Construction of Escherichia coli Mutant with Decreased Endotoxic Activity by Modifying Lipid A Structure
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, Media, and Growth Conditions
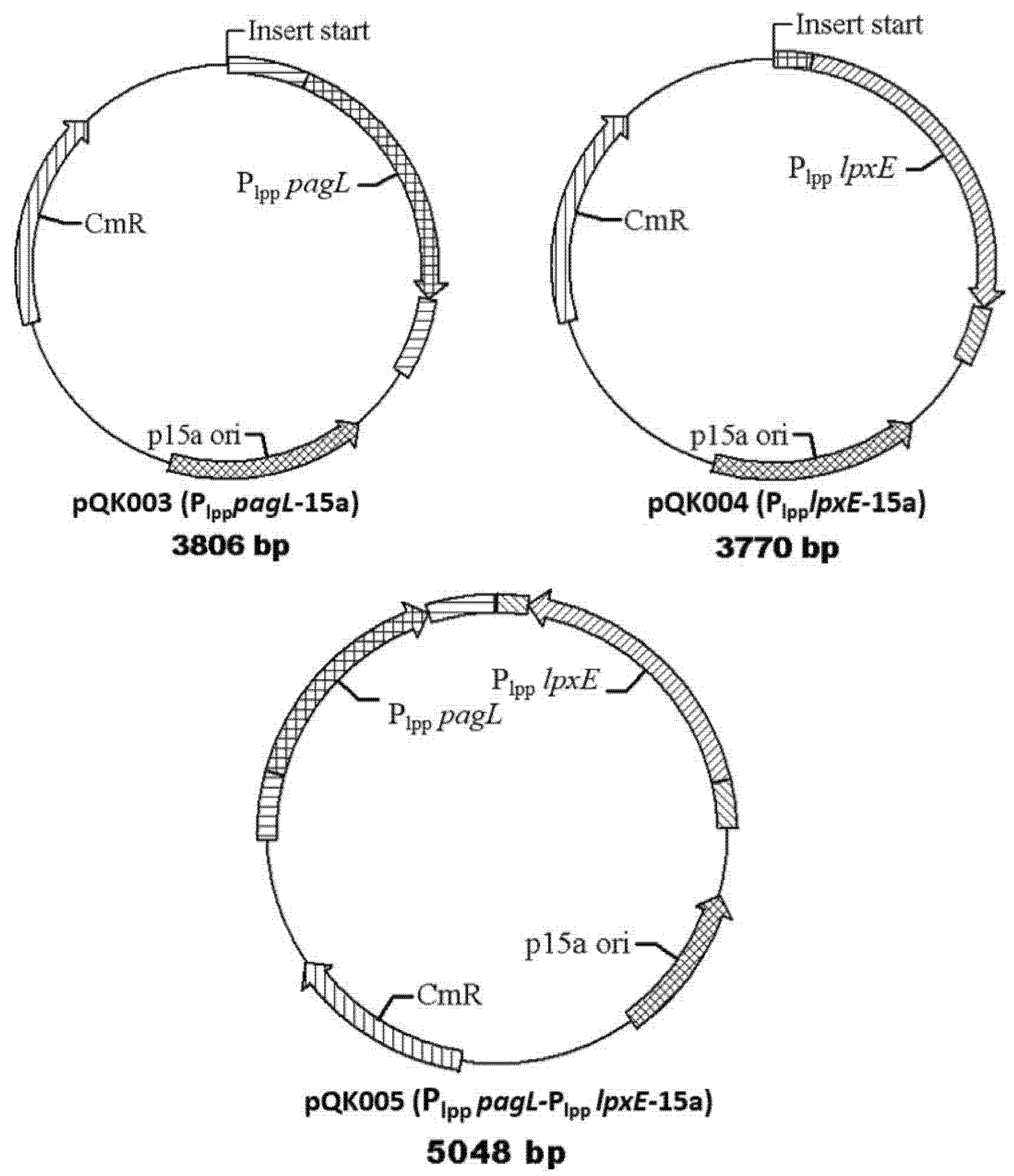
2.2. Construction of Plasmids and Bacterial Strains
| Primers | Sequences (5′–3′) | Function |
|---|---|---|
| msbB-1F | CAGTTCGACAATGTGGAAGAAG | For deletion of msbB by suicide plasmid |
| msbB-1R | ACCTGCAGGATGCGGCCGCGG GCCTCTCGCGAGG | |
| msbB-2F | CCGCGGCCGCATCCTGCAGGT GCTTTTCCAGTTTCGG | |
| msbB-2R | GCGTTATATGCACTTGCGC | |
| pagP-1F | CCTTGATTGCATTTTGTCAT | For deletion of pagP by suicide plasmid |
| pagP-1R | GTCTCACCCGGGCCTGCAGGTTGTGACCATAAAACATTTA | |
| pagP-2F | CACAACCTGCAGGCCCGGGTGAGACAAATGAAGTTTTAGT | |
| pagP-2R | TGCTGCCGTCTTCCGGAGTA | |
| pagPscreenF | AAACGCCGTTAACCCGATA | Insert Plpp lpxE to p15a vector |
| pagPscreenR | TAGACACAAATGCTGCTGTGTCG | |
| lpxRscreenF | CAGGGGGTGTCAGTATTTGGCG | Insert Plpp pagL to p15a vector |
| lpxRscreenR | CGTGAAGCCAATAATTTCTCGCAC | |
| phoP-F | TCATATGCGAATTTTATTAATAGAATATG | Insert phoP gene to pET-32a for expression phoP protein from Pasteurella |
| phoP-R | TGGATCCTTAAGCCATTTCATCATTTTTTCC |
| Strains or Plasmids | Description | Source |
|---|---|---|
| Strains | ||
| E. coli BL21 (DE3) | Expression protein strain | NEB |
| S001 | BL21 (DE3) ∆msbB28 ∆pagP38 | This work |
| S002 | S001 with pQK003 | This work |
| S003 | S001 with pQK004 | This work |
| S004 | S001 with pQK005 | This work |
| χ7232 | endA1 hsdR17 (rK−, mK+) supE44 thi-1 recA1 gyrArelA1Δ (lacZYA-argF) U169λpirdeoR (φ80dlac Δ (lacZ ) M15) | [37] |
| χ7213 | thi-1 thr-1 leuB6 glnV44 tonA21 lacY1 recA1 RP4-2-Tc:μλpir ΔasdA4 Δzhf-2:Tn 10 | [37] |
| Plasmids | ||
| p15a T-vector | Low copy expression plasmid | [38] |
| pET-32a | Protein expression | Novagen |
| pQK001 | For deletion of msbB in BL21 | This work |
| pQK002 | For deletion of pagP in BL21 | This work |
| pYA4291 | Plpp pagL in pRE112 | Lab collection |
| pYA4295 | Plpp lpxE (codon-optimized) in pRE112 | [39] |
| pQK003 | Insert Plpp pagL to p15a and express PagL for deacylation of lipid A | This work |
| pQK004 | Insert Plpp lpxE to p15a and express lpxE to remove the 1-phosphate group of lipid A | This work |
| pQK005 | Insert Plpp pagL and Plpp lpxE to p15a Express both pagL and lpxE to modify the lipid A | This work |
| pQK006 | Expression of PhoP protein of Pasteurella multocida | Lab collection |
