Google Scholar]
- Pettit, GR; Herald, CL; Doubek, DL; Herald, DL; Arnold, E; Clardy, J. Isolation and structure of bryostatin 1. J Am Chem Soc 1982, 104, 6846–6848. [Google Scholar]
- Zhang, HP; Kamano, Y; Ichihara, Y; Kizu, H; Komiyama, K; Itokawa, H; Pettit, GR. Isolation and structure of convolutamydines B-D from marine broyzoan Amathia convoluta. Tetrahedron 1995, 51, 5523–5528. [Google Scholar]
- Blackman, AJ; Walls, JT. Bryozoan secondary metabolites and their chemical ecology. In Studies in Natural Products Chemistry; Atta-ur-Rahman, Ed.; Elsevier: Amsterdam, The Netherlands, 1995; pp. 73–112. [Google Scholar]
- Lin, HW; Yi, YH; Li, WL; Yao, XS; Wu, HM. Bryostatin 19: A new antineoplastic component from Bugula neritina in the South China Sea. J Chin Mar Drugs 1998, 65, 1–3. [Google Scholar]
- Yang, F; Zhang, H-J; Chen, JT; Tang, HF; Piao, SJ; Chen, WS; Lin, HW. New cytotoxic oxygenated sterols from marine bryozoan Bugula neritina. Nat Prod Res 2010, 2, 1–7. [Google Scholar]
- Tian, XR; Tang, HF; Li, YS; Lin, HW; Ma, N; Zhang, W; Yao, MN. Ceramides and cerebrosides from the marine bryozoan Bugula neritina inbating South China Sea. J Asian Nat Prod Res 2009, 11, 1005–1012. [Google Scholar]
- Tian, XR; Tang, HF; Li, YS; Lin, HW; Ma, N; Zhang, W. Sterols from marine bryozoan Bugula neritina. Biochem Syst Ecol 2010, 38, 435–437. [Google Scholar]
- Harte, RA; Yeaman, SJ; McEIhinney, J; Suckling, CJ; Jackson, B; Suckling, KE. Effects of novel synthetic sterol probes on enzymes of cholesterol metabolism in cell-free and cellular systems. Chem Phys Lipids 1996, 83, 45–59. [Google Scholar]
- Fürst, A; Labler, L; Meier, W. Neue synthesen von 1α,25-dihydroxycholesterin. Helv Chim Acta 1982, 65, 1499–1521. [Google Scholar]
- Koizumi, N; Ishiguro, M; Yasuda, M; Ikekawa, N. Stereoselective introduction of hydroxyl groups into the cholesterol side chain. Preparation of (24R)- and (24S)-24,25-dihydroxy- and (25R)- and (25S)-25,26-dihydroxyvitamin D3 by asymmetric synthesis. J Chem Soc Perkin Trans 1983, 1, 1401–1410. [Google Scholar]
- Wilson, WK; Sumpter, RM; Warren, JJ; Rogers, PS; Ruan, B; Schroepfer, GJ, Jr. Analysis of unsaturated C27 sterols by nuclear magnetic resonance spectroscopy. J Lipid Res 1996, 37, 1529–1555. [Google Scholar]
- Calderon, GJ; Castellanos, L; Duque, C; Echigo, S; Hara, N; Fujimoto, Y. Ophirasterol, a new C31 sterol form the marine sponge Topsentia ophiraphidites. Steroids 2004, 69, 93–100. [Google Scholar]
- Sheu, JH; Huang, SY; Duh, CY. Cytotoxic oxygenated desmosterols of the red alge Galaxaura marginata. J Nat Prod 1996, 59, 23–26. [Google Scholar]
- Fattorusso, E; Magno, S; Santacroce, C; Sica, D; Impellizzeri, G; Mangiafico, S; Oriente, G; Piattelli, M; Sciuto, S. Sterols of some red algae. Phytochemistry 1975, 14, 1579–1582. [Google Scholar]