2.3. BL21 (DE3) Mutant Strain Construction
2.4. Analysis of Characteristics of BL21 (DE3) Mutant Strains
2.5. Lipid A Isolation and Mass Spectrometry Procedures
2.6. Purification and Concentration Measurement of Lipopolysaccharide (LPS) from BL21 (DE3) and Its Mutants
2.7. LPS Stimulation of the Murine Macrophage Cell RAW264.7 and Human Monocyte Cell THP1
3. Results
3.1. Construction of BL21 (DE3) ∆msbB28 ∆pagP38 Mutant and Its Derivatives
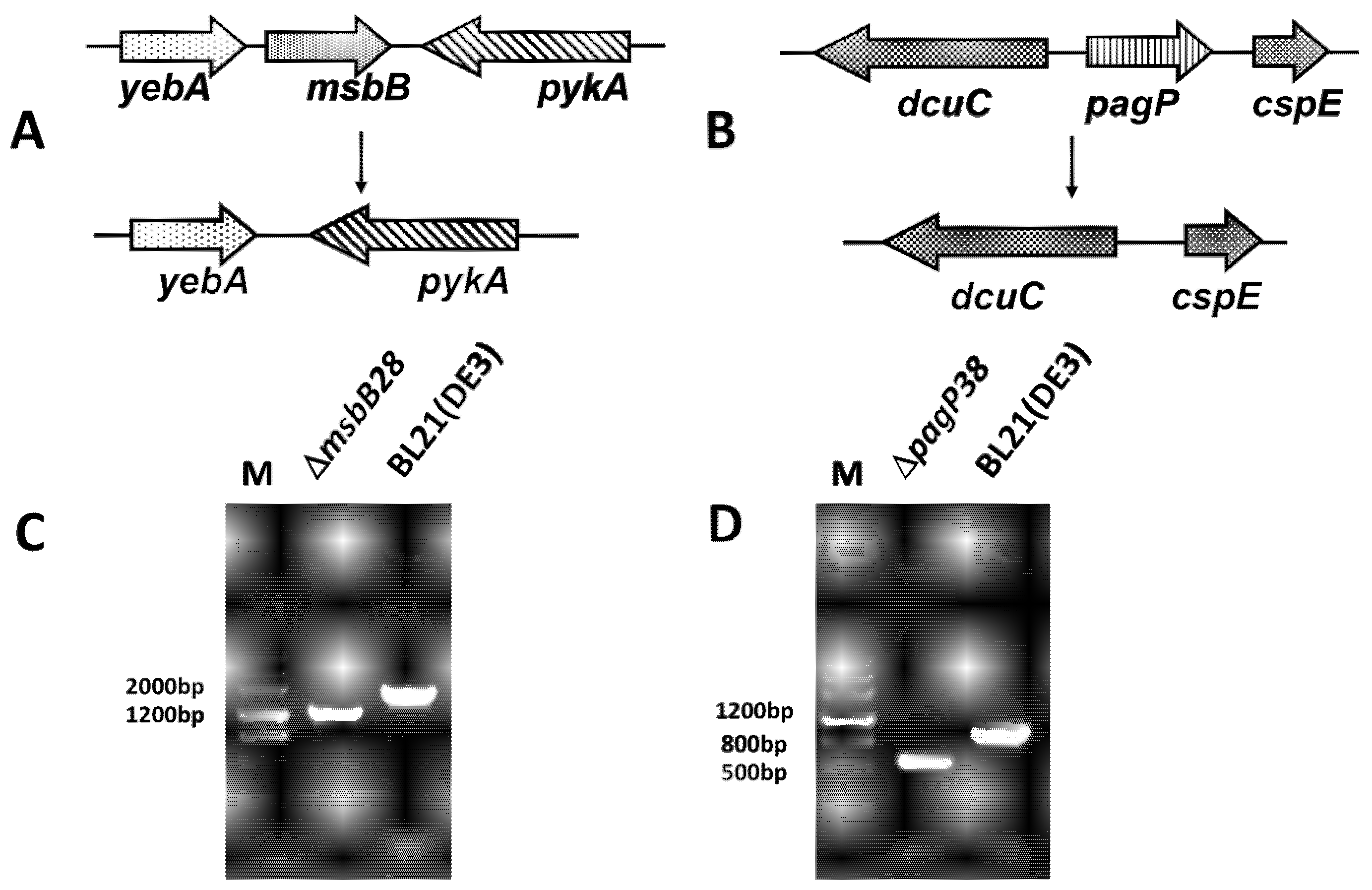
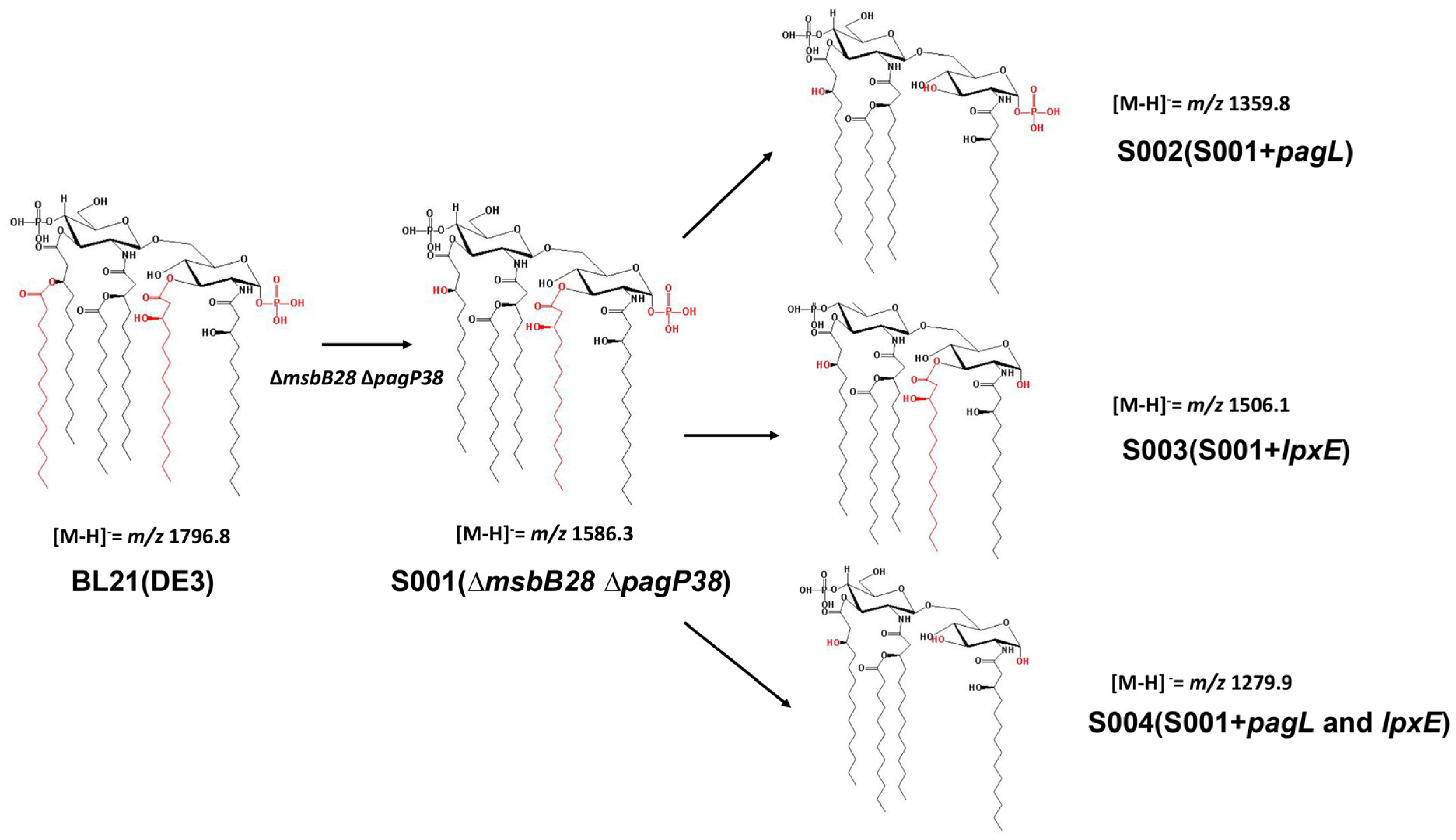
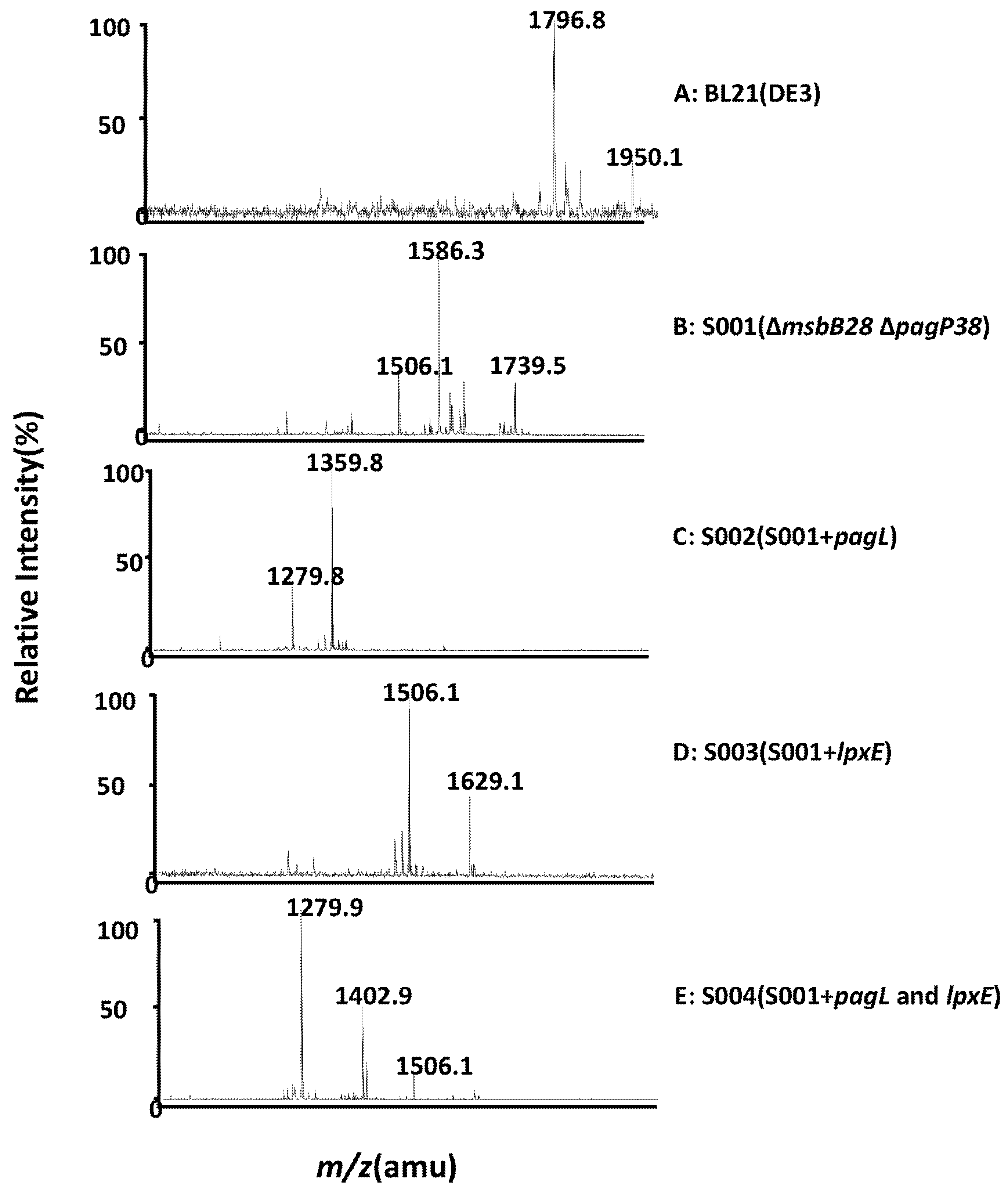
3.2. Structure Analysis of Lipid A Extracted From BL21(DE3) and Its Derivatives
3.3. Comparison of the Endotoxic Activities of LPS Isolated from BL21 (DE3) and Its Derivatives
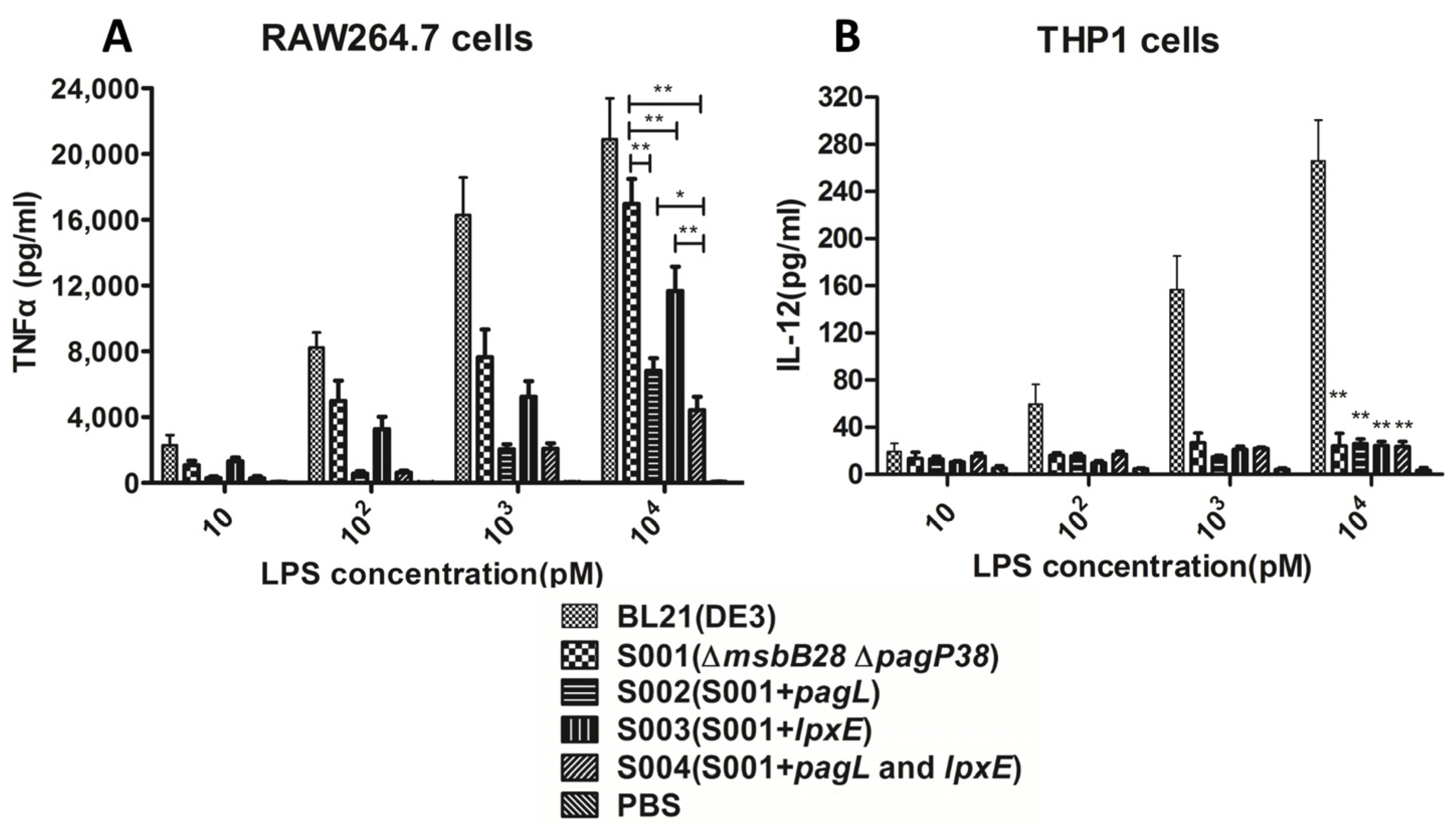
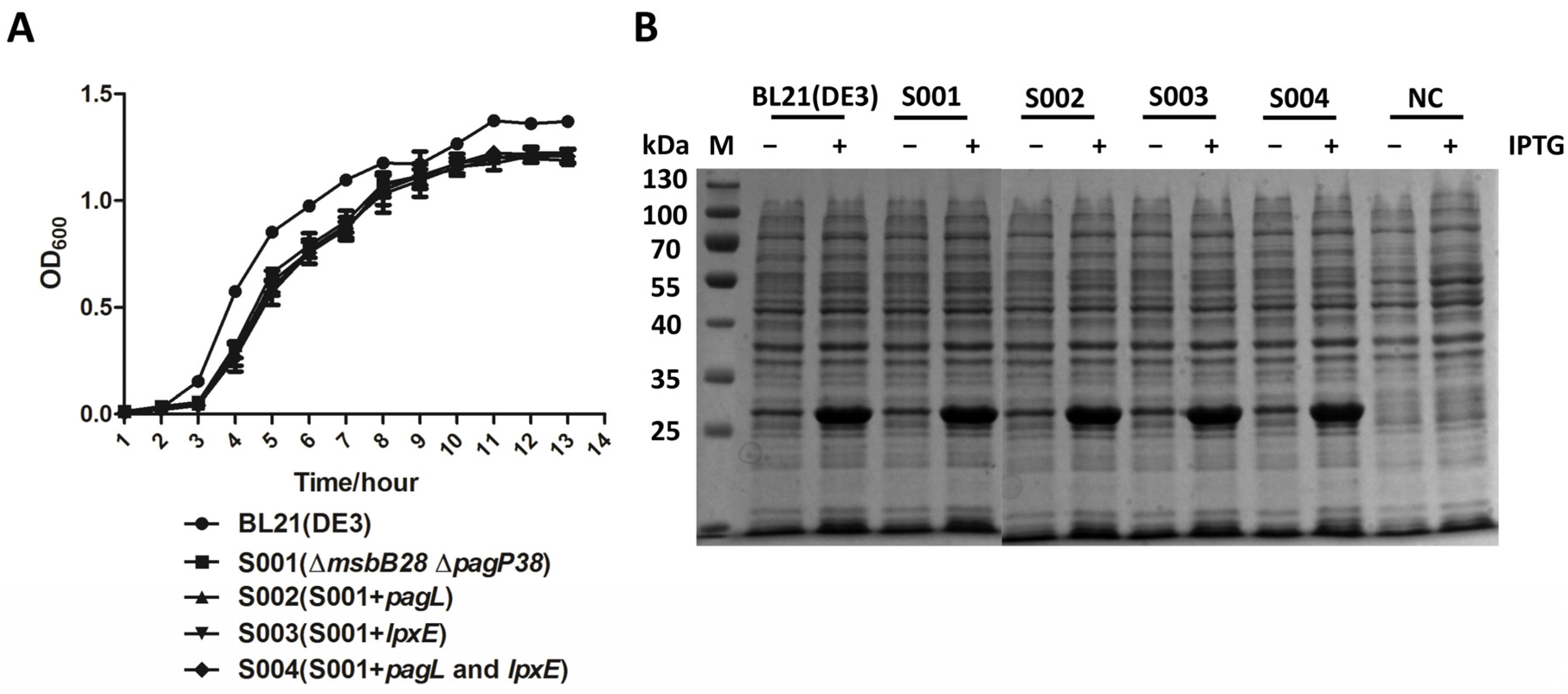
3.4. Lipid A Modification Did Not Change the Growth Kinetics of the BL21 and Its Derivatives and They Retain Ability to Produce Recombinant Proteins
4. Discussion
5. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflict of Interest
References
- Kong, Q.; Yang, J.; Liu, Q.; Alamuri, P.; Roland, K.L.; Curtiss, R. Effect of deletion of genes involved in lipopolysaccharide core and O-antigen synthesis on virulence and immunogenicity of Salmonella enterica serovarTyphimurium. Infect. Immun. 2011, 79, 4227–4239. [Google Scholar] [CrossRef] [PubMed]
- Shiloach, J.; Kaufman, J.; Guillard, A.; Fass, R. Effect of glucose supply strategy on acetate accumulation, growth, and recombinant protein production by Escherichia coli BL21 (λDE3) and Escherichia coli JM109. Biotechnol. Bioeng. 1996, 49, 421–428. [Google Scholar] [CrossRef]
- Colucci, M.; Balconi, G.; Lorenzet, R.; Pietra, A.; Locati, D.; Donati, M.; Semeraro, N. Cultured human endothelial cells generate tissue factor in response to endotoxin. J. Clin. Investig. 1983, 71, 1893–1896. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, E.J.; Fisher, C.J., Jr.; Sprung, C.L.; Straube, R.C.; Sadoff, J.C.; Foulke, G.E.; Wortel, C.H.; Fink, M.P.; Dellinger, R.P.; Teng, N.N. Treatment of Gram-negative bacteremia and septic shock with HA-1A human monoclonal antibody against endotoxin: A randomized, double-blind, placebo-controlled trial. N. Engl. J. Med. 1991, 324, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Petsch, D.; Anspach, F.B. Endotoxin removal from protein solutions. J. Biotechnol. 2000, 76, 97–119. [Google Scholar] [CrossRef]
- Galanos, C.; Lüderitz, O.; Rietschel, E.T.; Westphal, O.; Brade, H.; Brade, L.; Freudenberg, M.; Schade, U.; Imoto, M.; Yoshimura, H. Synthetic and natural Escherichia coli free lipid A express identical endotoxic activities. Eur. J. Biochem. 1985, 148, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.; Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef] [PubMed]
- Needham, B.D.; Carroll, S.M.; Giles, D.K.; Georgiou, G.; Whiteley, M.; Trent, M.S. Modulating the innate immune response by combinatorial engineering of endotoxin. Proc. Natl. Acad. Sci. USA 2013, 110, 1464–1469. [Google Scholar] [CrossRef] [PubMed]
- Piya, M.K.; McTernan, P.G.; Kumar, S. Adipokine inflammation and insulin resistance: The role of glucose, lipids and endotoxin. J. Endocrinol. 2013, 216. [Google Scholar] [CrossRef] [PubMed]
- Xia, D.; Samols, D. Transgenic mice expressing rabbit C-reactive protein are resistant to endotoxemia. Proc. Natl. Acad. Sci. USA 1997, 94, 2575–2580. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.; Guan, Z.; Ingram, B.O.; Six, D.A.; Song, F.; Wang, X.; Zhao, J. Discovery of new biosynthetic pathways: The lipid A story. J. Lipid Res. 2009, 50. [Google Scholar] [CrossRef] [PubMed]
- Trent, M.S.; Pabich, W.; Raetz, C.R.; Miller, S.I. A PhoP/PhoQ-induced lipase (PagL) that catalyzes 3-O-deacylation of lipid A precursors in membranes of Salmonella typhimurium. J. Biol. Chem. 2001, 276, 9083–9092. [Google Scholar] [CrossRef] [PubMed]
- Somerville, J.E.; Cassiano, L.; Darveau, R.P. Escherichia coli msbB gene as a virulence factor and a therapeutic target. Infect. Immun. 1999, 67, 6583–6590. [Google Scholar] [PubMed]
- Somerville, J.E., Jr.; Cassiano, L.; Bainbridge, B.; Cunningham, M.D.; Darveau, R.P. A novel Escherichia coli lipid A mutant that produces an antiinflammatory lipopolysaccharide. J. Clin. Investig. 1996, 97, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Coats, S.R.; Pham, T.-T.T.; Bainbridge, B.W.; Reife, R.A.; Darveau, R.P. MD-2 mediates the ability of tetra-acylated and penta-acylated lipopolysaccharides to antagonize Escherichia coli lipopolysaccharide at the TLR4 signaling complex. J. Immunol. 2005, 175, 4490–4498. [Google Scholar] [CrossRef] [PubMed]