- Notaro, G; Piccialli, V; Sica, D. New steroidal hydroxyketones and closely related diols from the marine sponge Cliona copiosa. J Nat Prod 1992, 55, 1588–1594. [Google Scholar]
- Kingston, JF; Fallis, AG. Marine natural products: highly functionalized steroids (12β-hydroxy-24-norcholesta-1,4,22-trien-3-one and 12β-acetoxy-24-norcholesta-1,4,22-trien-3-one) from the sea raspberry Gersemia rubiformis. Can J Chem 1982, 60, 820–824. [Google Scholar]
- Riccardis, F; Minale, L; Iorizzi, M; Debitus, C; Levi, C. Marine sterols. Side-chain-oxygenated sterols. Possibly of abiotic origin, from the New Caledonian sponge Stelodoryx chlorophylla. J Nat Prod 1993, 56, 282–287. [Google Scholar]
- Li, GQ; Deng, ZW; Guan, HS; Guo, DA; Lin, WH. Polyhydroxy sterols from soft coral Dendronephthya gigantea from the South Chin Sea. J Chin Mar Drugs 2004, 1, 1–5. [Google Scholar]
- Yang, F; Zhang, HJ; Liu, XF; Chen, WS; Tang, HF; Lin, HW. Oxygenated steroids from marine bryozoan Biflustra grandicella. Biochem Syst Ecol 2009, 37, 686–689. [Google Scholar]
- Supino, R. MTT Assay. In The ERGATT/FRAME Data Bank of in Vitro Techniques in Toxicology, Invittox Protocol Number 17; INVITTOX: Nottingham, UK, 1990. [Google Scholar]
| Position | 1 | 2 | ||
|---|---|---|---|---|
| δc, mult. | δH, (int., mult., J in Hz) | δc, mult. | δH, (int., mult., J in Hz) | |
| 1 | 37.4 t | α 1.84 (1H, m), β 1.14 (1H, m) | 36.9 t | α 1.82 (1H, m), β 1.16 (1H, m) |
| 2 | 31.8 t | α 1.83 (1H, m), β 1.49 (1H, m) | 31.6 t | α 1.84 (1H, m), β 1.51 (1H, m) |
| 3 | 71.9 d | 3.52 (1H, m) | 71.6 d | 3.61 (1H, m) |
| 4 | 42.4 t | α 2.28 (1H, m), β 2.23 (1H, m) | 42.5 t | α 2.33 (1H, m), β 2.29 (1H, m) |
| 5 | 140.9 s | – | 146.2 s | – |
| 6 | 121.8 d | 5.35 (1H, t, 2.8) | 120.9 d | 5.73 (1H, dd, 5.0, 1.7) |
| 7 | 32.0 t | α 1.48 (1H, m), β 1.97 (1H, m) | 74.1 d | 3.27 (1H, m) |
| 8 | 32.0 d | 1.46 (1H, m) | 37.3 d | 1.50 (1H, m) |
| 9 | 50.2 d | 0.93 (1H, m) | 42.9 d | 1.31 (1H, m) |
| 10 | 36.6 s | – | 37.6 s | – |
| 11 | 21.2 t | α 1.00 (1H, m), β 1.47 (1H, m) | 20.9 t | α 1.02 (1H, m), β 1.49 (1H, m) |
| 12 | 39.9 t | α 1.15 (1H, m), β 1.99 (1H, m) | 39.1 t | α 1.19 (1H, m), β 1.94 (1H, dt, 12.6, 3.6) |
| 13 | 42.5 s | – | 42.2 s | – |
| 14 | 56.9 d | 0.98 (1H, m) | 49.3 d | 1.51 (1H, m) |
| 15 | 24.5 t | α 1.59 (1H, m), β 1.08 (1H, m) | 24.4 t | α 1.59 (1H, m), β 1.07 (1H, m) |
| 16 | 28.4 t | α 1.84 (1H, m), β 1.28 (1H, m) | 28.8 t | α 1.74 (1H, m), β 1.26 (1H, m) |
| 17 | 56.0 d | 1.10 (1H, m) | 55.8 d | 1.21 (1H, m) |
| 18 | 12.1 q | 0.69 (3H, s) | 11.8 q | 0.67 (3H, s) |
| 19 | 19.5 q | 1.00 (3H, s) | 18.4 q | 0.98 (3H, s) |
| 20 | 36.2 d | 1.47 (1H, m) | 40.3 d | 2.05 (1H, m) |
| 21 | 18.9 q | 0.91 (3H, d, 6.6) | 21.0 q | 1.01 (3H, d, 6.6) |
| 22 | 39.3 t | a 2.17 (1H, m), b 1.78 (1H, m) | 138.4 d | 5.22 (1H, dd, 15.2, 8.1) |
| 23 | 128.8 d | 5.50 (1H, m) | 126.3 d | 5.27 (1H, m) |