- Cognet, I.; de Coignac, A.B.; Magistrelli, G.; Jeannin, P.; Aubry, J.-P.; Maisnier-Patin, K.; Caron, G.; Chevalier, S.; Humbert, F.; Nguyen, T. Expression of recombinant proteins in a lipid A mutant of Escherichia coli BL21 with a strongly reduced capacity to induce dendritic cell activation and maturation. J. Immunol. Methods 2003, 272, 199–210. [Google Scholar] [CrossRef]
- Low, K.B.; Ittensohn, M.; Le, T.; Platt, J.; Sodi, S.; Amoss, M.; Carmichael, E.; Chakraborty, A.; Ash, O.; Fischer, J. Lipid A mutant Salmonella with suppressed virulence and TNFα induction retain tumor-targeting in vivo. Nat. Biotechnol. 1999, 17, 37–41. [Google Scholar] [PubMed]
- Bishop, R.E.; Gibbons, H.S.; Guina, T.; Trent, M.S.; Miller, S.I.; Raetz, C.R. Transfer of palmitate from phospholipids to lipid A in outer membranes of Gram-negative bacteria. EMBO J. 2000, 19, 5071–5080. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Lim, K.B.; Poduje, C.M.; Daniel, M.; Gunn, J.S.; Hackett, M.; Miller, S.I. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell 1998, 95, 189–198. [Google Scholar] [CrossRef]
- Bishop, R.E. The lipid A palmitoyltransferase PagP: Molecular mechanisms and role in bacterial pathogenesis. Mol. Microbiol. 2005, 57, 900–912. [Google Scholar] [CrossRef] [PubMed]
- Hwang, P.M.; Choy, W.-Y.; Lo, E.I.; Chen, L.; Forman-Kay, J.D.; Raetz, C.R.; Privé, G.G.; Bishop, R.E.; Kay, L.E. Solution structure and dynamics of the outer membrane enzyme PagP by NMR. Proc. Natl. Acad. Sci. USA 2002, 99, 13560–13565. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.B.; Muszyński, A.; Salas, O.; Speed, K.; Carlson, R.W. Elucidation of the 3-O-deacylase gene, pagL, required for the removal of primary β-hydroxy fatty acid from the lipid A in the nitrogen-fixing endosymbiont Rhizobium etli CE3. J. Biol. Chem. 2013, 288, 12004–12013. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, K.; Teramoto, M.; Tatsui, R.; Amamoto, S. Lipid A 3′-O-deacylation by Salmonella outer membrane enzyme LpxR modulates the ability of lipid A to stimulate Toll-like receptor 4. Biochem. Biophys. Res. Commun. 2012, 428, 343–347. [Google Scholar] [CrossRef] [PubMed]
- Geurtsen, J.; Steeghs, L.; Hamstra, H.-J.; ten Hove, J.; de Haan, A.; Kuipers, B.; Tommassen, J.; van der Ley, P. Expression of the lipopolysaccharide-modifying enzymes PagP and PagL modulates the endotoxic activity of Bordetella pertussis. Infect. Immun. 2006, 74, 5574–5585. [Google Scholar] [CrossRef] [PubMed]
- Geurtsen, J.; Steeghs, L.; ten Hove, J.; van der Ley, P.; Tommassen, J. Dissemination of lipid A deacylases (PagL) among Gram-negative bacteria identification of active-site histidine and serine residues. J. Biol. Chem. 2005, 280, 8248–8259. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Karbarz, M.J.; McGrath, S.C.; Cotter, R.J.; Raetz, C.R. MsbA Transporter-dependent lipid A 1-Dephosphorylation on the periplasmic surface of the inner membrane topography of Francisella novicida LpxE expressed in Escherichia coli. J. Biol. Chem. 2004, 279, 49470–49478. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.; Six, D.A.; Roland, K.L.; Liu, Q.; Gu, L.; Reynolds, C.M.; Wang, X.; Raetz, C.R.; Curtiss, R. Salmonella synthesizing 1-monophosphorylated lipopolysaccharide exhibits low endotoxic activity while retaining its immunogenicity. J. Immunol. 2011, 187, 412–423. [Google Scholar] [CrossRef] [PubMed]
- Kayagaki, N.; Wong, M.T.; Stowe, I.B.; Ramani, S.R.; Gonzalez, L.C.; Akashi-Takamura, S.; Miyake, K.; Zhang, J.; Lee, W.P.; Muszyński, A. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science 2013, 341, 1246–1249. [Google Scholar] [CrossRef] [PubMed]
- Park, B.S.; Song, D.H.; Kim, H.M.; Choi, B.-S.; Lee, H.; Lee, J.-O. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 2009, 458, 1191–1195. [Google Scholar] [CrossRef] [PubMed]
- Rietschel, E.T.; Kirikae, T.; Schade, F.U.; Mamat, U.; Schmidt, G.; Loppnow, H.; Ulmer, A.J.; Zähringer, U.; Seydel, U.; di Padova, F. Bacterial endotoxin: Molecular relationships of structure to activity and function. FASEB J. 1994, 8, 217–225. [Google Scholar] [PubMed]
- Rietschel, E.T.; Kirikae, T.; Schade, F.U.; Ulmer, A.J.; Holst, O.; Brade, H.; Schmidt, G.; Mamat, U.; Grimmecke, H.-D.; Kusumoto, S. The chemical structure of bacterial endotoxin in relation to bioactivity. Immunobiology 1993, 187, 169–190. [Google Scholar] [CrossRef]
- Brandenburg, K.; Wiese, A. Endotoxins: Relationships between structure, function, and activity. Curr. Top. Med. Chem. 2004, 4, 1127–1146. [Google Scholar] [CrossRef] [PubMed]
- Miroux, B.; Walker, J.E. Over-production of proteins in Escherichia coli: Muztant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 1996, 260, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, K.; Kelly, S.M.; Curtiss, R. Construction of an ASD+ expression-cloning vector: Stable maintenance and high level expression of cloned genes in a Salmonella vaccine strain. Nat. Biotechnol. 1988, 6, 693–697. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W.; Russell, D.W. Molecular Cloning: A Laboratory Manual (3-Volume Set); Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Edwards, R.A.; Keller, L.H.; Schifferli, D.M. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 1998, 207, 149–157. [Google Scholar] [CrossRef]