| 24 | 136.8 d | 5.38 (1H, d, 15.8) | 42.1 t | 1.83 (2H, m) |
| 25 | 75.0 s | – | 28.7 d | 1.58 (1H, m) |
| 26 | 25.9 q | 1.25 (3H, s) | 22.4 q | 0.86 (3H, d, 1.9) |
| 27 | 26.3 q | 1.25 (3H, s) | 22.5 q | 0.83 (3H, d, 1.9) |
| 7−OCH3 | – | – | 56.9 q | 3.35 (3H, s) |
| 25−OCH3 | 50.4 q | 3.15 (3H, s) | – | – |
| Position | 3 | 4 | 5 | 6 |
|---|---|---|---|---|
| δC, mult. | δC, mult. | δC, mult. | δC, mult. | |
| 1 | 36.9 t | 37.4 t | 37.4 t | 37.4 t |
| 2 | 31.6 t | 31.8 t | 31.8 t | 31.8 t |
| 3 | 71.6 d | 71.9 d | 71.9 d | 71.9 d |
| 4 | 42.5 t | 42.5 t | 42.4 t | 42.4 t |
| 5 | 146.2 s | 140.9 s | 140.9 s | 140.9 s |
| 6 | 120.9 d | 121.8 d | 121.8 d | 121.9 d |
| 7 | 74.1 t | 32.0 t | 32.0 t | 32.1 t |
| 8 | 37.3 d | 32.1 d | 32.0 d | 32.1 d |
| 9 | 42.9 d | 50.2 d | 50.3 d | 50.3 d |
| 10 | 37.6 s | 36.6 s | 36.7 s | 36.7 s |
| 11 | 20.9 t | 21.2 t | 21.2 t | 21.2 t |
| 12 | 39.2 t | 39.8 t | 39.9 t | 39.9 t |
| 13 | 42.2 s | 42.6 s | 42.5 s | 42.5 s |
| 14 | 49.2 d | 56.8 d | 56.9 d | 56.9 d |
| 15 | 24.4 t | 24.4 t | 24.4 t | 24.4 t |
| 16 | 28.8 t | 28.5 t | 28.3 t | 28.4 t |
| 17 | 55.9 d | 55.9 d | 56.0 d | 56.0 d |
| 18 | 11.6 q | 12.0 q | 12.0 q | 12.0 q |
| 19 | 18.4 q | 19.5 q | 19.8 q | 19.6 q |
| 20 | 36.0 d | 36.0 d | 35.7 d | 35.7 d |
| 21 | 18.9 q | 19.2 q | 18.9 q | 18.9 q |
| 22 | 36.3 t | 39.5 t | 31.8 t | 31.8 t |
| 23 | 23.9 t | 147.6 d | 31.4 t | 31.5 t |
| 24 | 39.7 t | 132.8 d | 76.9 d | 76.5 d |
| 25 | 28.2 d | 198.7 s | 147.6 d | 147.9 d |
| 26 | 22.7 q | 27.1 q | 111.5 t | 111.0 t |
| 27 | 23.0 q | – | 17.4 q | 17.8 q |
| 7−OCH3 | 56.9 q | – | – | – |
| Compound | IC50 (μg/mL) | Compound | IC50 (μg/mL) |
|---|---|---|---|
| 1 | 17.91 | 10 | 15.12 |
| 2 | 21.30 | 11 | 14.73 |
| 3 | 22.11 | 12 | NA |
| 4 | 15.05 | 13 | NA |
| 7 | 18.28 | Adriamycin | 2.50 |
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Tian, X.-R.; Tang, H.-F.; Li, Y.-S.; Lin, H.-W.; Chen, X.-L.; Ma, N.; Yao, M.-N.; Zhang, P.-H. New Cytotoxic Oxygenated Sterols from the Marine Bryozoan Cryptosula pallasiana. Mar. Drugs 2011, 9, 162-183. https://doi.org/10.3390/md9020162
Tian X-R, Tang H-F, Li Y-S, Lin H-W, Chen X-L, Ma N, Yao M-N, Zhang P-H. New Cytotoxic Oxygenated Sterols from the Marine Bryozoan Cryptosula pallasiana. Marine Drugs. 2011; 9(2):162-183. https://doi.org/10.3390/md9020162
Chicago/Turabian StyleTian, Xiang-Rong, Hai-Feng Tang, Yu-Shan Li, Hou-Wen Lin, Xiao-Li Chen, Ning Ma, Min-Na Yao, and Ping-Hu Zhang. 2011. "New Cytotoxic Oxygenated Sterols from the Marine Bryozoan Cryptosula pallasiana" Marine Drugs 9, no. 2: 162-183. https://doi.org/10.3390/md9020162
APA StyleTian, X.-R., Tang, H.-F., Li, Y.-S., Lin, H.-W., Chen, X.-L., Ma, N., Yao, M.-N., & Zhang, P.-H. (2011). New Cytotoxic Oxygenated Sterols from the Marine Bryozoan Cryptosula pallasiana. Marine Drugs, 9(2), 162-183. https://doi.org/10.3390/md9020162