- Roland, K.; Curtiss, R., III; Sizemore, D. Construction and evaluation of a Δcya Δcrp Salmonella typhimurium strain expressing avian pathogenic Escherichia coli O78 LPS as a vaccine to prevent airsacculitis in chickens. Avian Dis. 1999, 43, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Kong, W.; Ashraf, S.; Curtiss, R. A one-plasmid system to generate influenza virus in cultured chicken cells for potential use in influenza vaccine. J. Virol. 2009, 83, 9296–9303. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.; Six, D.A.; Liu, Q.; Gu, L.; Roland, K.L.; Raetz, C.R.; Curtiss, R., III. Palmitoylation state impacts induction of innate and acquired immunity by the Salmonella enterica serovar Typhimurium msbB mutant. Infect. Immun. 2012, 80, 1302. [Google Scholar] [CrossRef]
- Kang, H.Y.; Dozois, C.M.; Tinge, S.A.; Lee, T.H.; Curtiss, R., III. Transduction-mediated transfer of unmarked deletion and point mutations through use of counterselectable suicide vectors. J. Bacteriol. 2002, 184, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Powell, D.A.; Shaffer, S.A.; Rasko, D.A.; Pelletier, M.R.; Leszyk, J.D.; Scott, A.J.; Masoudi, A.; Goodlett, D.R.; Wang, X. LPS remodeling is an evolved survival strategy for bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, 8716–8721. [Google Scholar] [CrossRef] [PubMed]
- Apicella, M.A.; Griffiss, J.; Schneider, H. Isolation and characterization of lipopolysaccharides, lipooligosaccharides, and lipid A. Methods Enzymol. 1994, 235, 242–252. [Google Scholar] [PubMed]
- Osborn, M.J. Studies on the Gram-negative cell wall, I. Evidence for the role of 2-keto-3-deoxyoctonate in the lipopolysaccharide of Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 1963, 50, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Trent, M.S.; Ribeiro, A.A.; Lin, S.; Cotter, R.J.; Raetz, C.R. An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-l-arabinose to lipid A induction in polymyxin-resistant mutants and role of novel lipid-linked donor. J. Biol. Chem. 2001, 276, 43122–43131. [Google Scholar] [CrossRef] [PubMed]
- Sims, K.; Haynes, C.A.; Kelly, S.; Allegood, J.C.; Wang, E.; Momin, A.; Leipelt, M.; Reichart, D.; Glass, C.K.; Sullards, M.C. Kdo2-lipid A, a TLR4-specific agonist, induces de novo sphingolipid biosynthesis in RAW264.7 macrophages, which is essential for induction of autophagy. J. Biol. Chem. 2010, 285, 38568–38579. [Google Scholar] [CrossRef] [PubMed]
- Bès-Houtmann, S.; Roche, R.; Hoareau, L.; Gonthier, M.-P.; Festy, F.; Caillens, H.; Gasque, P.; d’Hellencourt, C.L.; Cesari, M. Presence of functional TLR2 and TLR4 on human adipocytes. Histochem. Cell Biol. 2007, 127, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Hirschfeld, M.; Ma, Y.; Weis, J.H.; Vogel, S.N.; Weis, J.J. Cutting edge: Repurification of lipopolysaccharide eliminates signaling through both human and murine toll-like receptor 2. J. Immunol. 2000, 165, 618–622. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Everest, P.; Servos, S.; Foxwell, N.; Zähringer, U.; Brade, H.; Rietschel, E.T.; Dougan, G.; Charles, I.G.; Maskell, D.J. A lethal role for lipid A in Salmonella infections. Mol. Microbiol. 1998, 29, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; el Zoeiby, A.; Petruzziello, T.N.; Jayabalasingham, B.; Seyedirashti, S.; Bishop, R.E. Lipid trafficking controls endotoxin acylation in outer membranes of Escherichia coli. J. Biol. Chem. 2004, 279, 44966–44975. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.A.; Keegan, D.S.; Sowell, C.G.; Livesay, M.T.; Johnson, C.L.; Taubner, L.M.; Harris, A.; Myers, K.R.; Thompson, J.D.; Gustafson, G.L. 3-O-Desacyl monophosphoryl lipid A derivatives: Synthesis and immunostimulant activities. J. Med. Chem. 1999, 42, 4640–4649. [Google Scholar] [CrossRef] [PubMed]
- Imoto, M.; Yoshimura, H.; Shimamoto, T.; Sakaguchi, N.; Kusumoto, S.; Shiba, T. Total synthesis of Escherichia coli lipid A, the endotoxically active principle of cell-surface lipopolysaccharide. Bull. Chem. Soc. Jpn. 1987, 60, 2205–2214. [Google Scholar] [CrossRef]
- Tran, A.X.; Lester, M.E.; Stead, C.M.; Raetz, C.R.; Maskell, D.J.; McGrath, S.C.; Cotter, R.J.; Trent, M.S. Resistance to the antimicrobial peptide polymyxin requires myristoylation of Escherichia coli and Salmonella typhimurium lipid A. J. Biol. Chem. 2005, 280, 28186–28194. [Google Scholar] [CrossRef] [PubMed]
- Doerrler, W.T.; Gibbons, H.S.; Raetz, C.R. MsbA-dependent translocation of lipids across the inner membrane of Escherichia coli. J. Biol. Chem. 2004, 279, 45102–45109. [Google Scholar] [CrossRef] [PubMed]
- Van Der Ley, P.; Steeghs, L.; Hamstra, H.J.; ten Hove, J.; Zomer, B.; van Alphen, L. Modification of lipid A biosynthesis in Neisseria meningitidis lpxL mutants: Influence on lipopolysaccharide structure, toxicity, and adjuvant activity. Infect. Immun. 2001, 69, 5981–5990. [Google Scholar] [CrossRef] [PubMed]
- Morrison, D.C.; Silverstein, R.; Luchi, M.; Shnyra, A. Structure-function relationships of bacterial endotoxins: Contribution to microbial sepsis. Infect. Dis. Clin. N. Am. 1999, 13, 313–340. [Google Scholar] [CrossRef]
- Kong, Q.; Six, D.A.; Liu, Q.; Gu, L.; Wang, S.; Alamuri, P.; Raetz, C.R.; Curtiss, R. Phosphate groups of lipid A are essential for Salmonella enterica serovar Typhimurium virulence and affect innate and adaptive immunity. Infect. Immun. 2012, 80, 3215–3224. [Google Scholar] [CrossRef] [PubMed]
- Karow, M.; Georgopoulos, C. Isolation and characterization of the Escherichia coli msbB gene, a multicopy suppressor of null mutations in the high-temperature requirement gene htrB. J. Bacteriol. 1992, 174, 702–710. [Google Scholar] [PubMed]
- Kim, K.; Jeong, J.H.; Lim, D.; Hong, Y.; Yun, M.; Min, J.-J.; Kwak, S.-J.; Choy, H.E. A novel balanced-lethal host-vector system based on glmS. PLoS ONE 2013, 8, e60511. [Google Scholar] [CrossRef] [PubMed]
- Galán, J.E.; Nakayama, K.; Curtiss, R. Cloning and characterization of the asd gene of Salmonella typhimurium: Use in stable maintenance of recombinant plasmids in Salmonella vaccine strains. Gene 1990, 94, 29–35. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Q.; Li, Y.; Zhao, X.; Yang, X.; Liu, Q.; Kong, Q. Construction of Escherichia coli Mutant with Decreased Endotoxic Activity by Modifying Lipid A Structure. Mar. Drugs 2015, 13, 3388-3406. https://doi.org/10.3390/md13063388
Liu Q, Li Y, Zhao X, Yang X, Liu Q, Kong Q. Construction of Escherichia coli Mutant with Decreased Endotoxic Activity by Modifying Lipid A Structure. Marine Drugs. 2015; 13(6):3388-3406. https://doi.org/10.3390/md13063388
Chicago/Turabian StyleLiu, Qiong, Yanyan Li, Xinxin Zhao, Xue Yang, Qing Liu, and Qingke Kong. 2015. "Construction of Escherichia coli Mutant with Decreased Endotoxic Activity by Modifying Lipid A Structure" Marine Drugs 13, no. 6: 3388-3406. https://doi.org/10.3390/md13063388





